AI-Predicted mTOR Inhibitor Reduces Cancer Cell Proliferation and Extends the Lifespan of C. elegans
Abstract
1. Introduction
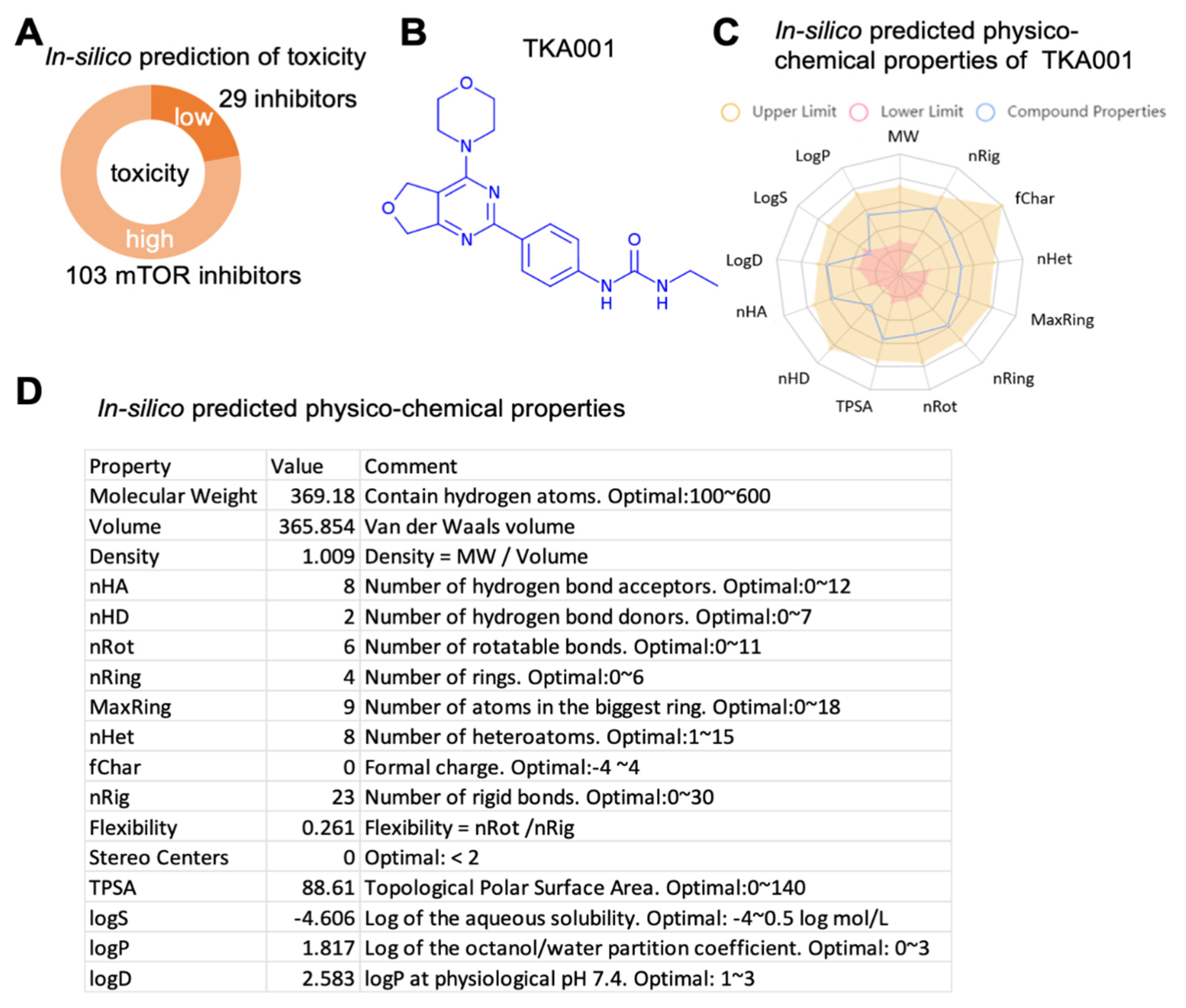
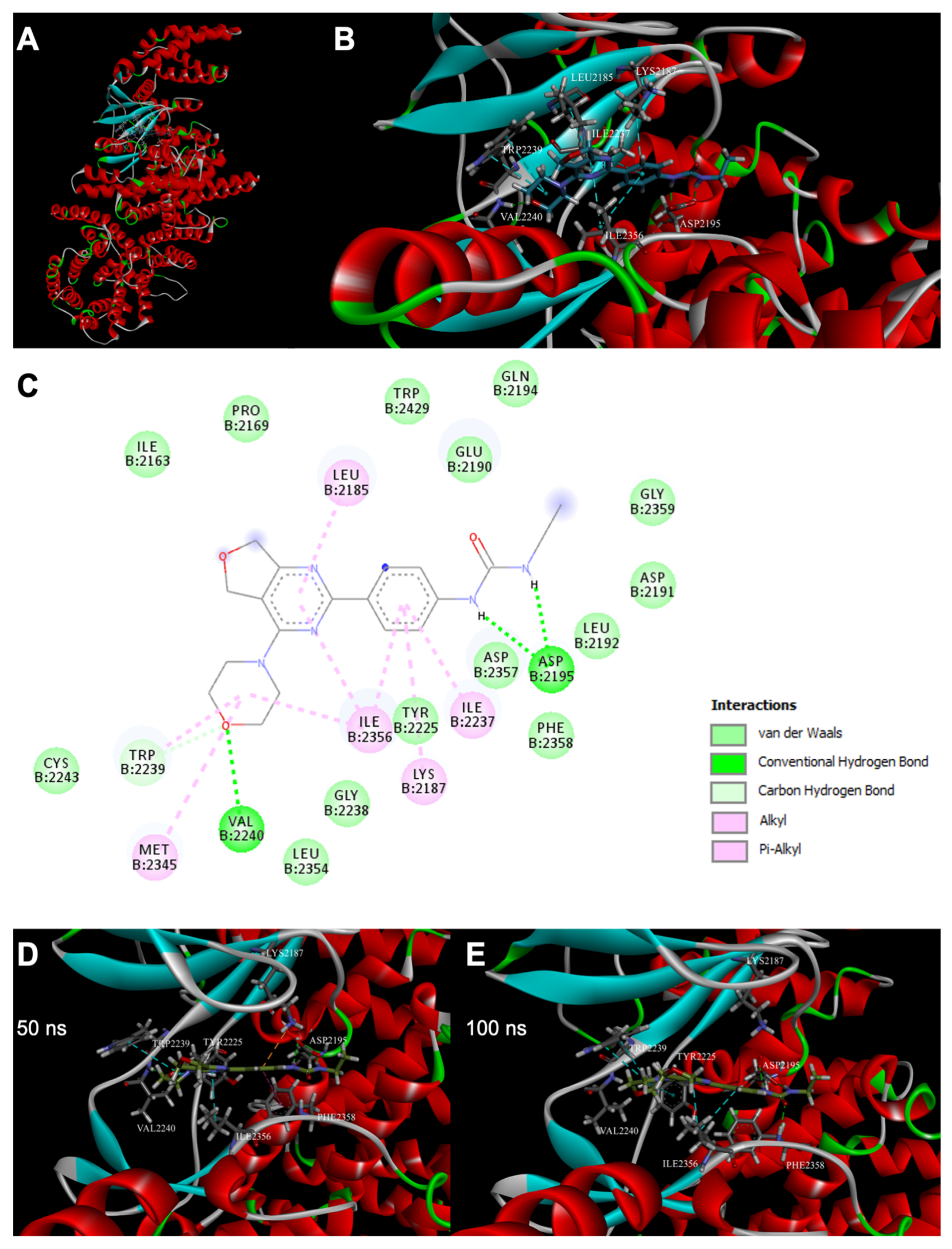
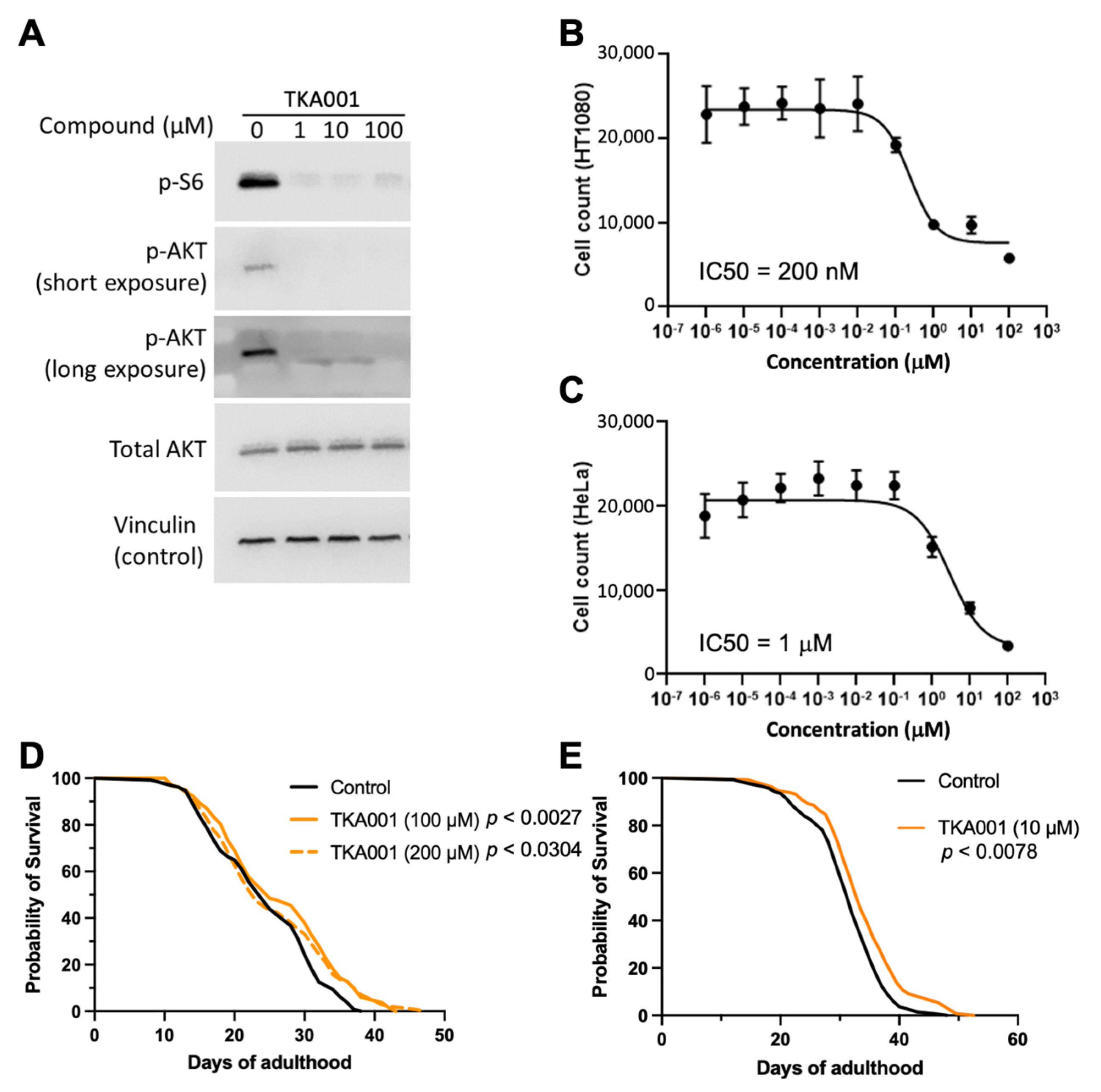
2. Results
3. Discussion
4. Materials and Methods
4.1. Reagents
4.2. Machine Learning
4.3. In Silico Prediction of the Mechanism of Action and Toxicity of Candidate Compounds
4.4. In Silico Predicted Physicochemical Properties of TKA001
4.5. Molecular Docking
4.6. Molecular Dynamics
4.7. Detailed Analysis of Molecular Docking and Molecular Dynamics
4.8. Human Cell Lines
4.9. Human Cell Proliferation
4.10. Western Blots
4.11. C. elegans Lifespan
4.12. C. elegans Lifespan Assessed with the Lifespan Machine
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Kapahi, P.; Zid, B.M.; Harper, T.; Koslover, D.; Sapin, V.; Benzer, S. Regulation of Lifespan in Drosophila by Modulation of Genes in the TOR Signaling Pathway. Curr. Biol. 2004, 14, 885–890. [Google Scholar] [CrossRef]
- Robida-Stubbs, S.; Glover-Cutter, K.; Lamming, D.W.; Mizunuma, M.; Narasimhan, S.D.; Neumann-Haefelin, E.; Sabatini, D.M.; Blackwell, T.K. TOR Signaling and Rapamycin Influence Longevity by Regulating SKN-1/Nrf and DAF-16/FoxO. Cell Metab. 2012, 15, 713–724. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Lanjuin, A.; Chowdhury, S.R.; Mistry, M.; Silva-García, C.G.; Weir, H.J.; Lee, C.-L.; Escoubas, C.C.; Tabakovic, E.; Mair, W.B. Neuronal TORC1 Modulates Longevity via AMPK and Cell Nonautonomous Regulation of Mitochondrial Dynamics in C. elegans. Elife 2019, 8, e49158. [Google Scholar] [CrossRef]
