Ion-Powered Rotary Motors: Where Did They Come from and Where They Are Going?
Abstract
1. Introduction
2. The Structure of the Bacterial Flagellar Motor
2.1. Ion Selectivity
2.2. Towards New Power-Sources for IRMs
3. The Role of Ancestral Sequence Reconstruction in Understanding the Evolution of Ion Selective Motor Proteins
3.1. Ancestral Sequence Reconstruction
3.2. The Use of ASR to Study Ion Selectivity in the BFM
3.3. Limitations of ASR
4. Concluding Remarks
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Iino, R.; Kinbara, K.; Bryant, Z. Introduction: Molecular Motors. Chem. Rev. 2020, 120, 1–4. [Google Scholar] [CrossRef] [PubMed]
- Herbert, K.M.; Greenleaf, W.J.; Block, S.M. Single-molecule studies of RNA polymerase: Motoring along. Annu. Rev. Biochem. 2008, 77, 149–176. [Google Scholar] [CrossRef] [PubMed]
- Bustamante, C.; Cheng, W.; Mejia, Y.X. Revisiting the central dogma one molecule at a time. Cell 2011, 144, 480–497. [Google Scholar] [CrossRef] [PubMed]
- Rayment, I.; Rypniewski, W.R.; Schmidt-Bäse, K.; Smith, R.; Tomchick, D.R.; Benning, M.M.; Winkelmann, D.A.; Wesenberg, G.; Holden, H.M. Three-dimensional structure of myosin subfragment-1: A molecular motor. Science 1993, 261, 50–58. [Google Scholar] [CrossRef]
- Schmidt, H.; Carter, A.P. Structure and mechanism of the dynein motor ATPase. Biopolymers 2016, 105, 557–567. [Google Scholar] [CrossRef]
- Berg, H.C. The Rotary Motor of Bacterial Flagella. Annu. Rev. Biochem. 2003, 72, 19–54. [Google Scholar] [CrossRef]
- Boyer, P.D. The Atp Synthase—A Splendid Molecular Machine. Annu. Rev. Biochem. 1997, 66, 717–749. [Google Scholar] [CrossRef]
- Forgac, M. Vacuolar ATPases: Rotary proton pumps in physiology and pathophysiology. Nat. Rev. Mol. Cell Biol. 2007, 8, 917–929. [Google Scholar] [CrossRef]
- Müller, V.; Grüber, G. ATP synthases: Structure, function and evolution of unique energy converters. Cell. Mol. Life Sci. CMLS 2003, 60, 474–494. [Google Scholar] [CrossRef]
- Kühlbrandt, W.; Davies, K.M. Rotary ATPases: A New Twist to an Ancient Machine. Trends Biochem. Sci. 2016, 41, 106–116. [Google Scholar] [CrossRef]
- Guo, H.; Rubinstein, J.L. Structure of ATP synthase under strain during catalysis. Nat. Commun. 2022, 13, 2232. [Google Scholar] [CrossRef]
- Santiveri, M.; Roa-Eguiara, A.; Kühne, C.; Wadhwa, N.; Hu, H.; Berg, H.C.; Erhardt, M.; Taylor, N.M.I. Structure and Function of Stator Units of the Bacterial Flagellar Motor. Cell 2020, 183, 244–257.e16. [Google Scholar] [CrossRef] [PubMed]
- Deme, J.C.; Johnson, S.; Vickery, O.; Aron, A.; Monkhouse, H.; Griffiths, T.; James, R.H.; Berks, B.C.; Coulton, J.W.; Stansfeld, P.J.; et al. Structures of the stator complex that drives rotation of the bacterial flagellum. Nat. Microbiol. 2020, 5, 1553–1564. [Google Scholar] [CrossRef] [PubMed]
- Hosking, E.R.; Vogt, C.; Bakker, E.P.; Manson, M.D. The Escherichia coli MotAB proton channel unplugged. J. Mol. Biol. 2006, 364, 921–937. [Google Scholar] [CrossRef]
- Yonekura, K.; Maki-Yonekura, S.; Homma, M. Structure of the flagellar motor protein complex PomAB: Implications for the torque-generating conformation. J. Bacteriol. 2011, 193, 3863–3870. [Google Scholar] [CrossRef][Green Version]
