The Road towards Gene Therapy for X-Linked Juvenile Retinoschisis: A Systematic Review of Preclinical Gene Therapy in Cell-Based and Rodent Models of XLRS
Abstract
1. Introduction
1.1. Clinical Features of Retinoschisis
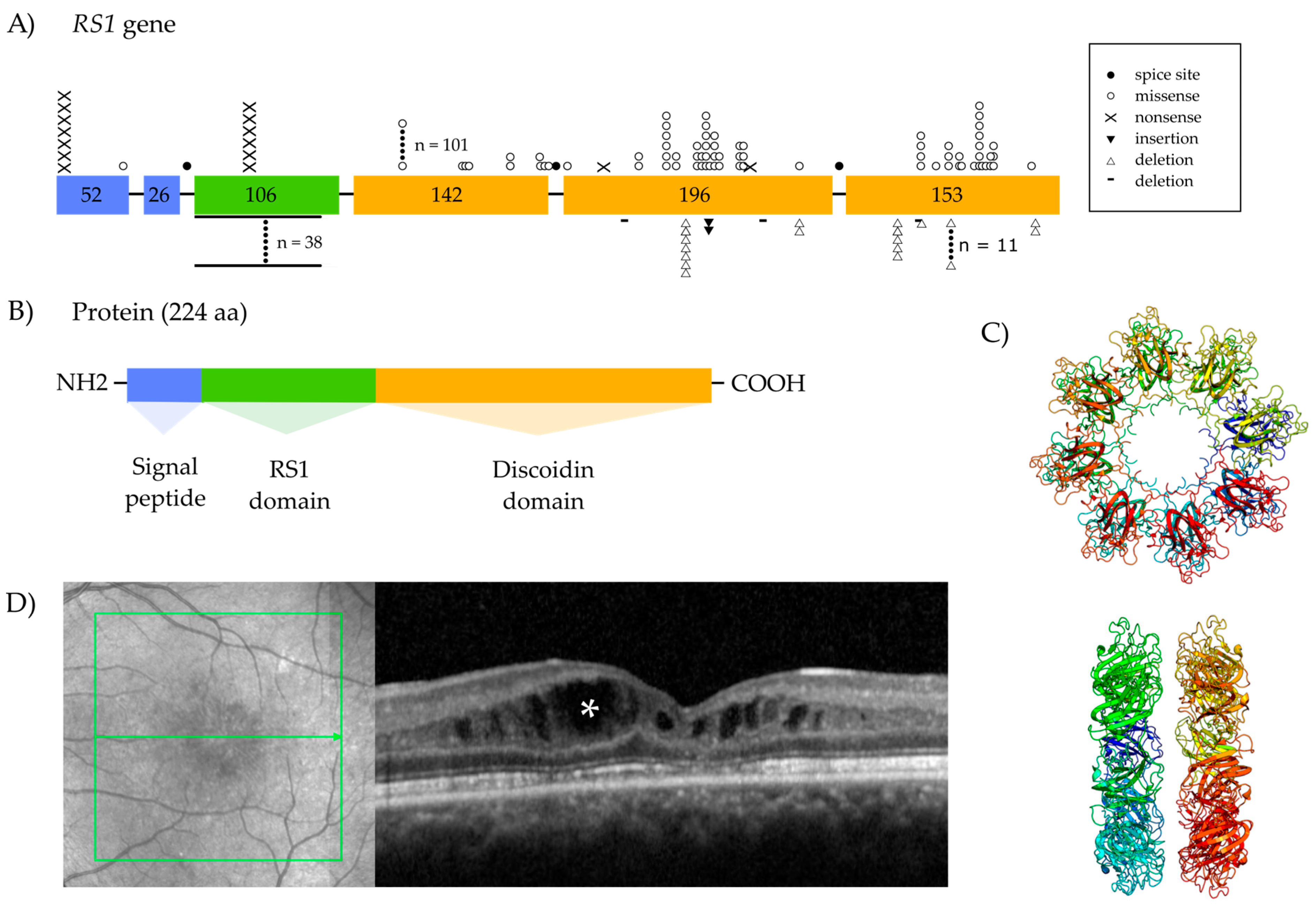
1.2. The Retinoschisin Gene and Its Expression
1.3. Retinoschisin Protein and Function
Localization
1.4. Treatment Options
1.4.1. Adeno-Associated Viral Vector-Mediated Gene Augmentation Therapy
1.4.2. Non-Viral Gene Augmentation or Gene Editing
2. Methods
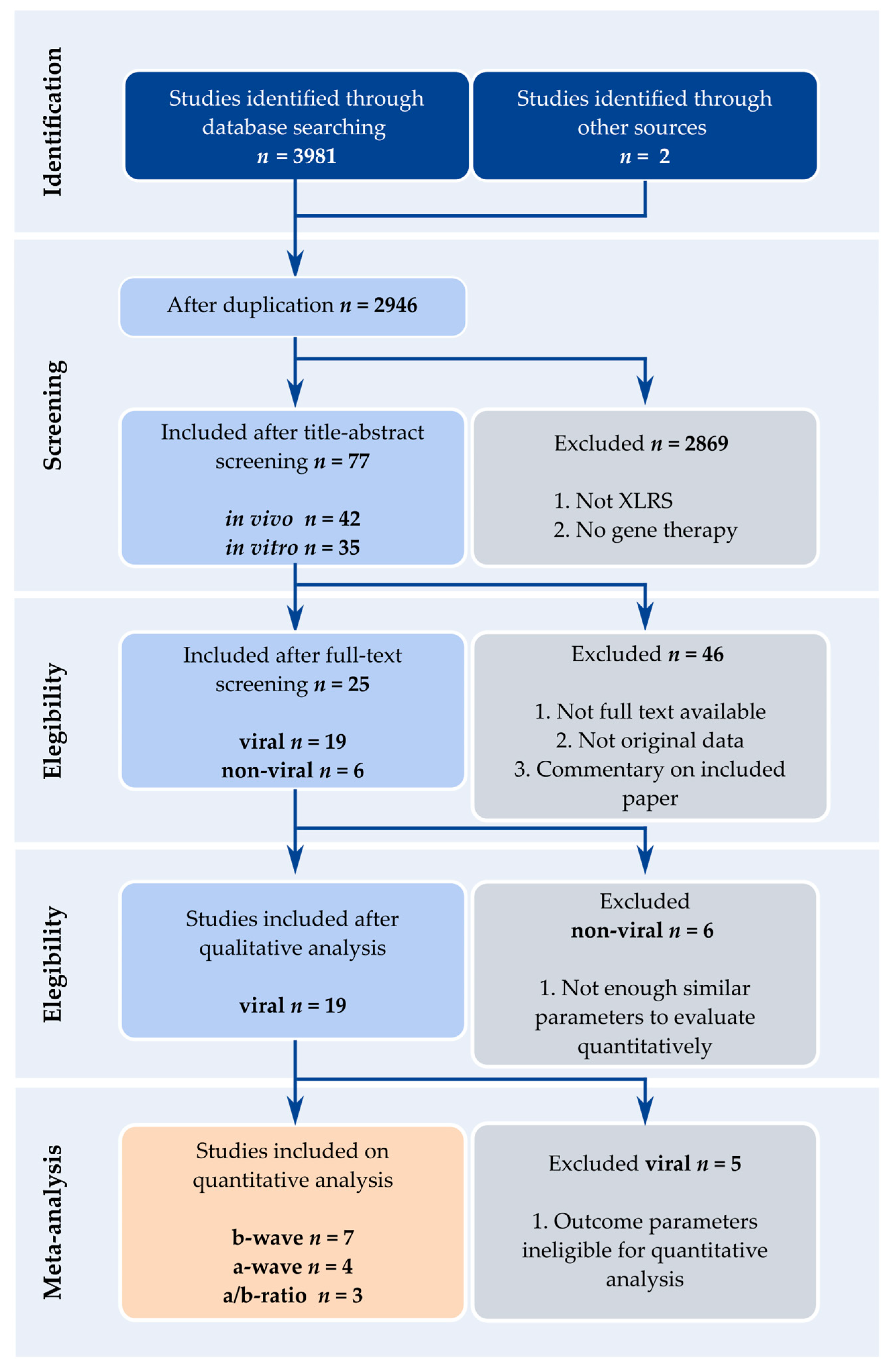
2.1. Literature Search
2.2. Eligibility and Exclusion Criteria
2.3. Data Extraction
2.4. Risk-of-Bias Assessment
2.5. Meta-Analyses
3. Results
3.1. Adeno-Associated Viral Vector-Mediated Gene Replacement Therapy in Rodent Models for X-Linked Juvenile Retinoschisis
3.1.1. Use of Rodent Models for X-Linked Juvenile Retinoschisis
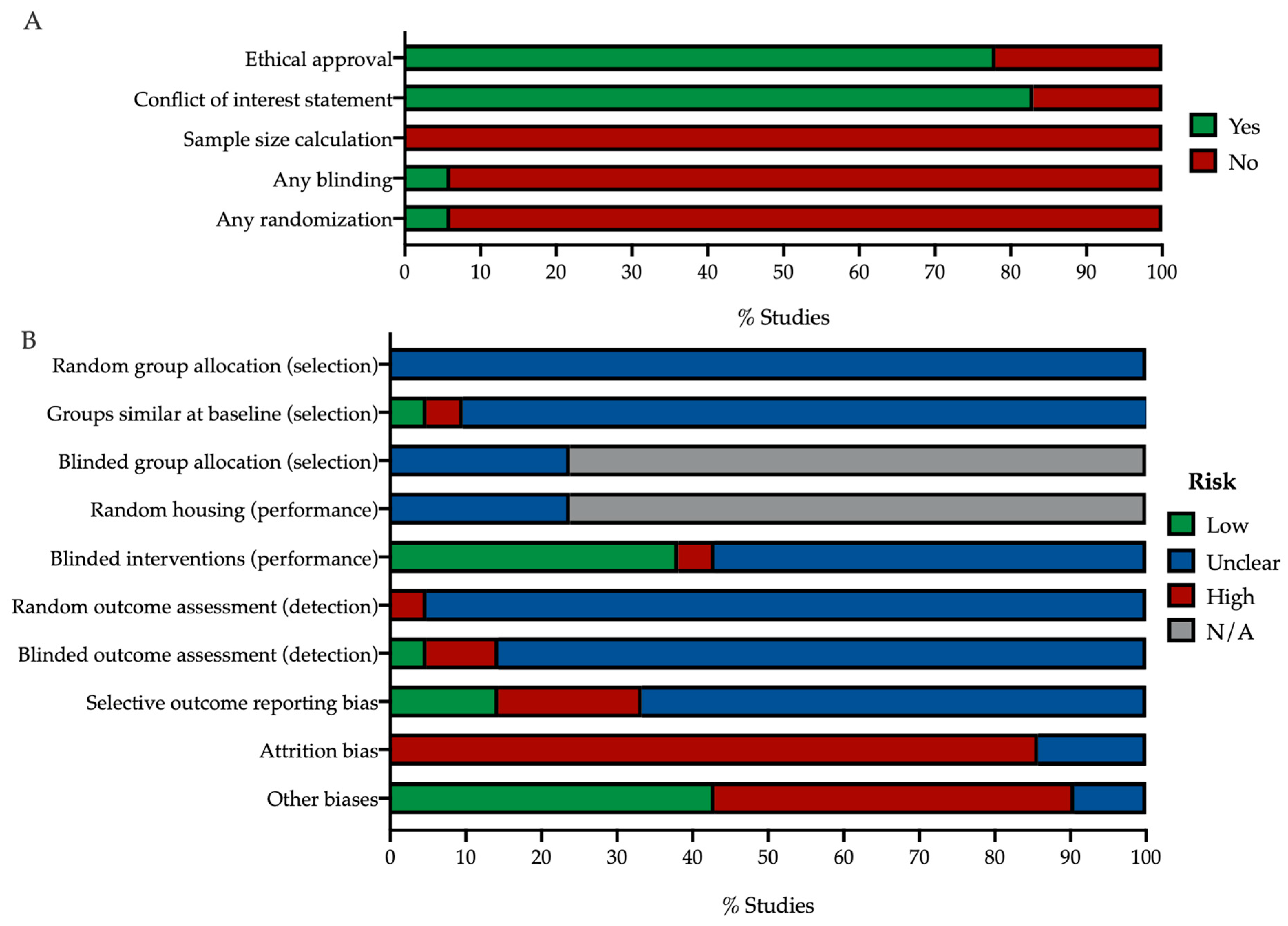
3.1.2. Risk of Bias
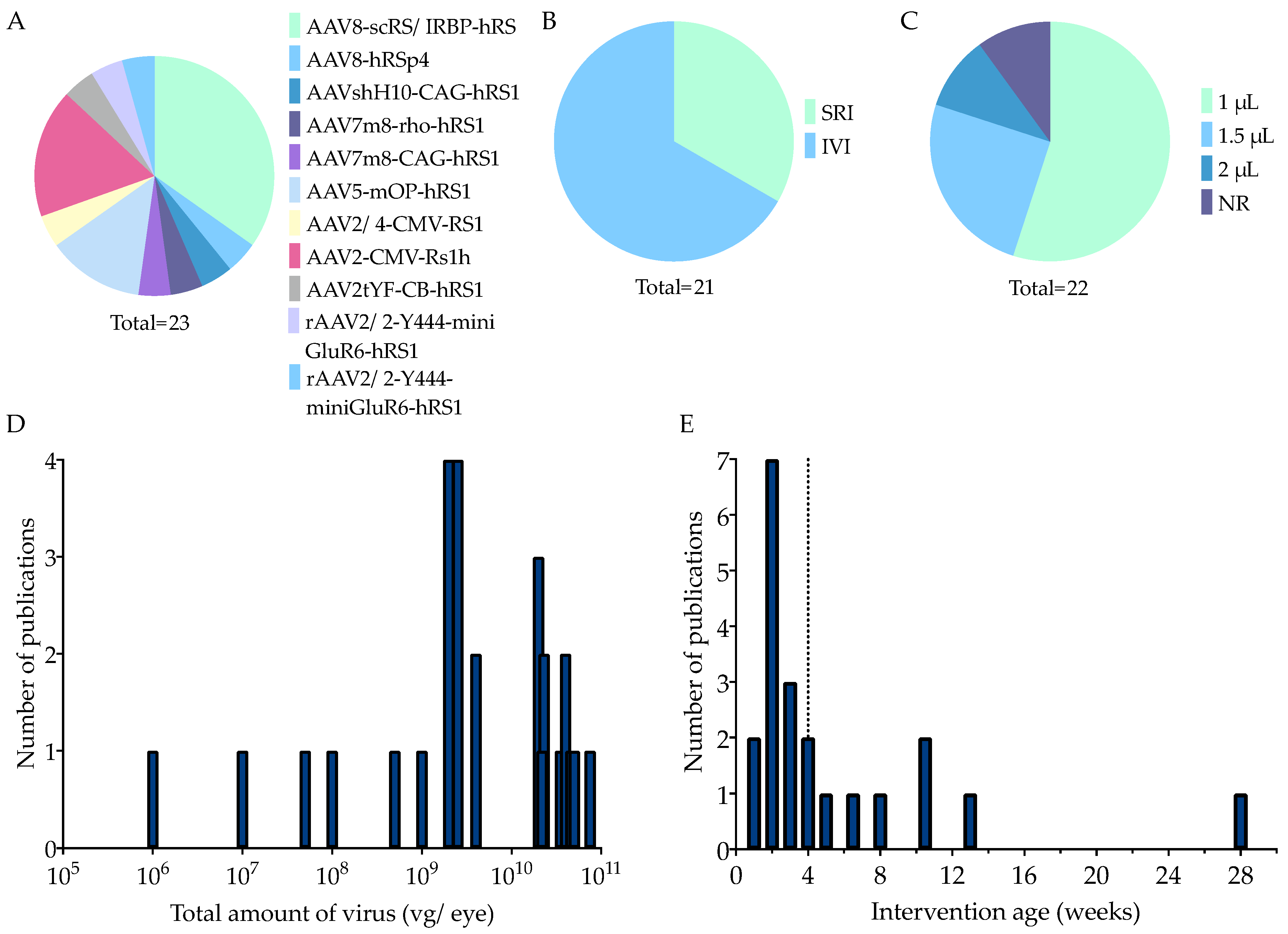
3.1.3. Main In Vivo Study Characteristics
Animal Characteristics
Therapy Characteristics
3.1.4. Outcome Parameters and Timepoints
3.1.5. Vision-Related Behavior
3.1.6. Morphological Read-Outs
Optical Coherence Tomography
Immunohistochemistry
3.1.7. Electroretinography
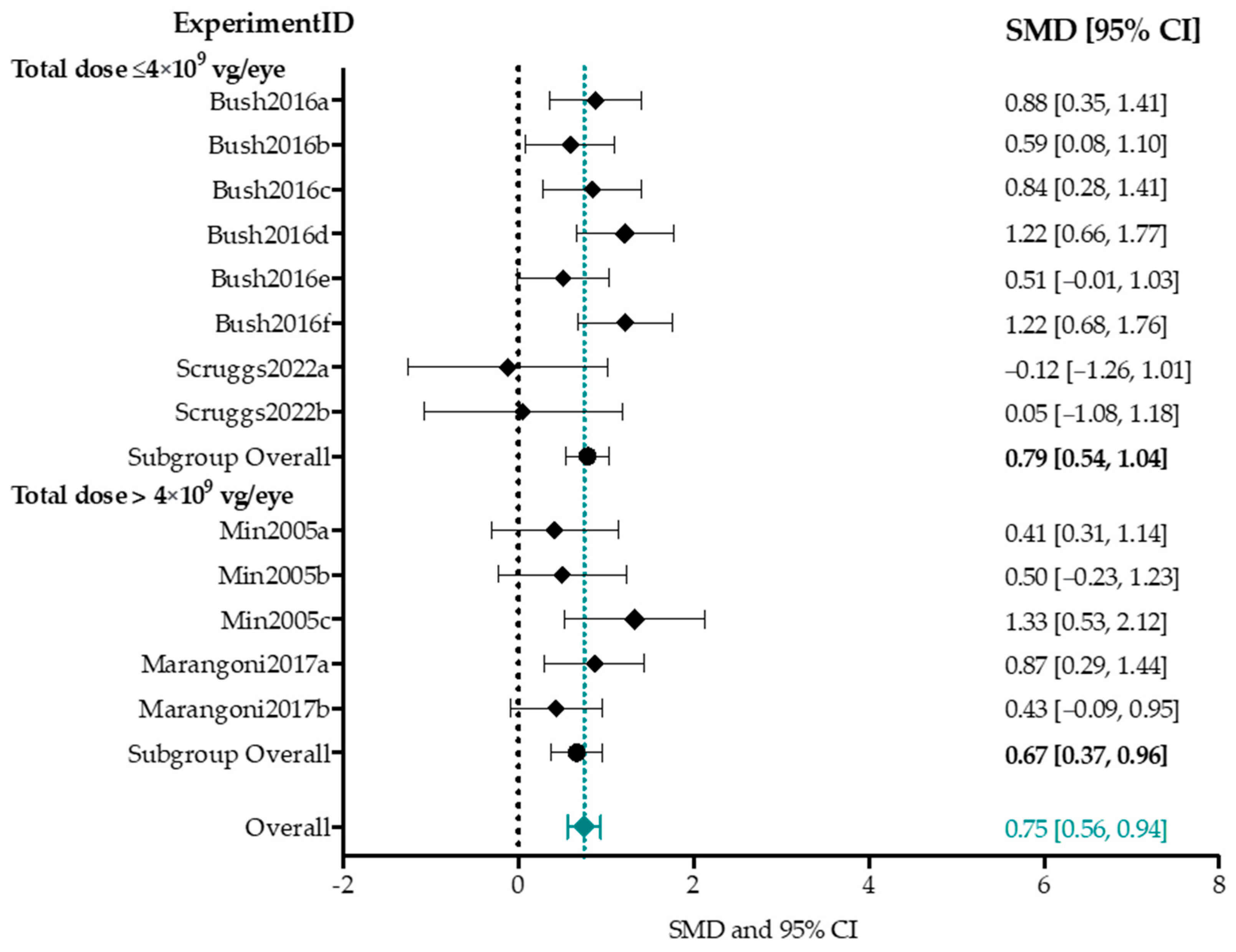
A-Wave Amplitude
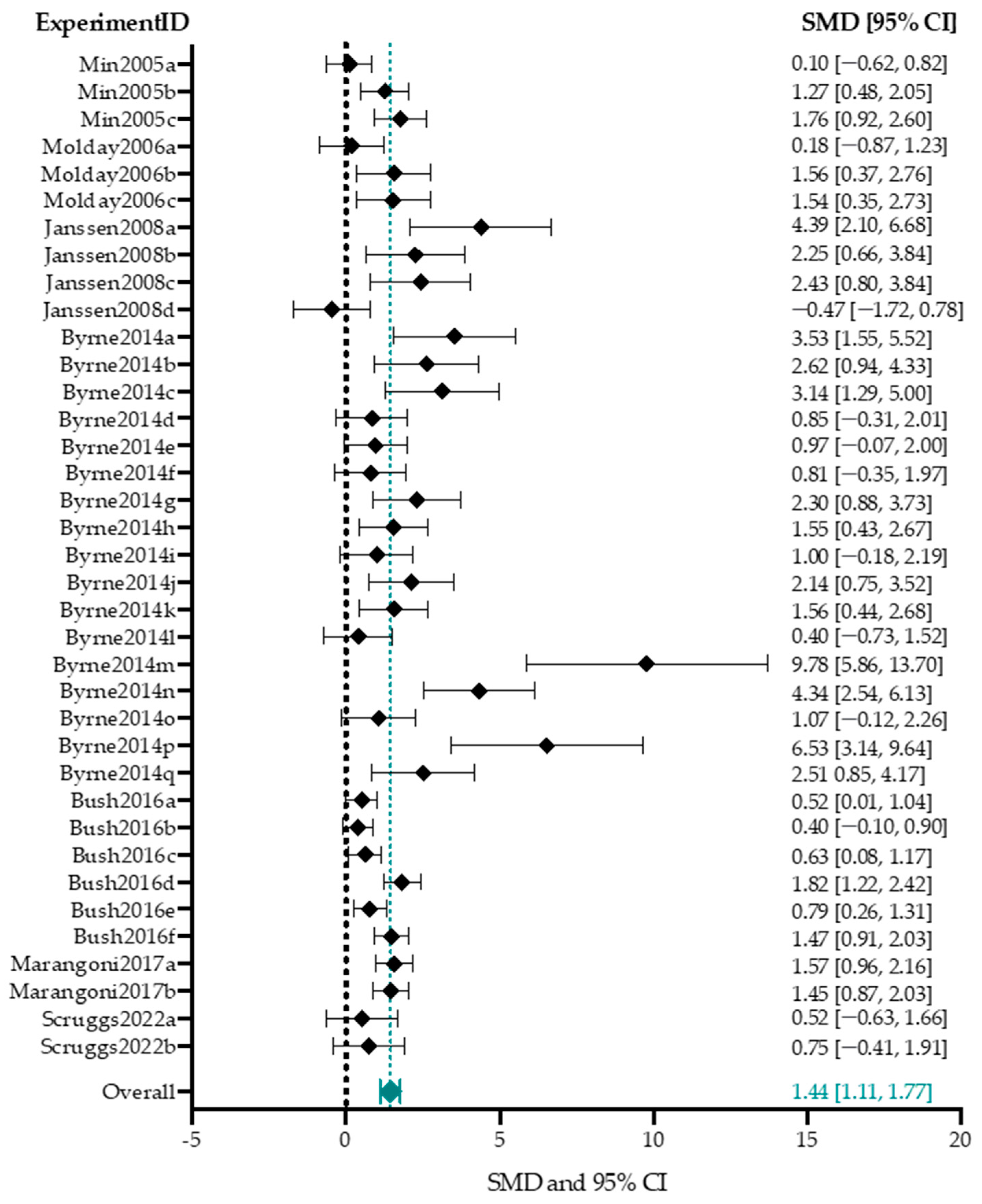
B-Wave Amplitude
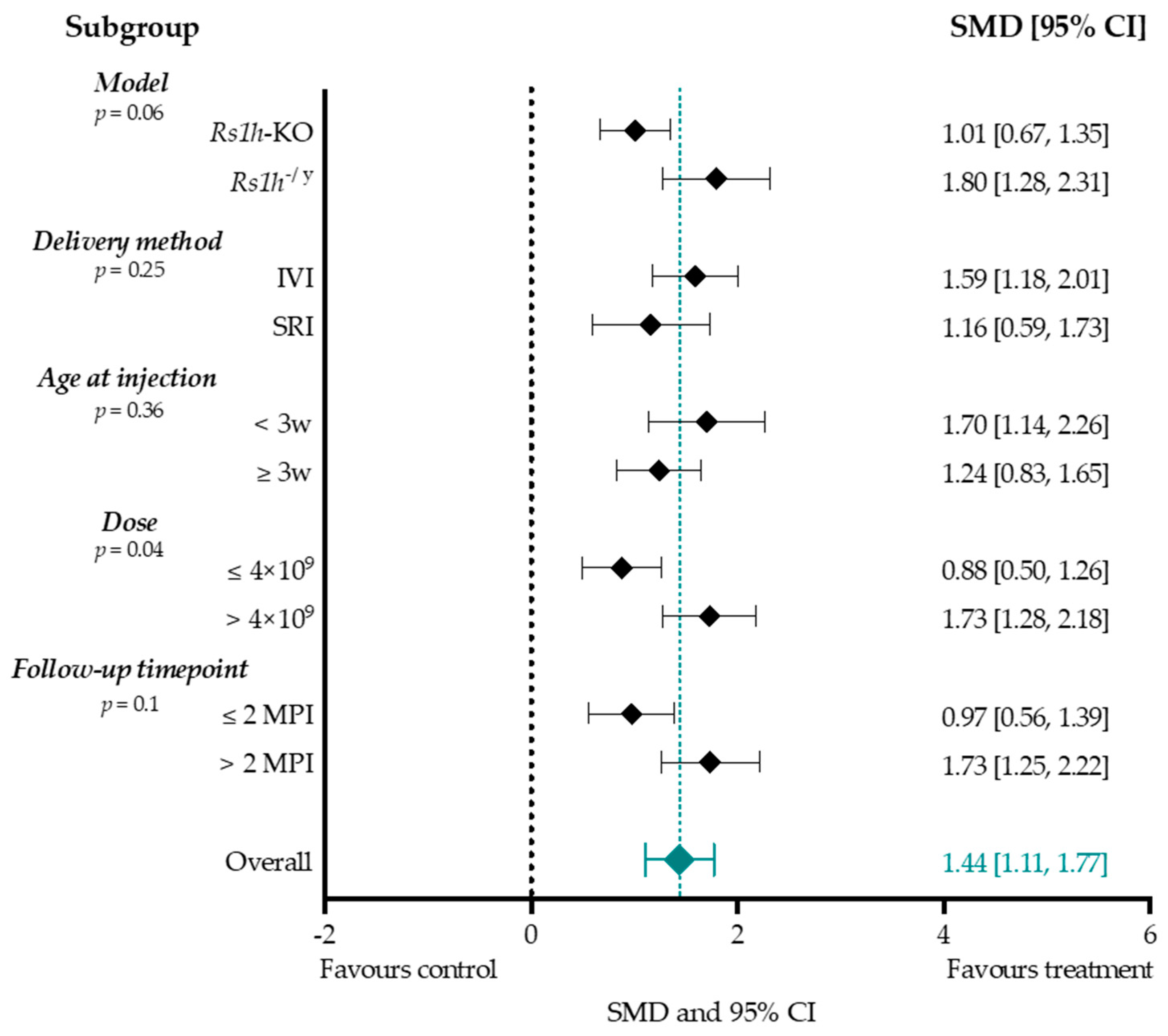
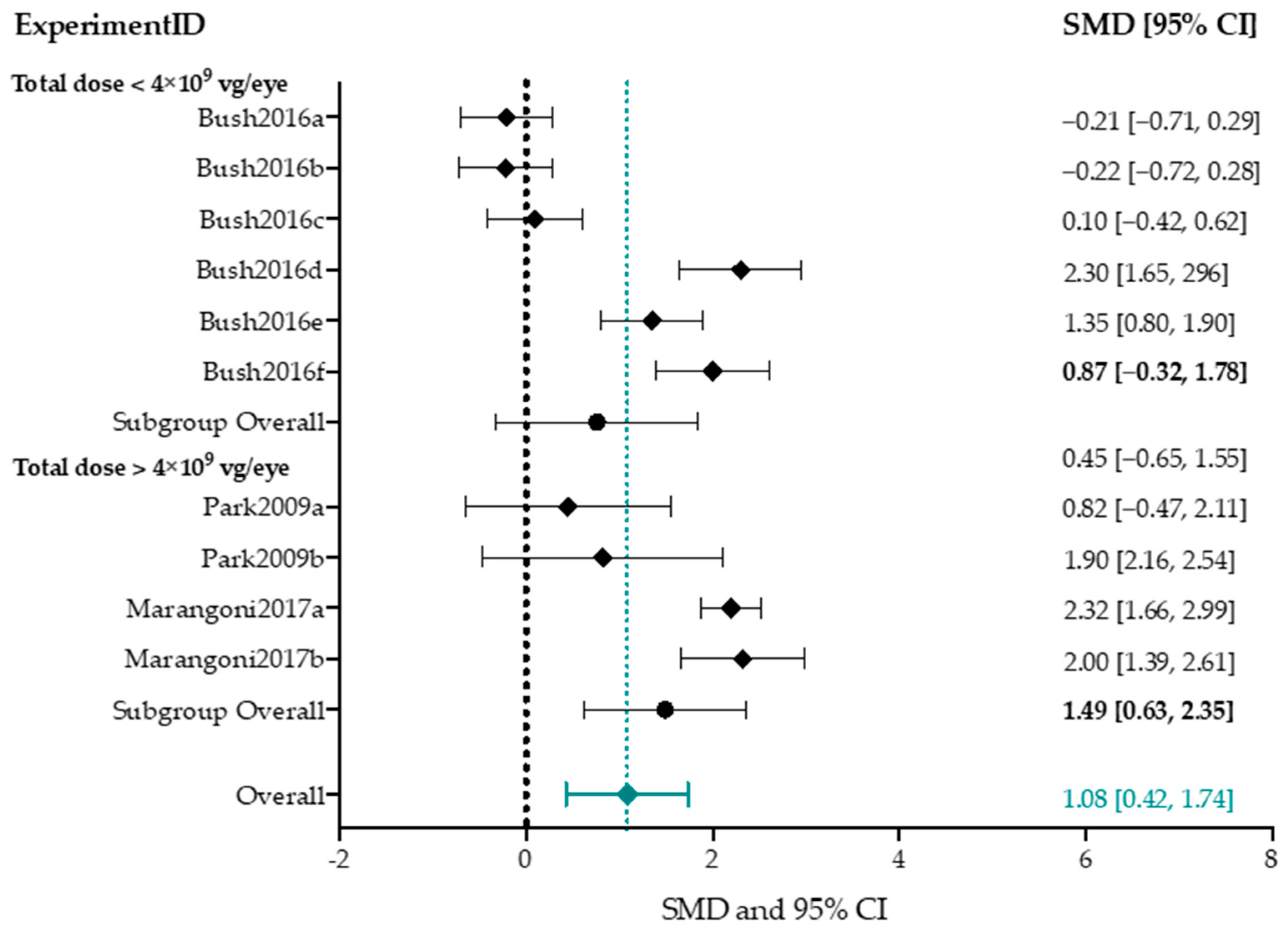
Subgroup Analysis b-Wave Amplitude
B/a-Wave Amplitude Ratio
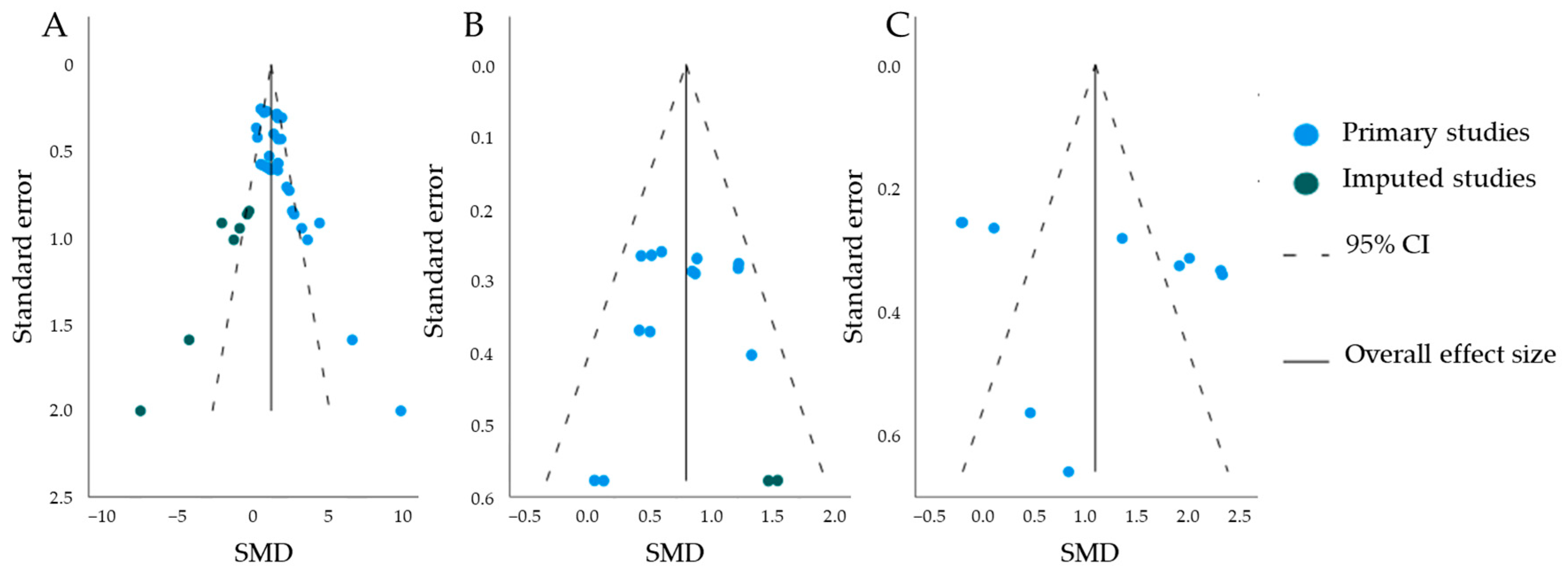
Publication Bias
3.2. Alternative Delivery and Non-Viral Delivery Methods to Target Retinoschisin in Cell-Based and Rodent Models
3.2.1. Use of Alternative Delivery and Non-Viral Methods for X-Linked Juvenile Retinoschisis
3.2.2. In-Depth Analysis of Exo-Adeno-Associated Viral Vector Delivery and Non-Viral Delivery Methods for Retinoschisin Integration
Exo-AAV Delivery Method
Non-Viral Chemical Methods
3.2.3. In Depth Analysis of Non-Viral Delivery Methods to Perform Gene Editing
Non-Viral Physical Method
Non-Viral Chemical Methods
3.2.4. In Vitro Readouts
3.2.5. In Vivo Readouts
4. Discussion
4.1. The Preferred Practices of In Vivo Study Characteristics
4.2. In Vivo Study Quality Assessment Reveals Insufficient Self-Reporting
4.3. In Vivo Qualitative Analysis Shows Variability in Outcome Parameters
4.4. Effect-Size Analyses Show a Significant Effect of Adeno-Associated Viral Vector-Mediated Gene Therapy on a- and b-Wave Amplitudes, and b/a-Ratio
4.4.1. Higher Titers Increase Therapy Effect on b-Wave Amplitude
4.4.2. Considerations When Choosing a Delivery Method
4.4.3. Considerations When Choosing an Intervention Age
4.4.4. Treatment Effect Size Was Increased When Evaluated at Two Months or Later
4.5. Discrepancies between Structural and Functional Readouts
4.6. Adeno-Associated Viral Vector Immunogenicity Considerations
4.7. Alternative Viral and Non-Viral Strategies as Novel Methods to Integrate or Edit Retinoschisin
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| AAV | adeno-associated viral vector |
| Ad | adamantane-grafted |
| b/a-ratio | ratio between the b-wave and a-wave amplitudes |
| Bp | base pair |
| C | cytosine |
| CAG | complex consisting of CMV early enhancer (C), chicken beta-actin promoter, first intron and first exon (A), and rabbit beta-globin splice adaptor (G) |
| CB | chicken beta-actin |
| CD-PEI | carbon dot- polyethylenimine |
| CI | confidence interval |
| CMV | cytomegalovirus |
| cND | carboxylated nanodiamond |
| CRISPR | clustered regularly interspaced palindromic repeats |
| D | days |
| EGFP | enhanced green fluorescent protein |
| ENU | N-ethyl-N-nitroso-ureum |
| ERG | electroretinography |
| EMA | European medicine agency |
| Exo-AAV | exosome-associated AAV |
| FDA | Food and drug administration |
| GFAP | glial-fibrillary protein |
| GFP | green fluorescent protein |
| H | hours |
| hiPSC | human induced pluripotent stem cell |
| hRSp4 | a complex consisting of the RS1 promoter, human RS1 cDNA with a truncated first intron, human beta-globin polyadenylation site and AAV2 inverted terminal repeats |
| H&E | haematoxylin and eosin |
| IHC | immunohistochemistry |
| ILM | inner limiting membrane |
| INL | inner nuclear layer |
| IRBP | interphotoreceptor retinoid-binding protein |
| IRD | inherited retinal disease |
| IS | inner segment |
| IVI | intravitreal injection |
| Kb | kilobase |
| KI | knock-in |
| KO | knock-out |
| M | months |
| Mini-mGluR6 | a shortened version of the ON bipolar cell-specific promoter mGlu6 |
| miRNA | microRNA |
| mOP | mouse opsin |
| MPI | months post-injection |
| mRNA | messenger ribonucleic acid |
| N | no |
| NR | not reported |
| N/A | not-applicable |
| OCT | optical coherence tomography |
| ONL | outer nuclear layer |
| OS | outer segment |
| P | postnatal day |
| PANAM | polyamidoamine |
| PBS | phosphate buffered saline |
| pCEP4 | vector encompassing the CMV promoter and a hygromycin resistance marker |
| pCMS | immediate early promoter of CMV |
| PEG | polyethylene glycol |
| Ple155 | human-DNA MiniPromoter specific to ON bipolar cells |
| pRS1HR-gRNA | plasmid including a 950-bp RS1 repair template and the gRNA expression cassette |
| PI | post-injection |
| PR | photoreceptor |
| Rho | rhodopsin |
| RPE | retinal pigment epithelium |
| RS1 | retinoschisin |
| Rs1h | RS1 homolog in rodents |
| scRS/IRBP | modified RS1 promoter coupled with an interphotoreceptor retinoid-binding protein |
| sgRNA | single guide RNA |
| shH10 | Müller-cell specific serotype derived from the AAV6 shuffled library |
| siRNA | small interfering RNA |
| SLN | solid lipid nanoparticles |
| SLO | scanning laser ophthalmoscopy |
| SMNP | supramolecular nanoparticle |
| SMD | standardized mean differences |
| SRI | subretinal injection |
| T | thymine |
| TAT | adenosine-5’-rp-alpha-thio-triphosphate |
| U6 | type III RNA polymerase promoter |
| VA | visual acuity |
| Vg/eye | vector genomes per eye |