- Statzer, C.; Meng, J.; Venz, R.; Bland, M.; Robida-Stubbs, S.; Patel, K.; Petrovic, D.; Emsley, R.; Liu, P.; Morantte, I.; et al. ATF-4 and Hydrogen Sulfide Signalling Mediate Longevity in Response to Inhibition of Translation or mTORC1. Nat. Commun. 2022, 13, 967. [Google Scholar] [CrossRef] [PubMed]
- Vellai, T.; Takacs-Vellai, K.; Zhang, Y.; Kovacs, A.L.; Orosz, L.; Müller, F. Genetics: Influence of TOR Kinase on Lifespan in C. elegans. Nature 2003, 426, 620. [Google Scholar] [CrossRef] [PubMed]
- Ewald, C.Y.; Landis, J.N.; Abate, J.P.; Murphy, C.T.; Blackwell, T.K. Dauer-Independent Insulin/IGF-1-Signalling Implicates Collagen Remodelling in Longevity. Nature 2015, 519, 97–101. [Google Scholar] [CrossRef] [PubMed]
- Wu, J.J.; Liu, J.; Chen, E.B.; Wang, J.J.; Cao, L.; Narayan, N.; Fergusson, M.M.; Rovira, I.I.; Allen, M.; Springer, D.A.; et al. Increased Mammalian Lifespan and a Segmental and Tissue-Specific Slowing of Aging after Genetic Reduction of mTOR Expression. Cell Rep. 2013, 4, 913–920. [Google Scholar] [CrossRef]
- Shindyapina, A.V.; Cho, Y.; Kaya, A.; Tyshkovskiy, A.; Castro, J.P.; Deik, A.; Gordevicius, J.; Poganik, J.R.; Clish, C.B.; Horvath, S.; et al. Rapamycin Treatment during Development Extends Life Span and Health Span of Male Mice and Daphnia Magna. Sci. Adv. 2022, 8, eabo5482. [Google Scholar] [CrossRef]
- Harrison, D.E.; Strong, R.; Sharp, Z.D.; Nelson, J.F.; Astle, C.M.; Flurkey, K.; Nadon, N.L.; Wilkinson, J.E.; Frenkel, K.; Carter, C.S.; et al. Rapamycin Fed Late in Life Extends Lifespan in Genetically Heterogeneous Mice. Nature 2009, 460, 392–395. [Google Scholar] [CrossRef]
- Strong, R.; Miller, R.A.; Bogue, M.; Fernandez, E.; Javors, M.A.; Libert, S.; Marinez, P.A.; Murphy, M.P.; Musi, N.; Nelson, J.F.; et al. Rapamycin-mediated Mouse Lifespan Extension: Late-life Dosage Regimes with Sex-specific Effects. Aging Cell 2020, 19, e13269. [Google Scholar] [CrossRef]