- Takekawa, N.; Nishiyama, M.; Kaneseki, T.; Kanai, T.; Atomi, H.; Kojima, S.; Homma, M. Sodium-driven energy conversion for flagellar rotation of the earliest divergent hyperthermophilic bacterium. Sci. Rep. 2015, 5, 12711. [Google Scholar] [CrossRef] [PubMed]
- Terahara, N.; Sano, M.; Ito, M. A Bacillus flagellar motor that can use both Na+ and K+ as a coupling ion is converted by a single mutation to use only Na+. PLoS ONE 2012, 7, e46248. [Google Scholar] [CrossRef] [PubMed]
- Imazawa, R.; Takahashi, Y.; Aoki, W.; Sano, M.; Ito, M. A novel type bacterial flagellar motor that can use divalent cations as a coupling ion. Sci. Rep. 2016, 6, 19773. [Google Scholar] [CrossRef]
- Onoe, S.; Yoshida, M.; Terahara, N.; Sowa, Y. Coupling Ion Specificity of the Flagellar Stator Proteins MotA1/MotB1 of Paenibacillus sp. TCA20. Biomolecules 2020, 10, 1078. [Google Scholar] [CrossRef]
- Celia, H.; Botos, I.; Ni, X.; Fox, T.; De Val, N.; Lloubes, R.; Jiang, J.; Buchanan, S.K. Cryo-EM structure of the bacterial Ton motor subcomplex ExbB–ExbD provides information on structure and stoichiometry. Commun. Biol. 2019, 2, 358. [Google Scholar] [CrossRef]
- Celia, H.; Noinaj, N.; Buchanan, S.K. Structure and Stoichiometry of the Ton Molecular Motor. Int. J. Mol. Sci. 2020, 21, 375. [Google Scholar] [CrossRef]
- Hennell James, R.; Deme, J.C.; Kjӕr, A.; Alcock, F.; Silale, A.; Lauber, F.; Johnson, S.; Berks, B.C.; Lea, S.M. Structure and mechanism of the proton-driven motor that powers type 9 secretion and gliding motility. Nat. Microbiol. 2021, 6, 221–233. [Google Scholar] [CrossRef] [PubMed]
- Doron, S.; Melamed, S.; Ofir, G.; Leavitt, A.; Lopatina, A.; Keren, M.; Amitai, G.; Sorek, R. Systematic discovery of antiphage defense systems in the microbial pangenome. Science 2018, 359, eaar4120. [Google Scholar] [CrossRef] [PubMed]
- Wadhwa, N.; Berg, H.C. Bacterial motility: Machinery and mechanisms. Nat. Rev. Microbiol. 2022, 20, 161–173. [Google Scholar] [CrossRef]
- Braun, V.; Ratliff, A.C.; Celia, H.; Buchanan, S.K. Energization of Outer Membrane Transport by the ExbB ExbD Molecular Motor. J. Bacteriol. 2023, e00035-23. [Google Scholar] [CrossRef] [PubMed]
- Sun, M.; Wartel, M.; Cascales, E.; Shaevitz, J.W.; Mignot, T. Motor-driven intracellular transport powers bacterial gliding motility. Proc. Natl. Acad. Sci. USA 2011, 108, 7559–7564. [Google Scholar] [CrossRef] [PubMed]
- Spence, M.A.; Kaczmarski, J.A.; Saunders, J.W.; Jackson, C.J. Ancestral sequence reconstruction for protein engineers. Curr. Opin. Struct. Biol. 2021, 69, 131–141. [Google Scholar] [CrossRef]
- Mascotti, M.L. Resurrecting Enzymes by Ancestral Sequence Reconstruction. Methods Mol. Biol. 2022, 2397, 111–136. [Google Scholar] [CrossRef]
- Thomson, R.E.; Carrera-Pacheco, S.E.; Gillam, E.M. Engineering functional thermostable proteins using ancestral sequence reconstruction. J. Biol. Chem. 2022, 298, 102435. [Google Scholar] [CrossRef]