| W | weeks |
| WB | Western blot |
| WT | wildtype |
| XLRS | X-linked juvenile retinoschisis |
| ZsGreen | Zoanthus sp. Green |
| 2tYF | AAV2-derived serotype with three tyrosine (Y) residues mutated to phenylalanine (F) |
References
- Khorrami-Nejad, M.; Sarabandi, A.; Akbari, M.R.; Askarizadeh, F. The Impact of Visual Impairment on Quality of Life. Med. Hypothesis Discov. Innov. Ophthalmol. 2016, 5, 96–103. [Google Scholar]
- Haas, J. Ueber das Zusammenvorkommen von Veraenderungen der Retina und Choroidea. Arch. Augenheilkd. 1898, 37, 343–348. [Google Scholar]
- George, N.D.; Yates, J.R.; Moore, A.T. X linked retinoschisis. Br. J. Ophthalmol. 1995, 79, 697–702. [Google Scholar] [CrossRef] [PubMed]
- Hahn, L.C.; van Schooneveld, M.J.; Wesseling, N.L.; Florijn, R.J.; Ten Brink, J.B.; Lissenberg-Witte, B.I.; Strubbe, I.; Meester-Smoor, M.A.; Thiadens, A.A.; Diederen, R.M.; et al. X-Linked Retinoschisis: Novel Clinical Observations and Genetic Spectrum in 340 Patients. Ophthalmology 2022, 129, 191–202. [Google Scholar] [CrossRef] [PubMed]
- Kirkby, J.; Halford, S.; Shanks, M.; Moore, A.; Gait, A.; Jenkins, L.; Clouston, P.; Patel, C.K.; Downes, S.M. A Carrier Female Manifesting an Unusual X-Linked Retinoschisis Phenotype Associated with the Pathogenic Variant c.266delA, p.(Tyr89LeufsTer37) in RS1, and Skewed X-Inactivation. Genes 2023, 14, 1193. [Google Scholar] [CrossRef] [PubMed]
- Kaplan, J.; Pelet, A.; Hentati, H.; Jeanpierre, M.; Briard, M.L.; Journel, H.; Munnich, A.; Dufier, J.L. Contribution to carrier detection and genetic counselling in X linked retinoschisis. J. Med. Genet. 1991, 28, 383–388. [Google Scholar] [CrossRef] [PubMed]
- Kim, L.S.; Seiple, W.; Fishman, G.A.; Szlyk, J.P. Multifocal ERG findings in carriers of X-linked retinoschisis. Doc. Ophthalmol. 2007, 114, 21–26. [Google Scholar] [CrossRef]
- Kjellström, S.; Vijayasarathy, C.; Ponjavic, V.; Sieving, P.A.; Andréasson, S. Long-term 12 year follow-up of X-linked congenital retinoschisis. Ophthalmic Genet. 2010, 31, 114–125. [Google Scholar] [CrossRef]
- Kellner, U.; Brümmer, S.; Foerster, M.H.; Wessing, A. X-linked congenital retinoschisis. Graefes Arch. Clin. Exp. Ophthalmol. 1990, 228, 432–437. [Google Scholar] [CrossRef]
- Tantri, A.; Vrabec, T.R.; Cu-Unjieng, A.; Frost, A.; Annesley, W.H., Jr.; Donoso, L.A. X-linked retinoschisis: A clinical and molecular genetic review. Surv. Ophthalmol. 2004, 49, 214–230. [Google Scholar] [CrossRef]
- Molday, R.S.; Kellner, U.; Weber, B.H. X-linked juvenile retinoschisis: Clinical diagnosis, genetic analysis, and molecular mechanisms. Prog. Retin. Eye Res. 2012, 31, 195–212. [Google Scholar] [CrossRef] [PubMed]
- Khan, N.W.; Jamison, J.A.; Kemp, J.A.; Sieving, P.A. Analysis of photoreceptor function and inner retinal activity in juvenile X-linked retinoschisis. Vis. Res. 2001, 41, 3931–3942. [Google Scholar] [CrossRef] [PubMed]
- Fairless, R.; Williams, S.K.; Katiyar, R.; Maxeiner, S.; Schmitz, F.; Diem, R. ERG Responses in Mice with Deletion of the Synaptic Ribbon Component RIBEYE. Investig. Ophthalmol. Vis. Sci. 2020, 61, 37. [Google Scholar] [CrossRef] [PubMed]
- Sauer, C.G.; Gehrig, A.; Warneke-Wittstock, R.; Marquardt, A.; Ewing, C.C.; Gibson, A.; Lorenz, B.; Jurklies, B.; Weber, B.H. Positional cloning of the gene associated with X-linked juvenile retinoschisis. Nat. Genet. 1997, 17, 164–170. [Google Scholar] [CrossRef] [PubMed]
- Sikkink, S.K.; Biswas, S.; Parry, N.R.; Stanga, P.E.; Trump, D. X-linked retinoschisis: An update. J. Med. Genet. 2007, 44, 225–232. [Google Scholar] [CrossRef] [PubMed]
- The Retinoschisis Consortium. Functional implications of the spectrum of mutations found in 234 cases with X-linked juvenile retinoschisis. Hum. Mol. Genet. 1998, 7, 1185–1192. [Google Scholar] [CrossRef]
- Vijayasarathy, C.; Zeng, Y.; Brooks, M.J.; Fariss, R.N.; Sieving, P.A. Genetic Rescue of X-Linked Retinoschisis Mouse (Rs1(-/y)) Retina Induces Quiescence of the Retinal Microglial Inflammatory State Following AAV8-RS1 Gene Transfer and Identifies Gene Networks Underlying Retinal Recovery. Hum. Gene Ther. 2021, 32, 667–681. [Google Scholar] [CrossRef]
- Molday, L.L.; Hicks, D.; Sauer, C.G.; Weber, B.H.; Molday, R.S. Expression of X-linked retinoschisis protein RS1 in photoreceptor and bipolar cells. Investig. Ophthalmol. Vis. Sci. 2001, 42, 816–825. [Google Scholar]
- Takada, Y.; Fariss, R.N.; Muller, M.; Bush, R.A.; Rushing, E.J.; Sieving, P.A. Retinoschisin expression and localization in rodent and human pineal and consequences of mouse RS1 gene knockout. Mol. Vis. 2006, 12, 1108–1116. [Google Scholar]
- Sjöstedt, E.; Zhong, W.; Fagerberg, L.; Karlsson, M.; Mitsios, N.; Adori, C.; Oksvold, P.; Edfors, F.; Limiszewska, A.; Hikmet, F.; et al. An atlas of the protein-coding genes in the human, pig, and mouse brain. Science 2020, 367, eaay5947. [Google Scholar] [CrossRef]
- Wu, W.W.; Molday, R.S. Defective discoidin domain structure, subunit assembly, and endoplasmic reticulum processing of retinoschisin are primary mechanisms responsible for X-linked retinoschisis. J. Biol. Chem. 2003, 278, 28139–28146. [Google Scholar] [CrossRef] [PubMed]
- Wu, W.W.; Wong, J.P.; Kast, J.; Molday, R.S. RS1, a discoidin domain-containing retinal cell adhesion protein associated with X-linked retinoschisis, exists as a novel disulfide-linked octamer. J. Biol. Chem. 2005, 280, 10721–10730. [Google Scholar] [CrossRef] [PubMed]
- Tolun, G.; Vijayasarathy, C.; Huang, R.; Zeng, Y.; Li, Y.; Steven, A.C.; Sieving, P.A.; Heymann, J.B. Paired octamer rings of retinoschisin suggest a junctional model for cell-cell adhesion in the retina. Proc. Natl. Acad. Sci. USA 2016, 113, 5287–5292. [Google Scholar] [CrossRef] [PubMed]
- Vogel, W. Discoidin domain receptors: Structural relations and functional implications. Faseb J. 1999, 13, S77–S82. [Google Scholar] [CrossRef] [PubMed]