- Miller, R.A.; Harrison, D.E.; Astle, C.M.; Baur, J.A.; Boyd, A.R.; de Cabo, R.; Fernandez, E.; Flurkey, K.; Javors, M.A.; Nelson, J.F.; et al. Rapamycin, But Not Resveratrol or Simvastatin, Extends Life Span of Genetically Heterogeneous Mice. J. Gerontol. Ser. 2011, 66A, 191–201. [Google Scholar] [CrossRef] [PubMed]
- Apelo, S.I.A.; Pumper, C.P.; Baar, E.L.; Cummings, N.E.; Lamming, D.W. Intermittent Administration of Rapamycin Extends the Life Span of Female C57BL/6J Mice. J. Gerontol. Ser. 2016, 71, 876–881. [Google Scholar] [CrossRef]
- Jia, K.; Chen, D.; Riddle, D.L. The TOR Pathway Interacts with the Insulin Signaling Pathway to Regulate C. elegans Larval Development, Metabolism and Life Span. Development 2004, 131, 3897–3906. [Google Scholar] [CrossRef] [PubMed]
- Torrence, M.E.; MacArthur, M.R.; Hosios, A.M.; Valvezan, A.J.; Asara, J.M.; Mitchell, J.R.; Manning, B.D. The mTORC1-Mediated Activation of ATF4 Promotes Protein and Glutathione Synthesis Downstream of Growth Signals. Elife 2021, 10, e63326. [Google Scholar] [CrossRef] [PubMed]
- Mannick, J.B.; Giudice, G.D.; Lattanzi, M.; Valiante, N.M.; Praestgaard, J.; Huang, B.; Lonetto, M.A.; Maecker, H.T.; Kovarik, J.; Carson, S.; et al. MTOR Inhibition Improves Immune Function in the Elderly. Sci. Transl. Med. 2014, 6, 268ra179. [Google Scholar] [CrossRef] [PubMed]
- Chung, C.L.; Lawrence, I.; Hoffman, M.; Elgindi, D.; Nadhan, K.; Potnis, M.; Jin, A.; Sershon, C.; Binnebose, R.; Lorenzini, A.; et al. Topical Rapamycin Reduces Markers of Senescence and Aging in Human Skin: An Exploratory, Prospective, Randomized Trial. Geroscience 2019, 41, 861–869. [Google Scholar] [CrossRef]
- DiMasi, J.A.; Grabowski, H.G.; Hansen, R.W. Innovation in the Pharmaceutical Industry: New Estimates of R&D Costs. J. Health Econ. 2016, 47, 20–33. [Google Scholar] [CrossRef]