- Vinde, M.H.; Cao, D.; Chesterfield, R.J.; Yoneyama, K.; Gumulya, Y.; Thomson, R.E.; Matila, T.; Ebert, B.E.; Beveridge, C.A.; Vickers, C.E. Ancestral sequence reconstruction of the CYP711 family reveals functional divergence in strigolactone biosynthetic enzymes associated with gene duplication events in monocot grasses. New Phytol. 2022, 235, 1900–1912. [Google Scholar] [CrossRef]
- Nakamura, S.; Minamino, T. Flagella-Driven Motility of Bacteria. Biomolecules 2019, 9, 279. [Google Scholar] [CrossRef]
- Chang, Y.; Zhang, K.; Carroll, B.L.; Zhao, X.; Charon, N.W.; Norris, S.J.; Motaleb, M.A.; Li, C.; Liu, J. Molecular mechanism for rotational switching of the bacterial flagellar motor. Nat. Struct. Mol. Biol. 2020, 27, 1041–1047. [Google Scholar] [CrossRef] [PubMed]
- Minamino, T.; Imada, K. The bacterial flagellar motor and its structural diversity. Trends Microbiol. 2015, 23, 267–274. [Google Scholar] [CrossRef] [PubMed]
- Kato, T.; Makino, F.; Miyata, T.; Horváth, P.; Namba, K. Structure of the native supercoiled flagellar hook as a universal joint. Nat. Commun. 2019, 10, 5295. [Google Scholar] [CrossRef]
- Kubori, T.; Okumura, M.; Kobayashi, N.; Nakamura, D.; Iwakura, M.; Aizawa, S. Purification and characterization of the flagellar hook–basal body complex of Bacillus subtilis. Mol. Microbiol. 1997, 24, 399–410. [Google Scholar] [CrossRef] [PubMed]
- Zhu, S.; Nishikino, T.; Hu, B.; Kojima, S.; Homma, M.; Liu, J. Molecular architecture of the sheathed polar flagellum in Vibrio alginolyticus. Proc. Natl. Acad. Sci. USA 2017, 114, 10966–10971. [Google Scholar] [CrossRef]
- Terashima, H.; Kawamoto, A.; Morimoto, Y.V.; Imada, K.; Minamino, T. Structural differences in the bacterial flagellar motor among bacterial species. Biophys. Phys. 2017, 14, 191–198. [Google Scholar] [CrossRef]
- Merino, S.; Tomás, J.M. The FlgT Protein Is Involved in Aeromonas hydrophila Polar Flagella Stability and Not Affects Anchorage of Lateral Flagella. Front. Microbiol. 2016, 7, 1150. [Google Scholar] [CrossRef]
- Terashima, H.; Fukuoka, H.; Yakushi, T.; Kojima, S.; Homma, M. The Vibrio motor proteins, MotX and MotY, are associated with the basal body of Na-driven flagella and required for stator formation. Mol. Microbiol. 2006, 62, 1170–1180. [Google Scholar] [CrossRef]
- Eggenhofer, E.; Haslbeck, M.; Scharf, B. MotE serves as a new chaperone specific for the periplasmic motility protein, MotC, in Sinorhizobium meliloti. Mol. Microbiol. 2004, 52, 701–712. [Google Scholar] [CrossRef]
- Terahara, N.; Krulwich, T.A.; Ito, M. Mutations alter the sodium versus proton use of a Bacillus clausii flagellar motor and confer dual ion use on Bacillus subtilis motors. Proc. Natl. Acad. Sci. USA 2008, 105, 14359–14364. [Google Scholar] [CrossRef] [PubMed]
- Chun, S.Y.; Parkinson, J.S. Bacterial Motility: Membrane Topology of the Escherichia coli MotB Protein. Science 1988, 239, 276–278. [Google Scholar] [CrossRef] [PubMed]
- Leake, M.C.; Chandler, J.H.; Wadhams, G.H.; Bai, F.; Berry, R.M.; Armitage, J.P. Stoichiometry and turnover in single, functioning membrane protein complexes. Nature 2006, 443, 355–358. [Google Scholar] [CrossRef]