- Kotani, E.; Yamakawa, M.; Iwamoto, S.; Tashiro, M.; Mori, H.; Sumida, M.; Matsubara, F.; Taniai, K.; Kadono-Okuda, K.; Kato, Y.; et al. Cloning and expression of the gene of hemocytin, an insect humoral lectin which is homologous with the mammalian von Willebrand factor. Biochim. Biophys. Acta 1995, 1260, 245–258. [Google Scholar] [CrossRef]
- Takagi, S.; Kasuya, Y.; Shimizu, M.; Matsuura, T.; Tsuboi, M.; Kawakami, A.; Fujisawa, H. Expression of a cell adhesion molecule, neuropilin, in the developing chick nervous system. Dev. Biol. 1995, 170, 207–222. [Google Scholar] [CrossRef]
- Takagi, S.; Hirata, T.; Agata, K.; Mochii, M.; Eguchi, G.; Fujisawa, H. The A5 antigen, a candidate for the neuronal recognition molecule, has homologies to complement components and coagulation factors. Neuron 1991, 7, 295–307. [Google Scholar] [CrossRef]
- Kawakami, A.; Kitsukawa, T.; Takagi, S.; Fujisawa, H. Developmentally regulated expression of a cell surface protein, neuropilin, in the mouse nervous system. J. Neurobiol. 1996, 29, 1–17. [Google Scholar] [CrossRef]
- Stubbs, J.D.; Lekutis, C.; Singer, K.L.; Bui, A.; Yuzuki, D.; Srinivasan, U.; Parry, G. cDNA cloning of a mouse mammary epithelial cell surface protein reveals the existence of epidermal growth factor-like domains linked to factor VIII-like sequences. Proc. Natl. Acad. Sci. USA 1990, 87, 8417–8421. [Google Scholar] [CrossRef]
- Cripe, L.D.; Moore, K.D.; Kane, W.H. Structure of the gene for human coagulation factor V. Biochemistry 1992, 31, 3777–3785. [Google Scholar] [CrossRef]
- Elder, B.; Lakich, D.; Gitschier, J. Sequence of the murine factor VIII cDNA. Genomics 1993, 16, 374–379. [Google Scholar] [CrossRef] [PubMed]
- Vijayasarathy, C.; Sui, R.; Zeng, Y.; Yang, G.; Xu, F.; Caruso, R.C.; Lewis, R.A.; Ziccardi, L.; Sieving, P.A. Molecular mechanisms leading to null-protein product from retinoschisin (RS1) signal-sequence mutants in X-linked retinoschisis (XLRS) disease. Hum. Mutat. 2010, 31, 1251–1260. [Google Scholar] [CrossRef] [PubMed]
- Takada, Y.; Fariss, R.N.; Tanikawa, A.; Zeng, Y.; Carper, D.; Bush, R.; Sieving, P.A. A retinal neuronal developmental wave of retinoschisin expression begins in ganglion cells during layer formation. Investig. Ophthalmol. Vis. Sci. 2004, 45, 3302–3312. [Google Scholar] [CrossRef] [PubMed]
- Vijayasarathy, C.; Zeng, Y.; Marangoni, D.; Dong, L.; Pan, Z.H.; Simpson, E.M.; Fariss, R.N.; Sieving, P.A. Targeted Expression of Retinoschisin by Retinal Bipolar Cells in XLRS Promotes Resolution of Retinoschisis Cysts Sans RS1 From Photoreceptors. Investig. Ophthalmol. Vis. Sci. 2022, 63, 8. [Google Scholar] [CrossRef] [PubMed]
- Reid, S.N.; Akhmedov, N.B.; Piriev, N.I.; Kozak, C.A.; Danciger, M.; Farber, D.B. The mouse X-linked juvenile retinoschisis cDNA: Expression in photoreceptors. Gene 1999, 227, 257–266. [Google Scholar] [CrossRef] [PubMed]
- Sen, P.; Agarwal, A.; Bhende, P.; Gopal, L.; Bhende, M.; Rishi, P.; Sharma, T.; Jain, M. Outcome of vitreoretinal surgery for rhegmatogenous retinal detachment in X-linked juvenile retinoschisis. Indian J. Ophthalmol. 2018, 66, 1825–1831. [Google Scholar] [CrossRef]
- Apushkin, M.A.; Fishman, G.A. Use of dorzolamide for patients with X-linked retinoschisis. Retina 2006, 26, 741–745. [Google Scholar] [CrossRef]
- Verbakel, S.K.; van de Ven, J.P.; Le Blanc, L.M.; Groenewoud, J.M.; de Jong, E.K.; Klevering, B.J.; Hoyng, C.B. Carbonic Anhydrase Inhibitors for the Treatment of Cystic Macular Lesions in Children With X-Linked Juvenile Retinoschisis. Investig. Ophthalmol. Vis. Sci. 2016, 57, 5143–5147. [Google Scholar] [CrossRef]
- Testa, F.; Di Iorio, V.; Gallo, B.; Marchese, M.; Nesti, A.; De Rosa, G.; Melillo, P.; Simonelli, F. Carbonic anhydrase inhibitors in patients with X-linked retinoschisis: Effects on macular morphology and function. Ophthalmic Genet. 2019, 40, 207–212. [Google Scholar] [CrossRef]
- Genead, M.A.; Fishman, G.A.; Walia, S. Efficacy of sustained topical dorzolamide therapy for cystic macular lesions in patients with X-linked retinoschisis. Arch. Ophthalmol. 2010, 128, 190–197. [Google Scholar] [CrossRef]
- Khandhadia, S.; Trump, D.; Menon, G.; Lotery, A.J. X-linked retinoschisis maculopathy treated with topical dorzolamide, and relationship to genotype. Eye 2011, 25, 922–928. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L.; Reyes, R.; Lee, W.; Chen, C.L.; Chan, L.; Sujirakul, T.; Chang, S.; Tsang, S.H. Rapid resolution of retinoschisis with acetazolamide. Doc. Ophthalmol. 2015, 131, 63–70. [Google Scholar] [CrossRef] [PubMed]
- Hensman, J.; Hahn, L.C.; van Schooneveld, M.J.; Diederen, R.M.H.; Ten Brink, J.B.; Florijn, R.J.; Bergen, A.A.; Strubbe, I.; Heutinck, P.; van Genderen, M.M.; et al. Efficacy of carbonic anhydrase inhibitors on cystoid fluid collections and visual acuity in patients with X-linked retinoschisis. Ophthalmol. Retina 2023. [Google Scholar] [CrossRef]
- Wang, D.; Tai, P.W.L.; Gao, G. Adeno-associated virus vector as a platform for gene therapy delivery. Nat. Rev. Drug Discov. 2019, 18, 358–378. [Google Scholar] [CrossRef] [PubMed]
- Qu, Y.; Liu, Y.; Noor, A.F.; Tran, J.; Li, R. Characteristics and advantages of adeno-associated virus vector-mediated gene therapy for neurodegenerative diseases. Neural Regen. Res. 2019, 14, 931–938. [Google Scholar] [CrossRef]
- Chien, Y.; Hsiao, Y.J.; Chou, S.J.; Lin, T.Y.; Yarmishyn, A.A.; Lai, W.Y.; Lee, M.S.; Lin, Y.Y.; Lin, T.W.; Hwang, D.K.; et al. Nanoparticles-mediated CRISPR-Cas9 gene therapy in inherited retinal diseases: Applications, challenges, and emerging opportunities. J. Nanobiotechnol. 2022, 20, 511. [Google Scholar] [CrossRef]
- Stingl, K.; Stingl, K.; Schwartz, H.; Reid, M.W.; Kempf, M.; Dimopoulos, S.; Kortuem, F.; Borchert, M.S.; Lee, T.C.; Nagiel, A. Full-field Scotopic Threshold Improvement after Voretigene Neparvovec-rzyl Treatment Correlates with Chorioretinal Atrophy. Ophthalmology 2023, 130, 764–770. [Google Scholar] [CrossRef] [PubMed]