- Lagunin, A.; Stepanchikova, A.; Filimonov, D.; Poroikov, V. PASS: Prediction of Activity Spectra for Biologically Active Substances. Bioinformatics 2000, 16, 747–748. [Google Scholar] [CrossRef]
- Loewith, R.; Jacinto, E.; Wullschleger, S.; Lorberg, A.; Crespo, J.L.; Bonenfant, D.; Oppliger, W.; Jenoe, P.; Hall, M.N. Two TOR Complexes, Only One of Which Is Rapamycin Sensitive, Have Distinct Roles in Cell Growth Control. Mol. Cell 2002, 10, 457–468. [Google Scholar] [CrossRef]
- Liu, G.Y.; Sabatini, D.M. MTOR at the Nexus of Nutrition, Growth, Ageing and Disease. Nat. Rev. Mol. Cell Biol. 2020, 21, 183–203. [Google Scholar] [CrossRef]
- Lagunin, A.A.; Dubovskaja, V.I.; Rudik, A.V.; Pogodin, P.V.; Druzhilovskiy, D.S.; Gloriozova, T.A.; Filimonov, D.A.; Sastry, N.G.; Poroikov, V.V. CLC-Pred: A Freely Available Web-Service for in Silico Prediction of Human Cell Line Cytotoxicity for Drug-like Compounds. PLoS ONE 2018, 13, e0191838. [Google Scholar] [CrossRef] [PubMed]
- Yang, W.; Soares, J.; Greninger, P.; Edelman, E.J.; Lightfoot, H.; Forbes, S.; Bindal, N.; Beare, D.; Smith, J.A.; Thompson, I.R.; et al. Genomics of Drug Sensitivity in Cancer (GDSC): A Resource for Therapeutic Biomarker Discovery in Cancer Cells. Nucleic Acids Res. 2013, 41, D955–D961. [Google Scholar] [CrossRef] [PubMed]
- Blackwell, T.K.; Sewell, A.K.; Wu, Z.; Han, M. TOR Signaling in Caenorhabditis elegans Development, Metabolism, and Aging. Genetics 2019, 213, 329–360. [Google Scholar] [CrossRef] [PubMed]
- Hansen, M.; Taubert, S.; Crawford, D.; Libina, N.; Lee, S.; Kenyon, C. Lifespan Extension by Conditions That Inhibit Translation in Caenorhabditis elegans. Aging Cell 2007, 6, 95–110. [Google Scholar] [CrossRef]