- Wadhwa, N.; Tu, Y.; Berg, H.C. Mechanosensitive remodeling of the bacterial flagellar motor is independent of direction of rotation. Proc. Natl. Acad. Sci. USA 2021, 118, e2024608118. [Google Scholar] [CrossRef] [PubMed]
- Berry, R.M.; Berg, H.C. Torque Generated by the Flagellar Motor of Escherichia coli while Driven Backward. Biophys. J. 1999, 76, 580–587. [Google Scholar] [CrossRef]
- Lo, C.-J.; Sowa, Y.; Pilizota, T.; Berry, R.M. Mechanism and kinetics of a sodium-driven bacterial flagellar motor. Proc. Natl. Acad. Sci. USA 2013, 110, E2544–E2551. [Google Scholar] [CrossRef]
- Zhou, J.; Sharp, L.L.; Tang, H.L.; Lloyd, S.A.; Billings, S.; Braun, T.F.; Blair, D.F. Function of Protonatable Residues in the Flagellar Motor of Escherichia coli: A Critical Role for Asp 32 of MotB. J. Bacteriol. 1998, 180, 2729–2735. [Google Scholar] [CrossRef]
- Tokárová, V.; Perumal, A.S.; Nayak, M.; Shum, H.; Kašpar, O.; Rajendran, K.; Mohammadi, M.; Tremblay, C.; Gaffney, E.A.; Martel, S.; et al. Patterns of bacterial motility in microfluidics-confining environments. Proc. Natl. Acad. Sci. USA 2021, 118, e2013925118. [Google Scholar] [CrossRef]
- Onoue, Y.; Iwaki, M.; Shinobu, A.; Nishihara, Y.; Iwatsuki, H.; Terashima, H.; Kitao, A.; Kandori, H.; Homma, M. Essential ion binding residues for Na+ flow in stator complex of the Vibrio flagellar motor. Sci. Rep. 2019, 9, 11216. [Google Scholar] [CrossRef]
- Hu, H.; Popp, P.; Santiveri, M.; Eguiara, A.; Yan, Y.; Liu, Z.; Wadhwa, N.; Wang, Y.; Erhardt, M.; Taylor, N.M. Mechanisms of ion selectivity and rotor coupling in the bacterial flagellar sodium-driven stator unit. bioRxiv 2022. [Google Scholar] [CrossRef]
- Nishikino, T.; Sagara, Y.; Terashima, H.; Homma, M.; Kojima, S. Hoop-like role of the cytosolic interface helix in Vibrio PomA, an ion-conducting membrane protein, in the bacterial flagellar motor. J. Biochem. 2022, 171, 443–450. [Google Scholar] [CrossRef] [PubMed]
- Asai, Y.; Kawagishi, I.; Sockett, R.E.; Homma, M. Hybrid motor with H+-and Na+-driven components can rotate Vibrio polar flagella by using sodium ions. J. Bacteriol. 1999, 181, 6332–6338. [Google Scholar] [CrossRef] [PubMed]
- Asai, Y.; Shoji, T.; Kawagishi, I.; Homma, M. Cysteine-scanning mutagenesis of the periplasmic loop regions of PomA, a putative channel component of the sodium-driven flagellar motor in Vibrio alginolyticus. J. Bacteriol. 2000, 182, 1001–1007. [Google Scholar] [CrossRef]
- Asai, Y.; Yakushi, T.; Kawagishi, I.; Homma, M. Ion-coupling determinants of Na+-driven and H+-driven flagellar motors. J. Mol. Biol. 2003, 327, 453–463. [Google Scholar] [CrossRef]
- Li, N.; Kojima, S.; Homma, M. Sodium-driven motor of the polar flagellum in marine bacteria Vibrio. Genes Cells 2011, 16, 985–999. [Google Scholar] [CrossRef] [PubMed]
- Nishihara, Y.; Kitao, A. Gate-controlled proton diffusion and protonation-induced ratchet motion in the stator of the bacterial flagellar motor. Proc. Natl. Acad. Sci. USA 2015, 112, 7737–7742. [Google Scholar] [CrossRef]
- Kojima, S.; Shoji, T.; Asai, Y.; Kawagishi, I.; Homma, M. A slow-motility phenotype caused by substitutions at residue Asp31 in the PomA channel component of a sodium-driven flagellar motor. J. Bacteriol. 2000, 182, 3314–3318. [Google Scholar] [CrossRef][Green Version]