- Bainbridge, J.W.; Smith, A.J.; Barker, S.S.; Robbie, S.; Henderson, R.; Balaggan, K.; Viswanathan, A.; Holder, G.E.; Stockman, A.; Tyler, N.; et al. Effect of gene therapy on visual function in Leber’s congenital amaurosis. N. Engl. J. Med. 2008, 358, 2231–2239. [Google Scholar] [CrossRef]
- Jacobson, S.G.; Cideciyan, A.V.; Ratnakaram, R.; Heon, E.; Schwartz, S.B.; Roman, A.J.; Peden, M.C.; Aleman, T.S.; Boye, S.L.; Sumaroka, A.; et al. Gene therapy for leber congenital amaurosis caused by RPE65 mutations: Safety and efficacy in 15 children and adults followed up to 3 years. Arch. Ophthalmol. 2012, 130, 9–24. [Google Scholar] [CrossRef]
- MacLaren, R.E.; Groppe, M.; Barnard, A.R.; Cottriall, C.L.; Tolmachova, T.; Seymour, L.; Clark, K.R.; During, M.J.; Cremers, F.P.; Black, G.C.; et al. Retinal gene therapy in patients with choroideremia: Initial findings from a phase 1/2 clinical trial. Lancet 2014, 383, 1129–1137. [Google Scholar] [CrossRef]
- Nuzbrokh, Y.; Ragi, S.D.; Tsang, S.H. Gene therapy for inherited retinal diseases. Ann. Transl. Med. 2021, 9, 1278. [Google Scholar] [CrossRef] [PubMed]
- Schultz, B.R.; Chamberlain, J.S. Recombinant adeno-associated virus transduction and integration. Mol. Ther. 2008, 16, 1189–1199. [Google Scholar] [CrossRef] [PubMed]
- Nieuwenhuis, B.; Haenzi, B.; Hilton, S.; Carnicer-Lombarte, A.; Hobo, B.; Verhaagen, J.; Fawcett, J.W. Optimization of adeno-associated viral vector-mediated transduction of the corticospinal tract: Comparison of four promoters. Gene Ther. 2021, 28, 56–74. [Google Scholar] [CrossRef] [PubMed]
- Byrne, L.C.; Oztürk, B.E.; Lee, T.; Fortuny, C.; Visel, M.; Dalkara, D.; Schaffer, D.V.; Flannery, J.G. Retinoschisin gene therapy in photoreceptors, Müller glia or all retinal cells in the Rs1h−/− mouse. Gene Ther. 2014, 21, 585–592. [Google Scholar] [CrossRef]
- Marangoni, D.; Bush, R.A.; Zeng, Y.; Wei, L.L.; Ziccardi, L.; Vijayasarathy, C.; Bartoe, J.T.; Palyada, K.; Santos, M.; Hiriyanna, S.; et al. Ocular and systemic safety of a recombinant AAV8 vector for X-linked retinoschisis gene therapy: GLP studies in rabbits and Rs1-KO mice. Mol. Ther. Methods Clin. Dev. 2016, 5, 16011. [Google Scholar] [CrossRef]
- Bainbridge, J.W.; Mehat, M.S.; Sundaram, V.; Robbie, S.J.; Barker, S.E.; Ripamonti, C.; Georgiadis, A.; Mowat, F.M.; Beattie, S.G.; Gardner, P.J.; et al. Long-term effect of gene therapy on Leber’s congenital amaurosis. N. Engl. J. Med. 2015, 372, 1887–1897. [Google Scholar] [CrossRef]
- Wang, C.; Pan, C.; Yong, H.; Wang, F.; Bo, T.; Zhao, Y.; Ma, B.; He, W.; Li, M. Emerging non-viral vectors for gene delivery. J. Nanobiotechnol. 2023, 21, 272. [Google Scholar] [CrossRef]
- Huang, X.; Chau, Y. Intravitreal nanoparticles for retinal delivery. Drug Discov. Today 2019, 24, 1510–1523. [Google Scholar] [CrossRef]
- Lan, T.; Que, H.; Luo, M.; Zhao, X.; Wei, X. Genome editing via non-viral delivery platforms: Current progress in personalized cancer therapy. Mol. Cancer 2022, 21, 71. [Google Scholar] [CrossRef]
- Collin, R.W.; Garanto, A. Applications of antisense oligonucleotides for the treatment of inherited retinal diseases. Curr. Opin. Ophthalmol. 2017, 28, 260–266. [Google Scholar] [CrossRef]
- Bondi, M.L.; Azzolina, A.; Craparo, E.F.; Lampiasi, N.; Capuano, G.; Giammona, G.; Cervello, M. Novel cationic solid-lipid nanoparticles as non-viral vectors for gene delivery. J. Drug Target. 2007, 15, 295–301. [Google Scholar] [CrossRef] [PubMed]
- Mellott, A.J.; Forrest, M.L.; Detamore, M.S. Physical non-viral gene delivery methods for tissue engineering. Ann. Biomed. Eng. 2013, 41, 446–468. [Google Scholar] [CrossRef] [PubMed]
- Apaolaza, P.S.; Del Pozo-Rodríguez, A.; Torrecilla, J.; Rodríguez-Gascón, A.; Rodríguez, J.M.; Friedrich, U.; Weber, B.H.; Solinís, M.A. Solid lipid nanoparticle-based vectors intended for the treatment of X-linked juvenile retinoschisis by gene therapy: In vivo approaches in Rs1h-deficient mouse model. J. Control Release 2015, 217, 273–283. [Google Scholar] [CrossRef] [PubMed]
- Davis, M.E. Non-viral gene delivery systems. Curr. Opin. Biotechnol. 2002, 13, 128–131. [Google Scholar] [CrossRef] [PubMed]
- Mintzer, M.A.; Simanek, E.E. Nonviral vectors for gene delivery. Chem. Rev. 2009, 109, 259–302. [Google Scholar] [CrossRef]
- Zu, H.; Gao, D. Non-viral Vectors in Gene Therapy: Recent Development, Challenges, and Prospects. Aaps J. 2021, 23, 78. [Google Scholar] [CrossRef]
- Kansara, V.; Muya, L.; Wan, C.R.; Ciulla, T.A. Suprachoroidal Delivery of Viral and Nonviral Gene Therapy for Retinal Diseases. J. Ocul. Pharmacol. Ther. 2020, 36, 384–392. [Google Scholar] [CrossRef]
- Cukras, C.; Wiley, H.E.; Jeffrey, B.G.; Sen, H.N.; Turriff, A.; Zeng, Y.; Vijayasarathy, C.; Marangoni, D.; Ziccardi, L.; Kjellstrom, S.; et al. Retinal AAV8-RS1 Gene Therapy for X-Linked Retinoschisis: Initial Findings from a Phase I/IIa Trial by Intravitreal Delivery. Mol. Ther. 2018, 26, 2282–2294. [Google Scholar] [CrossRef]
- Hooijmans, C.R.; Rovers, M.M.; de Vries, R.B.; Leenaars, M.; Ritskes-Hoitinga, M.; Langendam, M.W. SYRCLE’s risk of bias tool for animal studies. BMC Med. Res. Methodol. 2014, 14, 43. [Google Scholar] [CrossRef]
- Weber, B.H.; Schrewe, H.; Molday, L.L.; Gehrig, A.; White, K.L.; Seeliger, M.W.; Jaissle, G.B.; Friedburg, C.; Tamm, E.; Molday, R.S. Inactivation of the murine X-linked juvenile retinoschisis gene, Rs1h, suggests a role of retinoschisin in retinal cell layer organization and synaptic structure. Proc. Natl. Acad. Sci. USA 2002, 99, 6222–6227. [Google Scholar] [CrossRef]
- Zeng, Y.; Takada, Y.; Kjellstrom, S.; Hiriyanna, K.; Tanikawa, A.; Wawrousek, E.; Smaoui, N.; Caruso, R.; Bush, R.A.; Sieving, P.A. RS-1 Gene Delivery to an Adult Rs1h Knockout Mouse Model Restores ERG b-Wave with Reversal of the Electronegative Waveform of X-Linked Retinoschisis. Investig. Ophthalmol. Vis. Sci. 2004, 45, 3279–3285. [Google Scholar] [CrossRef] [PubMed]
- Jablonski, M.M.; Dalke, C.; Wang, X.; Lu, L.; Manly, K.F.; Pretsch, W.; Favor, J.; Pardue, M.T.; Rinchik, E.M.; Williams, R.W.; et al. An ENU-induced mutation in Rs1h causes disruption of retinal structure and function. Mol. Vis. 2005, 11, 569–581. [Google Scholar] [PubMed]
