Structure and Assembly of the Proteus mirabilis Flagellar Motor by Cryo-Electron Tomography
Abstract
1. Introduction
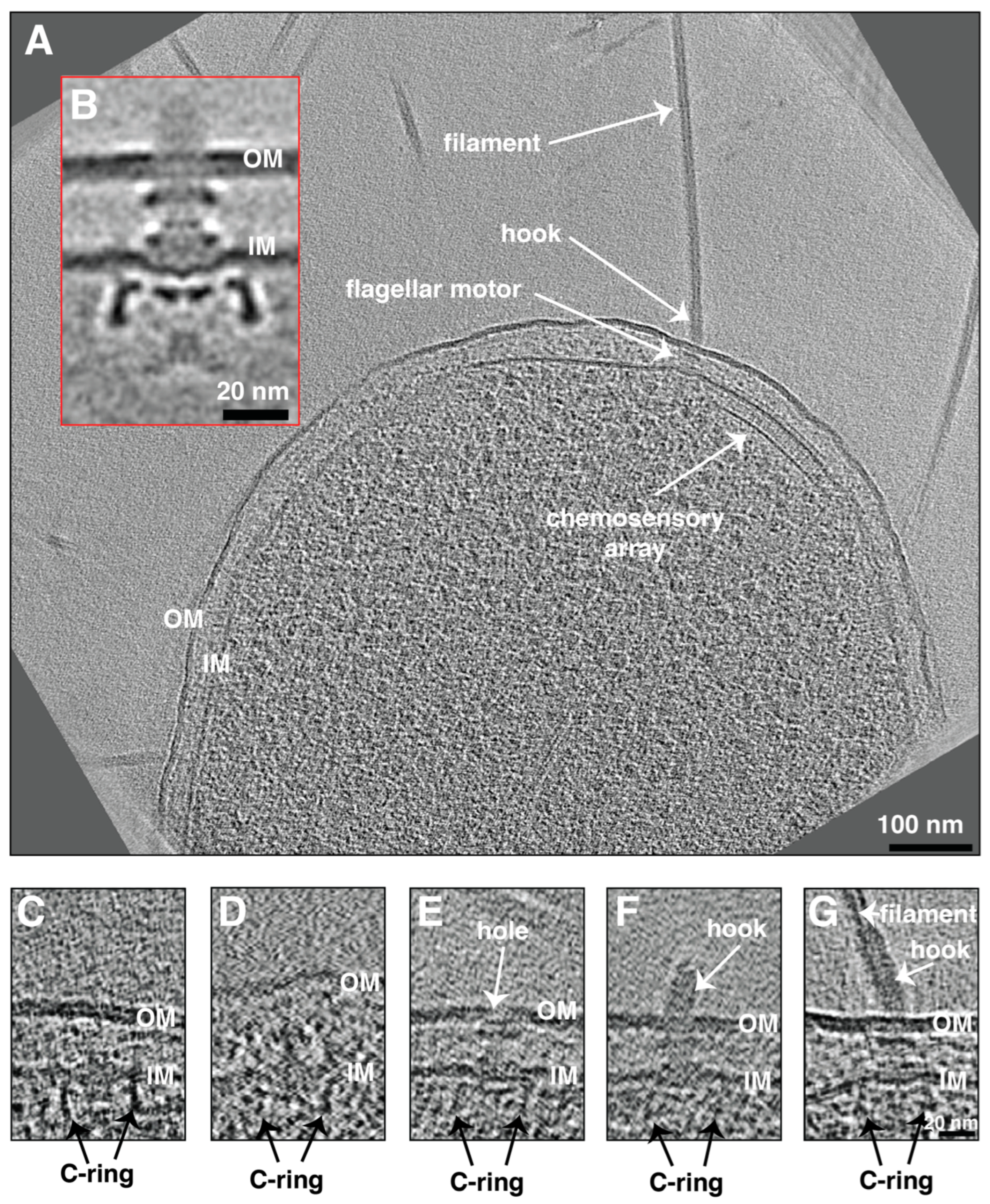
2. Results and Discussion
3. Materials and Methods
3.1. Strains and Growth Conditions
3.2. Cryo-ET Sample Preparation and Imaging
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Hoeniger, J.F.M. Development of Flagella by Proteus mirabilis. J. Gen. Microbiol. 1965, 40, 29–42. [Google Scholar] [CrossRef]
- Armbruster, C.E.; Mobley, H.L.T. Merging mythology and morphology: The multifaceted lifestyle of Proteus mirabilis. Nat. Rev. Microbiol. 2012, 10, 743–754. [Google Scholar] [CrossRef] [PubMed]
- O’Hara, C.M.; Brenner, F.W.; Miller, J.M. Classification, Identification, and Clinical Significance of Proteus, Providencia and Morganella. Clin. Microbiol. Rev. 2000, 13, 534–546. [Google Scholar] [CrossRef]
- Nicolle, L.E. Catheter-Related Urinary Tract Infection. Drugs Aging 2005, 22, 627–639. [Google Scholar] [CrossRef] [PubMed]
- Pearson, M.M.; Sebaihia, M.; Churcher, C.; Quail, M.A.; Seshasayee, A.S.; Luscombe, N.M.; Abdellah, Z.; Arrosmith, C.; Atkin, B.; Chillingworth, T.; et al. Complete Genome Sequence of Uropathogenic Proteus mirabilis, a Master of both Adherence and Motility. J. Bacteriol. 2008, 190, 4027–4037. [Google Scholar] [CrossRef] [PubMed]
- Jones, B.D.; Mobley, H.L. Proteus mirabilis urease: Genetic organization, regulation, and expression of structural genes. J. Bacteriol. 1988, 170, 3342–3349. [Google Scholar] [CrossRef]
- Jones, B.D.; Mobley, H.L. Proteus mirabilis urease: Nucleotide sequence determination and comparison with jack bean urease. J. Bacteriol. 1989, 171, 6414–6422. [Google Scholar] [CrossRef]
- Stickler, D.; Hughes, G. Ability of Proteus mirabilis to Swarm over Urethral Catheters. Eur. J. Clin. Microbiol. Infect. Dis. 1999, 18, 206–208. [Google Scholar] [CrossRef]
- Sabbuba, N.; Hughes, G.; Stickler, D.J. The migration of Proteus mirabilis and other urinary tract pathogens over Foley catheters: Migration of proteus mirabilis and other pathogens over foley catheters. BJU Int. 2008, 89, 55–60. [Google Scholar] [CrossRef]
- Macnab, R.M. How Bacteria Assemble Flagella. Annu. Rev. Microbiol. 2003, 57, 77–100. [Google Scholar] [CrossRef]
- Kaplan, M.; Sweredoski, M.J.; Rodrigues, J.P.G.L.M.; Tocheva, E.I.; Chang, Y.-W.; Ortega, D.R.; Beeby, M.; Jensen, G.J. Bacterial flagellar motor PL-ring disassembly subcomplexes are widespread and ancient. Proc. Natl. Acad. Sci. USA 2020, 117, 8941–8947. [Google Scholar] [CrossRef]