- Nishino, Y.; Onoue, Y.; Kojima, S.; Homma, M. Functional chimeras of flagellar stator proteins between E. coli MotB and Vibrio PomB at the periplasmic region in Vibrio or E. coli. Microbiologyopen 2015, 4, 323–331. [Google Scholar] [CrossRef]
- Ridone, P.; Ishida, T.; Lin, A.; Humphreys, D.T.; Giannoulatou, E.; Sowa, Y.; Baker, M.A. The rapid evolution of flagellar ion selectivity in experimental populations of E. coli. Sci. Adv. 2022, 8, eabq2492. [Google Scholar] [CrossRef]
- Kacar, B.; Guy, L.; Smith, E.; Baross, J. Resurrecting ancestral genes in bacteria to interpret ancient biosignatures. Philos. Trans. R. Soc. Math. Phys. Eng. Sci. 2017, 375, 20160352. [Google Scholar] [CrossRef]
- Merkl, R.; Sterner, R. Ancestral protein reconstruction: Techniques and applications. Biol. Chem. 2016, 397, 1–21. [Google Scholar] [CrossRef] [PubMed]
- Harms, M.J.; Thornton, J.W. Analyzing protein structure and function using ancestral gene reconstruction. Curr. Opin. Struct. Biol. 2010, 20, 360–366. [Google Scholar] [CrossRef] [PubMed]
- Thornton, J.W. Resurrecting ancient genes: Experimental analysis of extinct molecules. Nat. Rev. Genet. 2004, 5, 366–375. [Google Scholar] [CrossRef]
- Pauling, L.; Zuckerkandl, E.; Henriksen, T.; Lövstad, R. Chemical paleogenetics. Acta Chem. Scand. 1963, 17, S9–S16. [Google Scholar] [CrossRef]
- Cai, W.; Pei, J.; Grishin, N.V. Reconstruction of ancestral protein sequences and its applications. BMC Evol. Biol. 2004, 4, 33. [Google Scholar] [CrossRef] [PubMed]
- Gumulya, Y.; Gillam, E.M. Exploring the past and the future of protein evolution with ancestral sequence reconstruction: The ‘retro’ approach to protein engineering. Biochem. J. 2017, 474, 1–19. [Google Scholar] [CrossRef] [PubMed]
- Babkova, P.; Sebestova, E.; Brezovsky, J.; Chaloupkova, R.; Damborsky, J. Ancestral Haloalkane Dehalogenases Show Robustness and Unique Substrate Specificity. ChemBioChem 2017, 18, 1448–1456. [Google Scholar] [CrossRef]
- Siddiq, M.A.; Hochberg, G.K.; Thornton, J.W. Evolution of protein specificity: Insights from ancestral protein reconstruction. Curr. Opin. Struct. Biol. 2017, 47, 113–122. [Google Scholar] [CrossRef]
- Islam, M.I.; Lin, A.; Lai, Y.-W.; Matzke, N.J.; Baker, M.A. Ancestral sequence reconstructions of MotB are proton-motile and require MotA for motility. Front. Microbiol. 2020, 11, 625837. [Google Scholar] [CrossRef]
- García-López, R.; Rivera-Gutiérrez, X.; Rosales-Rivera, M.; Zárate, S.; Muñoz-Medina, J.E.; Roche, B.; Herrera-Estrella, A.; Gómez-Gil, B.; Sanchez-Flores, A.; Taboada, B. SARS-CoV-2 BW lineage, a fast-growing Omicron variant from southeast Mexico bearing relevant escape mutations. Infection 2023. [Google Scholar] [CrossRef]
- Chiang, C.-H.; Wymore, T.; Rodríguez Benítez, A.; Hussain, A.; Smith, J.L.; Brooks III, C.L.; Narayan, A.R. Deciphering the evolution of flavin-dependent monooxygenase stereoselectivity using ancestral sequence reconstruction. Proc. Natl. Acad. Sci. USA 2023, 120, e2218248120. [Google Scholar] [CrossRef]