- Pan, K.Z.; Palter, J.E.; Rogers, A.N.; Olsen, A.; Chen, D.; Lithgow, G.J.; Kapahi, P. Inhibition of mRNA Translation Extends Lifespan in Caenorhabditis elegans. Aging Cell 2007, 6, 111–119. [Google Scholar] [CrossRef] [PubMed]
- Wullschleger, S.; Loewith, R.; Hall, M.N. TOR Signaling in Growth and Metabolism. Cell 2006, 124, 471–484. [Google Scholar] [CrossRef]
- Sengupta, S.; Peterson, T.R.; Laplante, M.; Oh, S.; Sabatini, D.M. MTORC1 Controls Fasting-Induced Ketogenesis and Its Modulation by Ageing. Nature 2010, 468, 1100–1104. [Google Scholar] [CrossRef]
- Ham, D.J.; Börsch, A.; Lin, S.; Thürkauf, M.; Weihrauch, M.; Reinhard, J.R.; Delezie, J.; Battilana, F.; Wang, X.; Kaiser, M.S.; et al. The Neuromuscular Junction Is a Focal Point of MTORC1 Signaling in Sarcopenia. Nat. Commun. 2020, 11, 4510. [Google Scholar] [CrossRef]
- Papadopoli, D.; Boulay, K.; Kazak, L.; Pollak, M.; Mallette, F.A.; Topisirovic, I.; Hulea, L. MTOR as a Central Regulator of Lifespan and Aging. F1000research 2019, 8, 998. [Google Scholar] [CrossRef]
- Johnson, S.C.; Rabinovitch, P.S.; Kaeberlein, M. mTOR Is a Key Modulator of Ageing and Age-Related Disease. Nature 2013, 493, 338–345. [Google Scholar] [CrossRef]
- Cohen, F.; Bergeron, P.; Blackwood, E.; Bowman, K.K.; Chen, H.; DiPasquale, A.G.; Epler, J.A.; Koehler, M.F.T.; Lau, K.; Lewis, C.; et al. Potent, Selective, and Orally Bioavailable Inhibitors of Mammalian Target of Rapamycin (mTOR) Kinase Based on a Quaternary Substituted Dihydrofuropyrimidine. J. Med. Chem. 2011, 54, 3426–3435. [Google Scholar] [CrossRef]
- Sun, J.; Jeliazkova, N.; Chupakhin, V.; Golib-Dzib, J.-F.; Engkvist, O.; Carlsson, L.; Wegner, J.; Ceulemans, H.; Georgiev, I.; Jeliazkov, V.; et al. ExCAPE-DB: An Integrated Large Scale Dataset Facilitating Big Data Analysis in Chemogenomics. J. Cheminform. 2017, 9, 17. [Google Scholar] [CrossRef]