- Ferreira, J.L.; Gao, F.Z.; Rossmann, F.M.; Nans, A.; Brenzinger, S.; Hosseini, R.; Wilson, A.; Briegel, A.; Thormann, K.M.; Rosenthal, P.B.; et al. γ-proteobacteria eject their polar flagella under nutrient depletion, retaining flagellar motor relic structures. PLoS Biol. 2019, 17, e3000165. [Google Scholar] [CrossRef]
- Zhu, S.; Schniederberend, M.; Zhitnitsky, D.; Jain, R.; Galán, J.E.; Kazmierczak, B.I.; Liu, J. In Situ Structures of Polar and Lateral Flagella Revealed by Cryo-Electron Tomography. J. Bacteriol. 2019, 201, e00117-19. [Google Scholar] [CrossRef] [PubMed]
- Kaplan, M.; Wang, Y.; Chreifi, G.; Zhang, L.; Chang, Y.-W.; Jensen, G.J. Programmed flagellar ejection in Caulobacter crescentus leaves PL-subcomplexes. J. Mol. Biol. 2021, 443, 167004. [Google Scholar] [CrossRef]
- Kaplan, M.; Subramanian, P.; Ghosal, D.; Oikonomou, C.M.; Pirbadian, S.; Starwalt-Lee, R.; Mageswaran, S.K.; Ortega, D.R.; Gralnick, J.A.; El-Naggar, M.Y.; et al. In situ imaging of the bacterial flagellar motor disassembly and assembly processes. EMBO J. 2019, 38, e100957. [Google Scholar] [CrossRef]
- Berg, H.C. The Rotary Motor of Bacterial Flagella. Annu. Rev. Biochem. 2003, 72, 19–54. [Google Scholar] [CrossRef]
- Al-Otaibi, N.S.; Bergeron, J.R.C. Structure and Assembly of the Bacterial Flagellum. In Macromolecular Protein Complexes IV; Harris, J.R., Marles-Wright, J., Eds.; Springer International Publishing: Cham, Switzerland, 2022; pp. 395–420. [Google Scholar]
- Deme, J.C.; Johnson, S.; Vickery, O.; Aron, A.; Monkhouse, H.; Griffiths, T.; James, R.H.; Berks, B.C.; Coulton, J.W.; Stansfeld, P.J.; et al. Structures of the stator complex that drives rotation of the bacterial flagellum. Nat. Microbiol. 2020, 5, 1553–1564. [Google Scholar] [CrossRef] [PubMed]
- Santiveri, M.; Roa-Eguiara, A.; Kühne, C.; Wadhwa, N.; Hu, H.; Berg, H.C.; Erhardt, M.; Taylor, N.M.I. Structure and Function of Stator Units of the Bacterial Flagellar Motor. Cell 2020, 183, 244–257.e16. [Google Scholar] [CrossRef] [PubMed]
- Hu, H.; Santiveri, M.; Wadhwa, N.; Berg, H.C.; Erhardt, M.; Taylor, N.M.I. Structural basis of torque generation in the bi-directional bacterial flagellar motor. Trends Biochem. Sci. 2021, 47, 160–172. [Google Scholar] [CrossRef] [PubMed]
- Chang, Y.; Zhang, K.; Carroll, B.L.; Zhao, X.; Charon, N.W.; Norris, S.J.; Motaleb, M.A.; Li, C.; Liu, J. Molecular mechanism for rotational switching of the bacterial flagellar motor. Nat. Struct. Mol. Biol. 2020, 27, 1041–1047. [Google Scholar] [CrossRef]