- Steube, N.; Moldenhauer, M.; Weiland, P.; Saman, D.; Kilb, A.; Ramírez Rojas, A.A.; Garg, S.G.; Schindler, D.; Graumann, P.L.; Benesch, J.L. Fortuitously compatible protein surfaces primed allosteric control in cyanobacterial photoprotection. Nat. Ecol. Evol. 2023, 7, 756–767. [Google Scholar] [CrossRef]
- Chen, S.; Xu, Z.; Ding, B.; Zhang, Y.; Liu, S.; Cai, C.; Li, M.; Dale, B.E.; Jin, M. Big data mining, rational modification, and ancestral sequence reconstruction inferred multiple xylose isomerases for biorefinery. Sci. Adv. 2023, 9, eadd8835. [Google Scholar] [CrossRef]
- Ghorbani, A.; Khataeipour, S.J.; Solbakken, M.H.; Huebert, D.N.; Khoddami, M.; Eslamloo, K.; Collins, C.; Hori, T.; Jentoft, S.; Rise, M.L. Ancestral reconstruction reveals catalytic inactivation of activation-induced cytidine deaminase concomitant with cold water adaption in the Gadiformes bony fish. BMC Biol. 2022, 20, 293. [Google Scholar] [CrossRef] [PubMed]
- Su, W.; Harfoot, R.; Su, Y.C.; DeBeauchamp, J.; Joseph, U.; Jayakumar, J.; Crumpton, J.-C.; Jeevan, T.; Rubrum, A.; Franks, J. Ancestral sequence reconstruction pinpoints adaptations that enable avian influenza virus transmission in pigs. Nat. Microbiol. 2021, 6, 1455–1465. [Google Scholar] [CrossRef] [PubMed]
- Harris, K.L.; Thomson, R.E.; Gumulya, Y.; Foley, G.; Carrera-Pacheco, S.E.; Syed, P.; Janosik, T.; Sandinge, A.-S.; Andersson, S.; Jurva, U. Ancestral Sequence Reconstruction of a Cytochrome P450 Family Involved in Chemical Defense Reveals the Functional Evolution of a Promiscuous, Xenobiotic-Metabolizing Enzyme in Vertebrates. Mol. Biol. Evol. 2022, 39, msac116. [Google Scholar] [CrossRef] [PubMed]
- Joho, Y.; Vongsouthi, V.; Spence, M.A.; Ton, J.; Gomez, C.; Tan, L.L.; Kaczmarski, J.A.; Caputo, A.T.; Royan, S.; Jackson, C.J. Ancestral sequence reconstruction identifies structural changes underlying the evolution of Ideonella Sakaiensis PETase and variants with improved stability and activity. Biochemistry 2023, 62, 437–450. [Google Scholar] [CrossRef]
- Knight, K.A.; Coyle, C.W.; Radford, C.E.; Parker, E.T.; Fedanov, A.; Shields, J.M.; Szlam, F.; Purchel, A.; Chen, M.; Denning, G. Identification of coagulation factor IX variants with enhanced activity through ancestral sequence reconstruction. Blood Adv. 2021, 5, 3333–3343. [Google Scholar] [CrossRef]
- Cortez, L.M.; Morrison, A.J.; Garen, C.R.; Patterson, S.; Uyesugi, T.; Petrosyan, R.; Sekar, R.V.; Harms, M.J.; Woodside, M.T.; Sim, V.L. Probing the origin of prion protein misfolding via reconstruction of ancestral proteins. Protein Sci. 2022, 31, e4477. [Google Scholar] [CrossRef]
- Valenti, R.; Jabłońska, J.; Tawfik, D.S. Characterization of ancestral Fe/Mn superoxide dismutases indicates their cambialistic origin. Protein Sci. 2022, 31, e4423. [Google Scholar] [CrossRef]
- Lipovsek, M.; Fierro, A.; Pérez, E.G.; Boffi, J.C.; Millar, N.S.; Fuchs, P.A.; Katz, E.; Elgoyhen, A.B. Tracking the molecular evolution of calcium permeability in a nicotinic acetylcholine receptor. Mol. Biol. Evol. 2014, 31, 3250–3265. [Google Scholar] [CrossRef]