- Pedregosa, F.; Varoquaux, G.; Gramfort, A.; Michel, V.; Thirion, B.; Grisel, O.; Blondel, M.; Müller, A.; Nothman, J.; Louppe, G.; et al. Scikit-Learn: Machine Learning in Python. arXiv 2012, arXiv:1201.0490. [Google Scholar] [CrossRef]
- Xiong, G.; Wu, Z.; Yi, J.; Fu, L.; Yang, Z.; Hsieh, C.; Yin, M.; Zeng, X.; Wu, C.; Lu, A.; et al. ADMETlab 2.0: An Integrated Online Platform for Accurate and Comprehensive Predictions of ADMET Properties. Nucleic Acids Res. 2021, 49, gkab255. [Google Scholar] [CrossRef]
- Yang, H.; Rudge, D.G.; Koos, J.D.; Vaidialingam, B.; Yang, H.J.; Pavletich, N.P. mTOR Kinase Structure, Mechanism and Regulation. Nature 2013, 497, 217–223. [Google Scholar] [CrossRef]
- Pettersen, E.F.; Goddard, T.D.; Huang, C.C.; Couch, G.S.; Greenblatt, D.M.; Meng, E.C.; Ferrin, T.E. UCSF Chimera—A Visualization System for Exploratory Research and Analysis. J. Comput. Chem. 2004, 25, 1605–1612. [Google Scholar] [CrossRef]
- Grosdidier, A.; Zoete, V.; Michielin, O. SwissDock, a Protein-Small Molecule Docking Web Service Based on EADock DSS. Nucleic Acids Res. 2011, 39, W270–W277. [Google Scholar] [CrossRef]
- Grosdidier, A.; Zoete, V.; Michielin, O. Fast Docking Using the CHARMM Force Field with EADock DSS. J. Comput. Chem. 2011, 32, 2149–2159. [Google Scholar] [CrossRef]
- Abraham, M.J.; Murtola, T.; Schulz, R.; Páll, S.; Smith, J.C.; Hess, B.; Lindahl, E. GROMACS: High Performance Molecular Simulations through Multi-Level Parallelism from Laptops to Supercomputers. Softwarex 2015, 1, 19–25. [Google Scholar] [CrossRef]
- Best, R.B.; Zhu, X.; Shim, J.; Lopes, P.E.M.; Mittal, J.; Feig, M.; MacKerell, A.D. Optimization of the Additive CHARMM All-Atom Protein Force Field Targeting Improved Sampling of the Backbone ϕ, ψ and Side-Chain Χ1 and Χ2 Dihedral Angles. J. Chem. Theory Comput. 2012, 8, 3257–3273. [Google Scholar] [CrossRef]