- Chen, D.; Xu, T.; Tu, M.; Xu, J.; Zhou, C.; Cheng, L.; Yang, R.; Yang, T.; Zheng, W.; He, X.; et al. Recapitulating X-Linked Juvenile Retinoschisis in Mouse Model by Knock-In Patient-Specific Novel Mutation. Front. Mol. Neurosci. 2017, 10, 453. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Kinoshita, J.; Ivanova, E.; Sun, D.; Li, H.; Liao, T.; Cao, J.; Bell, B.A.; Wang, J.M.; Tang, Y.; et al. Mouse models of X-linked juvenile retinoschisis have an early onset phenotype, the severity of which varies with genotype. Hum. Mol. Genet. 2019, 28, 3072–3090. [Google Scholar] [CrossRef] [PubMed]
- Zeng, Y.; Qian, H.; Campos, M.M.; Li, Y.; Vijayasarathy, C.; Sieving, P.A. Rs1h(-/y) exon 3-del rat model of X-linked retinoschisis with early onset and rapid phenotype is rescued by RS1 supplementation. Gene Ther. 2022, 29, 431–440. [Google Scholar] [CrossRef] [PubMed]
- Ye, E.A.; Zeng, Y.; Thomas, S.; Sun, N.; Smit-McBride, Z.; Sieving, P.A. XLRS Rat with Rs1(-/Y) Exon-1-Del Shows Failure of Early Postnatal Outer Retina Development. Genes 2022, 13, 1995. [Google Scholar] [CrossRef] [PubMed]
- Min, S.H.; Molday, L.L.; Seeliger, M.W.; Dinculescu, A.; Timmers, A.M.; Janssen, A.; Tonagel, F.; Tanimoto, N.; Weber, B.H.; Molday, R.S.; et al. Prolonged recovery of retinal structure/function after gene therapy in an Rs1h-deficient mouse model of x-linked juvenile retinoschisis. Mol. Ther. 2005, 12, 644–651. [Google Scholar] [CrossRef]
- Molday, L.L.; Min, S.H.; Seeliger, M.W.; Wu, W.W.; Dinculescu, A.; Timmers, A.M.; Janssen, A.; Tonagel, F.; Hudl, K.; Weber, B.H.; et al. Disease mechanisms and gene therapy in a mouse model for X-linked retinoschisis. Adv. Exp. Med. Biol. 2006, 572, 283–289. [Google Scholar] [CrossRef]
- Kjellstrom, S.; Bush, R.A.; Zeng, Y.; Takada, Y.; Sieving, P.A. Retinoschisin gene therapy and natural history in the Rs1h-KO mouse: Long-term rescue from retinal degeneration. Investig. Ophthalmol. Vis. Sci. 2007, 48, 3837–3845. [Google Scholar] [CrossRef]
- Janssen, A.; Min, S.H.; Molday, L.L.; Tanimoto, N.; Seeliger, M.W.; Hauswirth, W.W.; Molday, R.S.; Weber, B.H. Effect of late-stage therapy on disease progression in AAV-mediated rescue of photoreceptor cells in the retinoschisin-deficient mouse. Mol. Ther. 2008, 16, 1010–1017. [Google Scholar] [CrossRef]
- Takada, Y.; Vijayasarathy, C.; Zeng, Y.; Kjellstrom, S.; Bush, R.A.; Sieving, P.A. Synaptic pathology in retinoschisis knockout (Rs1-/y) mouse retina and modification by rAAV-Rs1 gene delivery. Investig. Ophthalmol. Vis. Sci. 2008, 49, 3677–3686. [Google Scholar] [CrossRef]
- Park, T.K.; Wu, Z.; Kjellstrom, S.; Zeng, Y.; Bush, R.A.; Sieving, P.A.; Colosi, P. Intravitreal delivery of AAV8 retinoschisin results in cell type-specific gene expression and retinal rescue in the Rs1-KO mouse. Gene Ther. 2009, 16, 916–926. [Google Scholar] [CrossRef]
- Ou, J.; Vijayasarathy, C.; Ziccardi, L.; Chen, S.; Zeng, Y.; Marangoni, D.; Pope, J.G.; Bush, R.A.; Wu, Z.; Li, W.; et al. Synaptic pathology and therapeutic repair in adult retinoschisis mouse by AAV-RS1 transfer. J. Clin. Investig. 2015, 125, 2891–2903. [Google Scholar] [CrossRef]
- Ye, G.J.; Conlon, T.; Erger, K.; Sonnentag, P.; Sharma, A.K.; Howard, K.; Knop, D.R.; Chulay, J.D. Safety and Biodistribution Evaluation of rAAV2tYF-CB-hRS1, a Recombinant Adeno-Associated Virus Vector Expressing Retinoschisin, in RS1-Deficient Mice. Hum. Gene Ther. Clin. Dev. 2015, 26, 177–184. [Google Scholar] [CrossRef]
- Bush, R.A.; Zeng, Y.; Colosi, P.; Kjellstrom, S.; Hiriyanna, S.; Vijayasarathy, C.; Santos, M.; Li, J.; Wu, Z.; Sieving, P.A. Preclinical Dose-Escalation Study of Intravitreal AAV-RS1 Gene Therapy in a Mouse Model of X-linked Retinoschisis: Dose-Dependent Expression and Improved Retinal Structure and Function. Hum. Gene Ther. 2016, 27, 376–389. [Google Scholar] [CrossRef]
- Zeng, Y.; Petralia, R.S.; Vijayasarathy, C.; Wu, Z.; Hiriyanna, S.; Song, H.; Wang, Y.X.; Sieving, P.A.; Bush, R.A. Retinal Structure and Gene Therapy Outcome in Retinoschisin-Deficient Mice Assessed by Spectral-Domain Optical Coherence Tomography. Investig. Ophthalmol. Vis. Sci. 2016, 57, 277–287. [Google Scholar] [CrossRef]
- Marangoni, D.; Yong, Z.; Kjellström, S.; Vijayasarathy, C.; Sieving, P.A.; Bush, R.A. Rearing Light Intensity Affects Inner Retinal Pathology in a Mouse Model of X-Linked Retinoschisis but Does Not Alter Gene Therapy Outcome. Investig. Ophthalmol. Vis. Sci. 2017, 58, 1656–1664. [Google Scholar] [CrossRef]
- Scruggs, B.A.; Bhattarai, S.; Helms, M.; Cherascu, I.; Salesevic, A.; Stalter, E.; Laird, J.; Baker, S.A.; Drack, A.V. AAV2/4-RS1 gene therapy in the retinoschisin knockout mouse model of X-linked retinoschisis. PLoS ONE 2022, 17, e0276298. [Google Scholar] [CrossRef]
- Petrs-Silva, H.; Dinculescu, A.; Li, Q.; Deng, W.T.; Pang, J.J.; Min, S.H.; Chiodo, V.; Neeley, A.W.; Govindasamy, L.; Bennett, A.; et al. Novel properties of tyrosine-mutant AAV2 vectors in the mouse retina. Mol. Ther. 2011, 19, 293–301. [Google Scholar] [CrossRef]
- Khabou, H.; Desrosiers, M.; Winckler, C.; Fouquet, S.; Auregan, G.; Bemelmans, A.P.; Sahel, J.A.; Dalkara, D. Insight into the mechanisms of enhanced retinal transduction by the engineered AAV2 capsid variant -7m8. Biotechnol. Bioeng. 2016, 113, 2712–2724. [Google Scholar] [CrossRef]
- Wiley, L.A.; Burnight, E.R.; Kaalberg, E.E.; Jiao, C.; Riker, M.J.; Halder, J.A.; Luse, M.A.; Han, I.C.; Russell, S.R.; Sohn, E.H.; et al. Assessment of Adeno-Associated Virus Serotype Tropism in Human Retinal Explants. Hum. Gene Ther. 2018, 29, 424–436. [Google Scholar] [CrossRef]
- Klimczak, R.R.; Koerber, J.T.; Dalkara, D.; Flannery, J.G.; Schaffer, D.V. A novel adeno-associated viral variant for efficient and selective intravitreal transduction of rat Müller cells. PLoS ONE 2009, 4, e7467. [Google Scholar] [CrossRef] [PubMed]