- Chen, S.; Beeby, M.; Murphy, G.E.; Leadbetter, J.R.; Hendrixson, D.R.; Briegel, A.; Li, Z.; Shi, J.; Tocheva, E.I.; Müller, A.; et al. Structural diversity of bacterial flagellar motors: Structural diversity of bacterial flagellar motors. EMBO J. 2011, 30, 2972–2981. [Google Scholar] [CrossRef] [PubMed]
- Kaplan, M.; Ghosal, D.; Subramanian, P.; Oikonomou, C.M.; Kjaer, A.; Pirbadian, S.; Ortega, D.R.; Briegel, A.; El-Naggar, M.Y.; Jensen, G.J. The presence and absence of periplasmic rings in bacterial flagellar motors correlates with stator type. eLife 2019, 8, e43487. [Google Scholar] [CrossRef] [PubMed]
- Zhao, X.; Norris, S.J.; Liu, J. Molecular Architecture of the Bacterial Flagellar Motor in Cells. Biochemistry 2014, 53, 4323–4333. [Google Scholar] [CrossRef] [PubMed]
- Chaban, B.; Coleman, I.; Beeby, M. Evolution of higher torque in Campylobacter-type bacterial flagellar motors. Sci. Rep. 2018, 8, 97. [Google Scholar] [CrossRef] [PubMed]
- Beeby, M.; Ribardo, D.A.; Brennan, C.A.; Ruby, E.G.; Jensen, G.J.; Hendrixson, D.R. Diverse high-torque bacterial flagellar motors assemble wider stator rings using a conserved protein scaffold. Proc. Natl. Acad. Sci. USA 2016, 113, E1917–E1926. [Google Scholar] [CrossRef]
- Qin, Z.; Lin, W.; Zhu, S.; Franco, A.T.; Liu, J. Imaging the Motility and Chemotaxis Machineries in Helicobacter pylori by Cryo-Electron Tomography. J. Bacteriol. 2017, 199, e00695-16. [Google Scholar] [CrossRef]
- Lele, P.P.; Hosu, B.G.; Berg, H.C. Dynamics of mechanosensing in the bacterial flagellar motor. Proc. Natl. Acad. Sci. USA 2013, 110, 11839–11844. [Google Scholar] [CrossRef]
- Nord, A.L.; Gachon, E.; Perez-Carrasco, R.; Nirody, J.A.; Barducci, A.; Berry, R.M.; Pedaci, F. Catch bond drives stator mechanosensitivity in the bacterial flagellar motor. Proc. Natl. Acad. Sci. USA 2017, 114, 12952–12957. [Google Scholar] [CrossRef]
- Kaplan, M.; Nicolas, W.J.; Zhao, W.; Carter, S.D.; Metskas, L.A.; Chreifi, G.; Ghosal, D.; Jensen, G.J. In Situ Imaging and Structure Determination of Biomolecular Complexes Using Electron Cryo-Tomography. In cryoEM; Gonen, T., Nannenga, B.L., Eds.; Springer: New York, NY, USA, 2021; pp. 83–111. [Google Scholar]
- Ghosal, D.; Kaplan, M.; Chang, Y.-W.; Jensen, G.J. In Situ Imaging and Structure Determination of Bacterial Toxin Delivery Systems Using Electron Cryotomography. In Legionella; Buchrieser, C., Hilbi, H., Eds.; Springer: New York, NY, USA, 2019; pp. 249–265. [Google Scholar]