- Rozi, M.F.A.M.; Rahman, R.N.Z.R.A.; Leow, A.T.C.; Ali, M.S.M. Ancestral sequence reconstruction of ancient lipase from family I. 3 bacterial lipolytic enzymes. Mol. Phylogenet. Evol. 2022, 168, 107381. [Google Scholar] [CrossRef]
- Islam, M.I.; Ridone, P.; Lin, A.; Michie, K.A.; Matzke, N.J.; Hochberg, G.; Baker, M.A. Ancestral reconstruction of the MotA stator subunit reveals that conserved residues far from the pore are required to drive flagellar motility. MicroLife 2023, 4, uqad011. [Google Scholar] [CrossRef] [PubMed]
- Perez-Jimenez, R.; Inglés-Prieto, A.; Zhao, Z.-M.; Sanchez-Romero, I.; Alegre-Cebollada, J.; Kosuri, P.; Garcia-Manyes, S.; Kappock, T.J.; Tanokura, M.; Holmgren, A. Single-molecule paleoenzymology probes the chemistry of resurrected enzymes. Nat. Struct. Mol. Biol. 2011, 18, 592–596. [Google Scholar] [CrossRef] [PubMed]
- Gaucher, E.A.; Govindarajan, S.; Ganesh, O.K. Palaeotemperature trend for Precambrian life inferred from resurrected proteins. Nature 2008, 451, 704–707. [Google Scholar] [CrossRef] [PubMed]
- Williams, P.D.; Pollock, D.D.; Blackburne, B.P.; Goldstein, R.A. Assessing the accuracy of ancestral protein reconstruction methods. PLoS Comput. Biol. 2006, 2, e69. [Google Scholar] [CrossRef]
- Dube, N.; Khan, S.H.; Okafor, C.D. Ancestral sequence reconstruction for evolutionary characterization of proteins. Trends Biochem. Sci. 2022, 47, 98–99. [Google Scholar] [CrossRef]
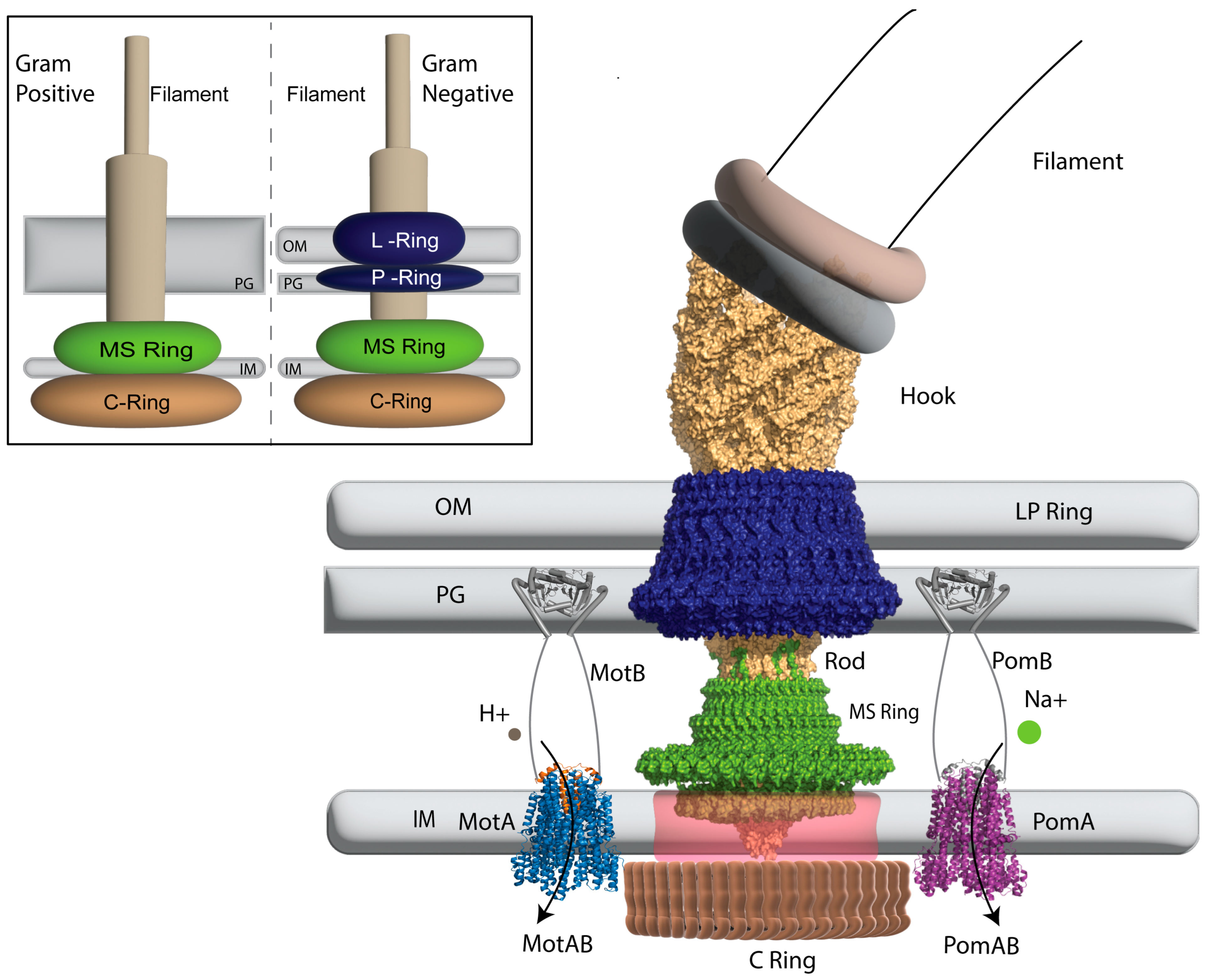
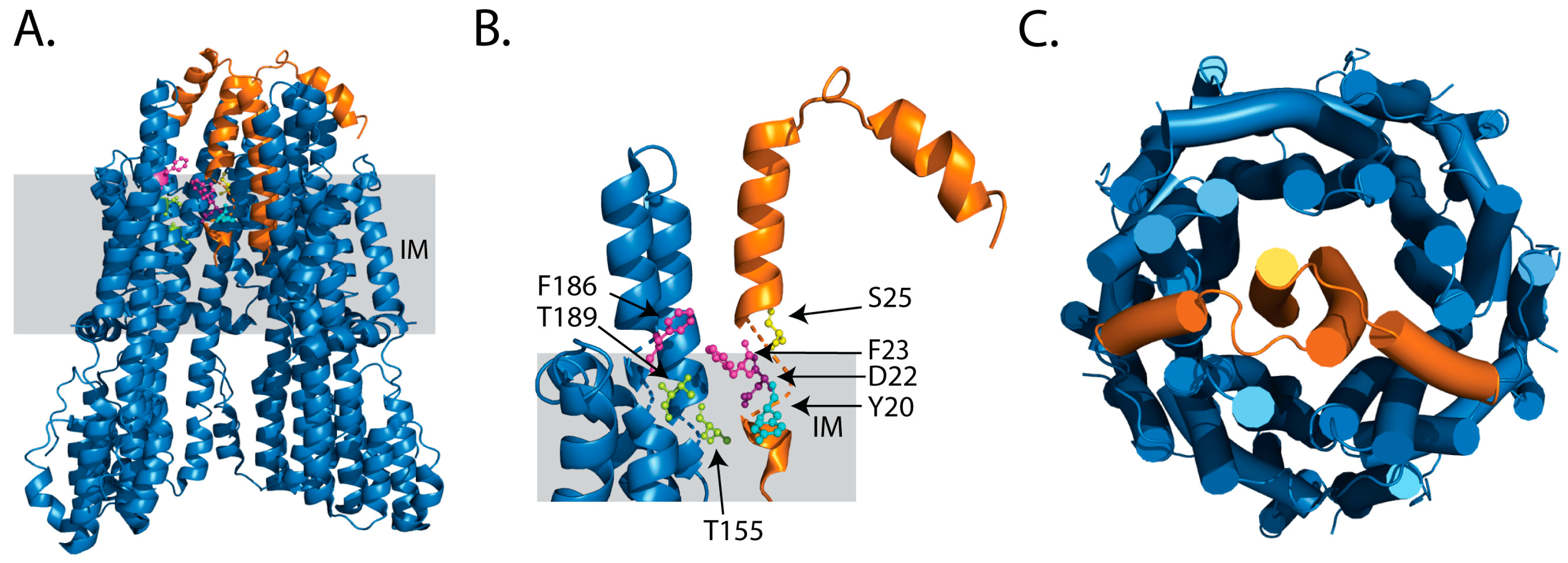

| Organism | Stator | Mutation | Ion Selectivity | Effect | Reference |
|---|---|---|---|---|---|
| Bacillus alcalophilus | MotPS | MotS-M33L | Na+/K+/Rb+ | Loss of K+/Rb+ coupling motility-E. coli | [17] |
| Bacillus clausii | MotAB | MotB-V37L, A40S, G42S | Na+ | Only Na+ selective | [41] |
| MotB-G42S, Q43S, Q46A | H+ | Only H+ selective | |||
| Vibrio algynolyticus | PomAB | PomA-S25C | Na+ | Reduce motility | [53] |
| PomA-D31N | Affect ion usage | [57] |
| Protein | Number of Aligned Sequences | Phylogenetic/Ancestral Reconstruction Software Used | Key Findings | Reference |
|---|---|---|---|---|
| MotB | 757 | Quicktree/PAML | Ancestral MotB proteins were Na+ independent and could interact with extant MotAs | [69] |
| SARS-CoV-2 | 14,164 | IQ-TREE (v2.1.2)/TreeTime (v0.9.5) | Analysis of mutants through ASR identified that strain BW.1 arose in the Yucatan Peninsula | [70] |
| Flavin-dependent monooxygenase | 276 | PhyML (v3.0)/PAML (v4.8) | Two mechanisms were identified to control stereochemical outcomes of the oxidative dearomatization reaction | [71] |
| Orange carotenoid protein (OCP) | 189 | PhyML (v3.1)/PAML (v4.9) | Showed that the ancestral OCP regulator was horizontally acquired by cyanobacteria then co-evolved and that pre-HGT protein could still interact | [72] |
| Xylose isomerases (XI) | 1042 | Mega11/PAMLX | Amino-terminal fragment of ancestral XIs were important for maintaining activity and high performing enzymes were found that could contribute to high ethanol titers | [73] |
| Activation-induced cytidine deaminase (AID) | 71 | RAxML (v8.2.9), MrBayes (v3.2.7)/ProtASR (v2.0 and 2.2) | Enzymatic inactivation of reconstructed AIDs took place recently in the Atlantic cod family and thus explains that lack of secondary immunity in cod | [74] |