- Wang, W.; Liu, J.; Yang, M.; Qiu, R.; Li, Y.; Bian, S.; Hao, B.; Lei, B. Intravitreal Injection of an Exosome-Associated Adeno-Associated Viral Vector Enhances Retinoschisin 1 Gene Transduction in the Mouse Retina. Hum. Gene Ther. 2021, 32, 707–716. [Google Scholar] [CrossRef]
- Hudry, E.; Martin, C.; Gandhi, S.; György, B.; Scheffer, D.I.; Mu, D.; Merkel, S.F.; Mingozzi, F.; Fitzpatrick, Z.; Dimant, H.; et al. Exosome-associated AAV vector as a robust and convenient neuroscience tool. Gene Ther. 2016, 23, 380–392. [Google Scholar] [CrossRef] [PubMed]
- Delgado, D.; del Pozo-Rodríguez, A.; Solinís, M.; Avilés-Triqueros, M.; Weber, B.H.; Fernández, E.; Gascón, A.R. Dextran and protamine-based solid lipid nanoparticles as potential vectors for the treatment of X-linked juvenile retinoschisis. Hum. Gene Ther. 2012, 23, 345–355. [Google Scholar] [CrossRef] [PubMed]
- Chou, S.J.; Yang, P.; Ban, Q.; Yang, Y.P.; Wang, M.L.; Chien, C.S.; Chen, S.J.; Sun, N.; Zhu, Y.; Liu, H.; et al. Dual Supramolecular Nanoparticle Vectors Enable CRISPR/Cas9-Mediated Knockin of Retinoschisin 1 Gene-A Potential Nonviral Therapeutic Solution for X-Linked Juvenile Retinoschisis. Adv. Sci. 2020, 7, 1903432. [Google Scholar] [CrossRef] [PubMed]
- Yang, T.C.; Chang, C.Y.; Yarmishyn, A.A.; Mao, Y.S.; Yang, Y.P.; Wang, M.L.; Hsu, C.C.; Yang, H.Y.; Hwang, D.K.; Chen, S.J.; et al. Carboxylated nanodiamond-mediated CRISPR-Cas9 delivery of human retinoschisis mutation into human iPSCs and mouse retina. Acta Biomater. 2020, 101, 484–494. [Google Scholar] [CrossRef]
- Duan, L.; Xu, L.; Xu, X.; Qin, Z.; Zhou, X.; Xiao, Y.; Liang, Y.; Xia, J. Exosome-mediated delivery of gene vectors for gene therapy. Nanoscale 2021, 13, 1387–1397. [Google Scholar] [CrossRef]
- del Pozo-Rodríguez, A.; Solinís, M.A.; Gascón, A.R.; Pedraz, J.L. Short- and long-term stability study of lyophilized solid lipid nanoparticles for gene therapy. Eur. J. Pharm. Biopharm. 2009, 71, 181–189. [Google Scholar] [CrossRef]
- Rees, H.A.; Liu, D.R. Base editing: Precision chemistry on the genome and transcriptome of living cells. Nat. Rev. Genet. 2018, 19, 770–788. [Google Scholar] [CrossRef]
- Tinwala, H.; Wairkar, S. Production, surface modification and biomedical applications of nanodiamonds: A sparkling tool for theranostics. Mater. Sci. Eng. C Mater. Biol. Appl. 2019, 97, 913–931. [Google Scholar] [CrossRef] [PubMed]
- Mishra, A.; Vijayasarathy, C.; Cukras, C.A.; Wiley, H.E.; Sen, H.N.; Zeng, Y.; Wei, L.L.; Sieving, P.A. Immune function in X-linked retinoschisis subjects in an AAV8-RS1 phase I/IIa gene therapy trial. Mol. Ther. 2021, 29, 2030–2040. [Google Scholar] [CrossRef] [PubMed]
- Pennesi, M.E.; Yang, P.; Birch, D.G.; Weng, C.Y.; Moore, A.T.; Iannaccone, A.; Comander, J.I.; Jayasundera, T.; Chulay, J. Intravitreal Delivery of rAAV2tYF-CB-hRS1 Vector for Gene Augmentation Therapy in Patients with X-Linked Retinoschisis: 1-Year Clinical Results. Ophthalmol. Retina 2022, 6, 1130–1144. [Google Scholar] [CrossRef] [PubMed]
- Vijayasarathy, C.; Sardar Pasha, S.P.B.; Sieving, P.A. Of men and mice: Human X-linked retinoschisis and fidelity in mouse modeling. Prog. Retin. Eye Res. 2022, 87, 100999. [Google Scholar] [CrossRef]
- Silverman, M.S.; Hughes, S.E. Photoreceptor rescue in the RCS rat without pigment epithelium transplantation. Curr. Eye Res. 1990, 9, 183–191. [Google Scholar] [CrossRef]
- Ross, M.; Ofri, R. The future of retinal gene therapy: Evolving from subretinal to intravitreal vector delivery. Neural Regen. Res. 2021, 16, 1751–1759. [Google Scholar] [CrossRef]
- Peng, Y.; Tang, L.; Zhou, Y. Subretinal Injection: A Review on the Novel Route of Therapeutic Delivery for Vitreoretinal Diseases. Ophthalmic Res. 2017, 58, 217–226. [Google Scholar] [CrossRef]
- Reichel, F.F.; Peters, T.; Wilhelm, B.; Biel, M.; Ueffing, M.; Wissinger, B.; Bartz-Schmidt, K.U.; Klein, R.; Michalakis, S.; Fischer, M.D. Humoral Immune Response After Intravitreal But Not After Subretinal AAV8 in Primates and Patients. Investig. Ophthalmol. Vis. Sci. 2018, 59, 1910–1915. [Google Scholar] [CrossRef]
- Ferrari, F.K.; Samulski, T.; Shenk, T.; Samulski, R.J. Second-strand synthesis is a rate-limiting step for efficient transduction by recombinant adeno-associated virus vectors. J. Virol. 1996, 70, 3227–3234. [Google Scholar] [CrossRef]
- Gehrig, A.; Janssen, A.; Horling, F.; Grimm, C.; Weber, B.H. The role of caspases in photoreceptor cell death of the retinoschisin-deficient mouse. Cytogenet. Genome Res. 2006, 115, 35–44. [Google Scholar] [CrossRef]
- Marangoni, D.; Wu, Z.; Wiley, H.E.; Zeiss, C.J.; Vijayasarathy, C.; Zeng, Y.; Hiriyanna, S.; Bush, R.A.; Wei, L.L.; Colosi, P.; et al. Preclinical safety evaluation of a recombinant AAV8 vector for X-linked retinoschisis after intravitreal administration in rabbits. Hum. Gene Ther. Clin. Dev. 2014, 25, 202–211. [Google Scholar] [CrossRef] [PubMed]
- Verdera, H.C.; Kuranda, K.; Mingozzi, F. AAV Vector Immunogenicity in Humans: A Long Journey to Successful Gene Transfer. Mol. Ther. 2020, 28, 723–746. [Google Scholar] [CrossRef] [PubMed]
- Oliveira, A.V.; Rosa da Costa, A.M.; Silva, G.A. Non-viral strategies for ocular gene delivery. Mater. Sci. Eng. C Mater. Biol. Appl. 2017, 77, 1275–1289. [Google Scholar] [CrossRef] [PubMed]
- Ramamoorth, M.; Narvekar, A. Non viral vectors in gene therapy- an overview. J. Clin. Diagn. Res. 2015, 9, Ge01-06. [Google Scholar] [CrossRef] [PubMed]
- Heymann, J.B.; Vijayasarathy, C.; Huang, R.K.; Dearborn, A.D.; Sieving, P.A.; Steven, A.C. Cryo-EM of retinoschisin branched networks suggests an intercellular adhesive scaffold in the retina. J. Cell Biol. 2019, 218, 1027–1038. [Google Scholar] [CrossRef] [PubMed]
- Koch, S.F.; Tsai, Y.T.; Duong, J.K.; Wu, W.H.; Hsu, C.W.; Wu, W.P.; Bonet-Ponce, L.; Lin, C.S.; Tsang, S.H. Halting progressive neurodegeneration in advanced retinitis pigmentosa. J. Clin. Investig. 2015, 125, 3704–3713. [Google Scholar] [CrossRef]