- Ferreira, J.L.; Coleman, I.; Addison, M.L.; Zachs, T.; Quigley, B.L.; Wuichet, K.; Beeby, M. The “Jack-of-all-Trades” Flagellum from Salmonella and E. coli Was Horizontally Acquired from an Ancestral β-Proteobacterium. Front. Microbiol. 2021, 12, 643180. [Google Scholar] [CrossRef]
- Latta, R.K.; Grondin, A.; Jarrell, H.C.; Nicholls, G.R.; Bérubé, L.R. Differential Expression of Nonagglutinating Fimbriae and MR/P Pili in Swarming Colonies of Proteus mirabilis. J. Bacteriol. 1999, 181, 3220–3225. [Google Scholar] [CrossRef]
- Mobley, H.L.; Belas, R.; Lockatell, V.; Chippendale, G.; Trifillis, A.L.; Johnson, D.E.; Warren, J.W. Construction of a flagellum-negative mutant of Proteus mirabilis: Effect on internalization by human renal epithelial cells and virulence in a mouse model of ascending urinary tract infection. Infect. Immun. 1996, 64, 5332–5340. [Google Scholar] [CrossRef] [PubMed]
- Pearson, M.M.; Mobley, H.L.T. Repression of motility during fimbrial expression: Identification of 14 mrpJ gene paralogues in Proteus mirabilis. Mol. Microbiol. 2008, 69, 548–558. [Google Scholar] [CrossRef] [PubMed]
- Li, X. Repression of bacterial motility by a novel fimbrial gene product. EMBO J. 2001, 20, 4854–4862. [Google Scholar] [CrossRef]
- Kaplan, M.; Oikonomou, C.M.; Wood, C.R.; Chreifi, G.; Subramanian, P.; Ortega, D.R.; Chang, Y.; Beeby, M.; Shaffer, C.L.; Jensen, G.J. Novel transient cytoplasmic rings stabilize assembling bacterial flagellar motors. EMBO J. 2022, 41, e109523. [Google Scholar] [CrossRef] [PubMed]
- Kubori, T.; Shimamoto, N.; Yamaguchi, S.; Namba, K.; Aizawa, S.-I. Morphological pathway of flagellar assembly in Salmonella typhimurium. J. Mol. Biol. 1992, 226, 433–446. [Google Scholar] [CrossRef] [PubMed]
- Jones, C.J.; Macnab, R.M. Flagellar assembly in Salmonella typhimurium: Analysis with temperature-sensitive mutants. J. Bacteriol. 1990, 172, 1327–1339. [Google Scholar] [CrossRef] [PubMed]
- Brown, P.N.; Terrazas, M.; Paul, K.; Blair, D.F. Mutational Analysis of the Flagellar Protein FliG: Sites of Interaction with FliM and Implications for Organization of the Switch Complex. J. Bacteriol. 2007, 189, 305–312. [Google Scholar] [CrossRef] [PubMed]
- Branch, R.W.; Sayegh, M.N.; Shen, C.; Nathan, V.S.J.; Berg, H.C. Adaptive Remodelling by FliN in the Bacterial Rotary Motor. J. Mol. Biol. 2014, 426, 3314–3324. [Google Scholar] [CrossRef]