| Influenza A | 3443 | RAxML (v8.0), Geneious (R9.0.3)/Lazarus (v2.0) | Analysis of reconstructed swine influenza viruses from 1979–1992 showed that transmission in piglets was enabled by changes in viral polymerase protein and nucleoprotein since 1983 | [75] |
| Cytochrome P450 family 1 enzymes (CYP1s) | 471 | Mega11/GRASP | Younger ancestors were shown to have activities toward xenobiotic and steroid substrates than older ancestors. Greater thermostability was seen in older ancestor CYP1s, however caffeine metabolism was shown to be a recently evolved trait | [76] |
| PETase | 914 | IQ-TREE2/GRASP and PAML | Two ASR candidates were shown to have higher catalytic activity and thermostability was also increased | [77] |
| Coagulation factor IX (FIX) | 59 | MrBAYES/PAML (v4.1) | Ancestral FIX variants were shown to have enhanced activity and that AAV-ancestral FIX Padua vectors had greater potency over AAV-human FIX Padua vectors in haemophilia B mice | [78] |
| Phytohormones-CYP711A | 346 | RAxML/GRASP | Reconstructed CYP11As accepted GR24 as a substrate and the monocot group 3 ancestor showed increased catalytic activity and high stereoselectivity towards GR24 | [30] |
| Prion Protein (PrP) | 161 | PhyML (v3.3.2)/PAML (v4) | Aggregation of the PrP from the oldest ancestor was observed however ancestral bird PrP could not be seeded with extant prions. Ancestral primate PrP could be converted with all prion seeds | [79] |
| Fe/Mn superoxide dismutases (SODs) | 738 | IQ-Tree/PAML and FastML | Fe/Mn SODs were shown to be able to bind to both Fe and Mn, whereas extant SODs have been shown to have specific affinity to one ion | [80] |
| Nicotinic acetylcholine receptor (α9α10) | 52 | Mega5/PAML | Three residues were found in the α9 subunit which increased Ca2+ permeability for mammalian receptors but not for avian receptors | [81] |
| Family I.3 lipase | 83 | RAxML/PhyML | There was a deletion of residues during the evolution of this protein. Mg2+, Rb+ and Zinc (Zn2+) ions were also able to increase the relative activity indicating greater promiscuity of the ancient protein | [82] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Nandel, V.; Scadden, J.; Baker, M.A.B. Ion-Powered Rotary Motors: Where Did They Come from and Where They Are Going? Int. J. Mol. Sci. 2023, 24, 10601. https://doi.org/10.3390/ijms241310601
Nandel V, Scadden J, Baker MAB. Ion-Powered Rotary Motors: Where Did They Come from and Where They Are Going? International Journal of Molecular Sciences. 2023; 24(13):10601. https://doi.org/10.3390/ijms241310601
Chicago/Turabian StyleNandel, Vibhuti, Jacob Scadden, and Matthew A. B. Baker. 2023. "Ion-Powered Rotary Motors: Where Did They Come from and Where They Are Going?" International Journal of Molecular Sciences 24, no. 13: 10601. https://doi.org/10.3390/ijms241310601
APA StyleNandel, V., Scadden, J., & Baker, M. A. B. (2023). Ion-Powered Rotary Motors: Where Did They Come from and Where They Are Going? International Journal of Molecular Sciences, 24(13), 10601. https://doi.org/10.3390/ijms241310601