- Apaolaza, P.S.; Delgado, D.; Pozo-Rodríguez, A.d.; Gascón, A.R.; Solinís, M.Á. A novel gene therapy vector based on hyaluronic acid and solid lipid nanoparticles for ocular diseases. Int. J. Pharm. 2014, 465, 413–426. [Google Scholar] [CrossRef]
- Peynshaert, K.; Devoldere, J.; Minnaert, A.K.; De Smedt, S.C.; Remaut, K. Morphology and Composition of the Inner Limiting Membrane: Species-Specific Variations and Relevance toward Drug Delivery Research. Curr. Eye Res. 2019, 44, 465–475. [Google Scholar] [CrossRef]
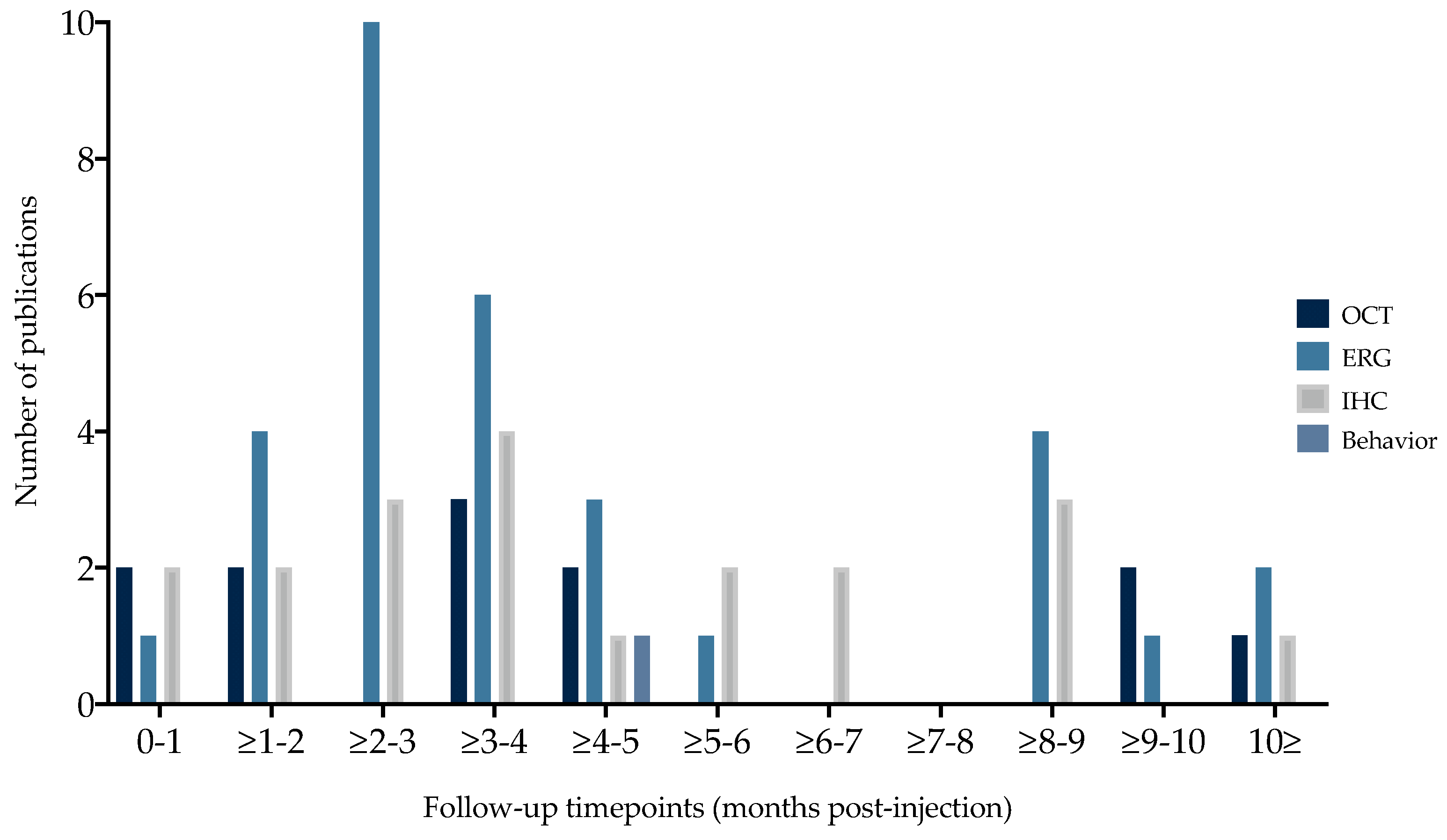

| Name [Ref.] | Species | Strain | Mutation | RS1 Protein Absence | Main Pathological Features | Age of Schisis Onset | Schisis Progression |
|---|---|---|---|---|---|---|---|
| Rs1h−/y [70] | mouse | C57BL/6 | Replacement of exons 3 and 4 and intron 3 by a lacZ-neo cassette. | Confirmed on IHC and WB | Schisis cavities in the inner retina are evenly dispersed throughout the central en and peripheral retina. Loss of laminar integrity in the OPL, INL and PR layer. Cell death is restricted to PRs. The a- and b-wave amplitudes are affected, b-wave more so. | P18 | Not reported |
| Rs1h-KO [71] | mouse | C57BL/6 | Replacement of exon 1 and 1.6 kb of intron 1 by a neomycin resistance cassette | Confirmed on IHC and WB | Schisis cavities are found throughout the retina. Lamination of the OPL, INL and PR layer is affected. The a- and b-wave amplitudes are diminished, and the b-wave is more strongly affected. | P14 | Cavities collapse from 4–12 months old |
| Rs1h−/y rat (1) [75] | rat | Long Evans | Deletion of 310 base pairs, encompassing exon 3 and parts of introns 2 and 3. | Confirmed on IHC and WB | Large cavities are observed at P15. Lamination is disrupted, there is cell loss in ONL, and OPL and IS/OS-zone are thinner. The a- and b-wave amplitudes are reduced, as is the b/a-ratio. | P15 | Cavities rapidly collapse at P21–P28 |
| Rs1h−/y rat (2) [76] | rat | Long Evans | Deletion of 769 bps, encompassing exon 1. | Confirmed on IHC and WB | Large cavities are observed at P15 that are resolved by P30. The ONL is thinner, IS/OS are shorter and OPL is disordered. B- and a-wave amplitudes are decreased. | P15 | Cavities have collapsed by P30 |
| Study ID | Species | Model | Treatment | Delivery Method | SLO | OCT | ERG | Behavior | IHC |
|---|---|---|---|---|---|---|---|---|---|
| Zeng2004 [71] | mouse | Rs1h-KO | AAV2-CMV-Rs1h | SRI | N | N | Y | N | N |
| Min2005 [77] | mouse | Rs1h−/y | AAV5-mOP-hRS1 | SRI | Y | N | Y | N | Y |
| Molday2006 [78] | mouse | Rs1h−/y | AAV5-mOP-hRS1 | SRI | N | N | Y | N | Y |
| Kjellstrom2007 [79] | mouse | Rs1h-KO | AAV2-CMV-Rs1h | IVI | N | N | Y | N | Y |
| Janssen2008 [80] | mouse | Rs1h−/y | AAV5-mOP-hRS1 | SRI | N | N | Y | N | Y |
| Takada2008 [81] | mouse | Rs1h-KO | AAV2-CMV-Rs1h | IVI | N | N | Y | N | Y |
| Park2009 [82] | mouse | Rs1h-KO | AAV2-CMV-Rs1h; | IVI | N | N | Y | N | Y |
| AAV8-CMV-hRSp4 | |||||||||
| Byrne2014 [54] | mouse | Rs1h−/y | AAV7m8-shH10-CAG-hRS1; | IVI | N | Y | Y | N | Y |
| AAV7m8-rho-hRS1; | |||||||||
| AAV7m8-CAG-hRS1 | |||||||||
| Ou2015 [83] | mouse | Rs1h-KO | AAV8-scRS/IRBP-hRS | IVI | N | N | Y | N | Y |
| Ye2015 [84] | mouse | Rs1h−/y | rAAV2tYF-CB-hRS1 | IVI | N | N | N | N | N |
| Bush2016 [85] | mouse | Rs1h-KO | AAV8-scRS/IRBP-hRS | IVI | N | Y | Y | N | Y |
| Marangoni2016 [55] | mouse | Rs1h-KO | AAV8-scRS/IRBP-hRS | IVI | N | N | Y | N | N |
| Zeng2016 [86] | mouse | Rs1h-KO | AAV8-scRS/IRBP-hRS | IVI | N | Y | N | N | N |
| Marangoni2017 [87] | mouse | Rs1h-KO | AAV8-scRS/IRBP-hRS | IVI | N | N | Y | N | N |
| Vijayasarathy2021 [17] | mouse | Rs1h-KO | AAV8-scRS/IRBP-hRS | SRI | N | Y | N | N | Y |
| Zeng2022 [75] | rat | Rs1h−/y rat (1) | AAV8-scRS/IRBP-hRS | IVI | N | Y | N | N | N |
| Ye2022 [76] | rat | Rs1h−/y rat (2) | AAV8-scRS/IRBP-hRS | IVI | N | Y | N | N | Y |
| Scruggs2022 [88] | mouse | Rs1h-KO | AAV2/4-CMV-RS1 | SRI and IVI | N | Y | Y | Y | Y |
| Vijayasarathy2022 [34] | mouse | Rs1h-KO | rAAV8-Ple155-RS1; | SRI | N | Y | Y | N | Y |
| rAAV2/2-Y444-miniGluR6-RS1 | IVI | N | N |
| Experiment ID | Timing (Post-Injection) | Cavities (Size and Amount) | Retinal Organization and Lamination | Retinal Thickness |
|---|---|---|---|---|
| Bush2016_1E6 [85] | 14 ± 2 w | No effect | Not reported | No effect |
| Bush2016_1E7 [85] | 14 ± 2 w | No effect | Not reported | No effect |
| Bush2016_5E7 [85] | 14 ± 2 w | No effect | Not reported | No effect |
| Bush2016_1E8 [85] | 14 ± 2 w | Reduced | Not reported | Increased |
| Bush2016_5E8 [85] | 14 ± 2 w | Reduced | Not reported | Increased |
| Bush2016_2.5E9 [85] | 14 ± 2 w | Reduced | Improved | Increased |
| Zeng2016 [86] | 12–15 w | Reduced | Improved | Increased |
| Vijayasarathy2021 [17] | 35 d | Reduced | Improved | Increased |
| Zeng2022 [75] | 12 w | Absent | Improved | Increased |
| Ye2022_10dpi [76] | 10 d | Reduced or absent | Improved | NR |
| Ye2022_25dpi [76] | 25 d | Absent | Improved | NR |
| Byrne2014_shH10 [54] | 4 m | No effect | No effect | No effect |
| Byrne2014_CAG [54] | 4 m | Reduced | Improved | Increased |
| Byrne2014_rho [54] | 4 m | Reduced | Improved | Increased |
| Byrne2014_15m [54] | 15 m | N/A | Improved | Increased |
| Scruggs2022_IVI [88] | 2 w | Reduced or absent | Improved | NR |
| Scruggs2022_SRI [88] | 2 w | Reduced or absent | Improved | NR |
| Vijayasarathy2022_Ple155 [34] | 5 w | Similar or reduced | Improved | NR |
| Outcomes Assessed | Timing Median <Range> | Number of Publications | Study IDs |
|---|---|---|---|
| Transgene expression | 11 w <1 w, 15 m> | 13 (25 experimental groups) | Min2005 [77], Molday2006 [78], Kjellstrom2007 [79], Janssen2008 [80], Takada2008 [81], Park2009 [82], Byrne2014 [54], Ou2015 [83], Bush2016 [85], Vijayasarathy2021 [17], Scruggs2022 [75], Ye2022 [76], Vijayasarathy2022 [34] |
| Laminar organization and integrity | 32 w <25 d–15 m> | 2 (4 experimental groups) | Byrne2014 [54], Ye2022 [76] |
| ONL cell count | 4 w <1 w–15 m> | 2 (7 experimental groups) | Janssen2008 [80], Byrne2014 [54] |