- Delalez, N.J.; Wadhams, G.H.; Rosser, G.; Xue, Q.; Brown, M.T.; Dobbie, I.M.; Berry, R.M.; Leake, M.C.; Armitage, J.P. Signal-dependent turnover of the bacterial flagellar switch protein FliM. Proc. Natl. Acad. Sci. USA 2010, 107, 11347–11351. [Google Scholar] [CrossRef]
- Fukuoka, H.; Inoue, Y.; Terasawa, S.; Takahashi, H.; Ishijima, A. Exchange of rotor components in functioning bacterial flagellar motor. Biochem. Biophys. Res. Commun. 2010, 394, 130–135. [Google Scholar] [CrossRef]
- Tuson, H.H.; Copeland, M.F.; Carey, S.; Sacotte, R.; Weibel, D.B. Flagellum Density Regulates Proteus mirabilis Swarmer Cell Motility in Viscous Environments. J. Bacteriol. 2013, 195, 368–377. [Google Scholar] [CrossRef] [PubMed]
- Tivol, W.F.; Briegel, A.; Jensen, G.J. An Improved Cryogen for Plunge Freezing. Microsc. Microanal. 2008, 14, 375–379. [Google Scholar] [CrossRef] [PubMed]
- Zheng, S.Q.; Keszthelyi, B.; Branlund, E.; Lyle, J.M.; Braunfeld, M.B.; Sedat, J.W.; Agard, D.A. UCSF tomography: An integrated software suite for real-time electron microscopic tomographic data collection, alignment, and reconstruction. J. Struct. Biol. 2007, 157, 138–147. [Google Scholar] [CrossRef] [PubMed]
- Kremer, J.R.; Mastronarde, D.N.; McIntosh, J.R. Computer visualization of three-dimensional image data using IMOD. J. Struct. Biol. 1996, 116, 71–76. [Google Scholar] [CrossRef] [PubMed]
- Nicastro, D. The Molecular Architecture of Axonemes Revealed by Cryoelectron Tomography. Science 2006, 313, 944–948. [Google Scholar] [CrossRef] [PubMed]

Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kaplan, M.; Yao, Q.; Jensen, G.J. Structure and Assembly of the Proteus mirabilis Flagellar Motor by Cryo-Electron Tomography. Int. J. Mol. Sci. 2023, 24, 8292. https://doi.org/10.3390/ijms24098292
Kaplan M, Yao Q, Jensen GJ. Structure and Assembly of the Proteus mirabilis Flagellar Motor by Cryo-Electron Tomography. International Journal of Molecular Sciences. 2023; 24(9):8292. https://doi.org/10.3390/ijms24098292
Chicago/Turabian StyleKaplan, Mohammed, Qing Yao, and Grant J. Jensen. 2023. "Structure and Assembly of the Proteus mirabilis Flagellar Motor by Cryo-Electron Tomography" International Journal of Molecular Sciences 24, no. 9: 8292. https://doi.org/10.3390/ijms24098292
APA StyleKaplan, M., Yao, Q., & Jensen, G. J. (2023). Structure and Assembly of the Proteus mirabilis Flagellar Motor by Cryo-Electron Tomography. International Journal of Molecular Sciences, 24(9), 8292. https://doi.org/10.3390/ijms24098292