| Presence of cavities | 5 w, 2 m and 27 w | 2 (3 experimental groups) | Ou2015 [83], Vijayasarathy2022 [34] |
| Microglial/Muller cell alterations | 25 d and 35 d | 2 | Vijayasarathy2021 [17], Ye2022 [76] |
| Synaptic pathology | 32 w <2 m, 15 m> | 3 (5 experimental groups) | Takada2008 [81], Byrne2014 [54], Ou2015 [83] |
| Study ID [Ref.] | Study Design | Cell Model | Plasmid |
|---|---|---|---|
| Delgado2012 [95] | RS1 delivery in SLNs | ARPE-19 cells | CEP4-RS1 |
| Apaolaza2015 [63] | RS1 delivery in SLNs | ARPE-19 cells | pCAG-GFP_CMV-RS1 |
| Wang2021 [93] | RS1 delivery in exo-AAV | HEK-293T cells/ARPE-19/Fibroblast cells | exo-AAV2-RS1-ZsGreen |
| Yang2020 [97] | cND-mediated CRISPR-Cas9 KO. c.625 C > T mutation | Human iPSCs | Cas9-GFP and RS1-sgRNA |
| Chou2020 [96] | Dual SMNP CRISPR/Cas9-mediated KI | B16 mouse melanoma cell line | Cas9/sgRNA-plasmid Donor-RS1/GFP-plasmid |
| Huang2019 [58] | CRISPR/Cas9 correction of disease-associated c.625 C > T mutation | Human iPSC differentiation into retinal organoids | pCas9_GFP and pRS1HR-gRNA/ pCas9_GFP, pgRNA and RS1 Oligo |
| Base editing correction of disease-associated c.625 C > T mutation | Human iPSC differentiation into retinal organoids | pCas9_GFP and pRS1HR-gRNA/ ABE7.10 |
| Study ID | Model | Strain | Delivery Method | Strategy | Administr-Ation Route | Injected Volume (µL) | Interven-Tion Age | Follow-Up | Persisting GFP Expression over Time |
|---|---|---|---|---|---|---|---|---|---|
| Apaolaza2015 [63] | Rs1h−/y mouse | C57BL/6 | SLN | RS1 integration | IVI, SRI | 0.75 μL | 14 w | 1 w, 2 w, and 2 m | NR |
| Wang2021 [93] | WT mouse | C57BL/6 | exo-AAV2 | RS1 integration | IVI | 1 μL | 4–5 w | 2 w | NR |
| Yang2020 [97] | WT mouse | C57BL/6 | cNDs | RS1 editing (KO (c.625C > T)) | IVI | 5 μL | 6–10 w | 2 w | 12 days |
| Chou2020 [96] | WT mouse | BALB/c | Dual SMNP | RS1 editing (KI) | IVI | 1 μL | NR | 2 w | 30 days |
| Outcomes Assessed (and Technique) | Evaluation Time Point (Post-Transfection) | Number of Publications | Study IDs |
|---|---|---|---|
| In vitro transfection capacity (quantification of GFP and RS1) | 72 h | 2 | Delgado2012 [95], Apaolaza2015 [63] |
| GFP and RS1 visualization | 72 h | 2 | Delgado2012 [95], Apaolaza2015 [63] |
| In vitro transfection capacity (GFP detection) | 72 h | 1 | Wang2021 [93] |
| In vitro transfection capacity (mRNA and protein expression) | 48 h and 72 h | 1 | Wang2021 [93] |
| Cellular uptake (Flow cytometry) | 2 h | 2 | Delgado2012 [95], Apaolaza2015 [63] |
| Cell viability (cell counting kit-8) | 72 h | 2 | Delgado2012 [95], Apaolaza2015 [63] |
| Outcomes Assessed (and Technique) | Evaluation Time Point (Post-Transfection) | Number of Publications | Study IDs |
|---|---|---|---|
| In vitro transfection capacity (% GFP-positive cells) | 21 d | 1 | Chou2020 [96] |
| RS1 visualization | 48 h | 1 | Chou2020 [96] |
| GFP visualization | 2, 3, 5, 7, 14 and 21 d | 1 | Chou2020 [96] |
| NR | 1 | Yang2020 [97] | |
| Cell viability (cell counting kit-8) | 24 h | 1 | Chou2020 [96] |
| 48 h | 2 | Yang2020 [97], Chou2020 [96] | |
| RS1 editing (qPCR, ddPCR) | 48 h (ddPCR) | 1 | Yang2020 [97] |
| 21 d (qPCR) | 1 | Chou2020 [96] | |
| NR | 1 | Huang2019 [58] | |
| RS1 editing (Sanger sequencing) | 21 d | 1 | Chou2020 [96] |
| NR | 1 | Huang2019 [58] |
| Outcomes Assessed (and Technique) | Evaluation Time Point (Post-Transfection) | Number of Publications | Study IDs |
|---|---|---|---|
| GFP and RS1 visualization (IHC) | 1 w, 2 m | 1 | Apaolaza2015 [63] |
| 2 w | 1 | Wang2021 [93] | |
| Layer organization | 2 w, 2 m | 1 | Apaolaza2015 [63] |
| PR loss | 2 w, 2 m | 1 | Apaolaza2015 [63] |
| Schisis (gaps) | 2 w, 2 m | 1 | Apaolaza2015 [63] |
| Retinal thickness (μm) | 2 w, 2 m | 1 | Apaolaza2015 [63] |
| ONL thickness (μm) | 2 w, 2 m | 1 | Apaolaza2015 [63] |
| In vivo transfection capacity (mRNA and protein expression) | 2 w | 1 | Wang2021 [93] |
| Outcomes Assessed (and Technique) | Evaluation Time Point (Post-Transfection) | Number of Publications | Study IDs |
|---|---|---|---|
| GFP and RS1 visualization (IHC) | 2 w | 2 | Yang2020 [97], Chou2020 [96] |
| GFP visualization (OCT) | 2 w | 1 | Chou2020 [96] |
| GFP visualization (SLO) | 2 w | 1 | Yang2020 [97] |
| Genome editing efficiency | 2 w | 2 | Yang2020 [97], Chou2020 [96] |
| Retinal thickness (μm) | 2 w | 1 | Yang2020 [97] |
| IS/OS thickness (μm) | 2 w | 1 | Yang2020 [97] |
| Retinal structure (OCT, H&E) | 2 w | 1 | Chou2020 [96] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
van der Veen, I.; Heredero Berzal, A.; Koster, C.; ten Asbroek, A.L.M.A.; Bergen, A.A.; Boon, C.J.F. The Road towards Gene Therapy for X-Linked Juvenile Retinoschisis: A Systematic Review of Preclinical Gene Therapy in Cell-Based and Rodent Models of XLRS. Int. J. Mol. Sci. 2024, 25, 1267. https://doi.org/10.3390/ijms25021267
van der Veen I, Heredero Berzal A, Koster C, ten Asbroek ALMA, Bergen AA, Boon CJF. The Road towards Gene Therapy for X-Linked Juvenile Retinoschisis: A Systematic Review of Preclinical Gene Therapy in Cell-Based and Rodent Models of XLRS. International Journal of Molecular Sciences. 2024; 25(2):1267. https://doi.org/10.3390/ijms25021267
Chicago/Turabian Stylevan der Veen, Isa, Andrea Heredero Berzal, Céline Koster, Anneloor L. M. A. ten Asbroek, Arthur A. Bergen, and Camiel J. F. Boon. 2024. "The Road towards Gene Therapy for X-Linked Juvenile Retinoschisis: A Systematic Review of Preclinical Gene Therapy in Cell-Based and Rodent Models of XLRS" International Journal of Molecular Sciences 25, no. 2: 1267. https://doi.org/10.3390/ijms25021267
APA Stylevan der Veen, I., Heredero Berzal, A., Koster, C., ten Asbroek, A. L. M. A., Bergen, A. A., & Boon, C. J. F. (2024). The Road towards Gene Therapy for X-Linked Juvenile Retinoschisis: A Systematic Review of Preclinical Gene Therapy in Cell-Based and Rodent Models of XLRS. International Journal of Molecular Sciences, 25(2), 1267. https://doi.org/10.3390/ijms25021267