- Šali, A.; Blundell, T.L. Comparative Protein Modelling by Satisfaction of Spatial Restraints. J. Mol. Biol. 1993, 234, 779–815. [Google Scholar] [CrossRef]
- Zoete, V.; Cuendet, M.A.; Grosdidier, A.; Michielin, O. SwissParam: A Fast Force Field Generation Tool for Small Organic Molecules. J. Comput. Chem. 2011, 32, 2359–2368. [Google Scholar] [CrossRef]
- Teuscher, A.C.; Statzer, C.; Pantasis, S.; Bordoli, M.R.; Ewald, C.Y. Assessing Collagen Deposition During Aging in Mammalian Tissue and in Caenorhabditis elegans. Methods Mol. Biol. 2019, 1944, 169–188. [Google Scholar] [CrossRef]
- Statzer, C.; Jongsma, E.; Liu, S.X.; Dakhovnik, A.; Wandrey, F.; Mozharovskyi, P.; Zülli, F.; Ewald, C.Y. Youthful and Age-related Matreotypes Predict Drugs Promoting Longevity. Aging Cell 2021, 20, e13441. [Google Scholar] [CrossRef]
- Ewald, C.Y.; Marfil, V.; Li, C. Alzheimer-related Protein APL-1 Modulates Lifespan through Heterochronic Gene Regulation in Caenorhabditis elegans. Aging Cell 2016, 15, 1051–1062. [Google Scholar] [CrossRef]
- Statzer, C.; Reichert, P.; Dual, J.; Ewald, C.Y. Longevity Interventions Temporally Scale Healthspan in Caenorhabditis elegans. iScience 2022, 25, 103983. [Google Scholar] [CrossRef]
- Stroustrup, N.; Ulmschneider, B.E.; Nash, Z.M.; López-Moyado, I.F.; Apfeld, J.; Fontana, W. The Caenorhabditis elegans Lifespan Machine. Nat. Methods 2013, 10, 665–670. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Vidovic, T.; Dakhovnik, A.; Hrabovskyi, O.; MacArthur, M.R.; Ewald, C.Y. AI-Predicted mTOR Inhibitor Reduces Cancer Cell Proliferation and Extends the Lifespan of C. elegans. Int. J. Mol. Sci. 2023, 24, 7850. https://doi.org/10.3390/ijms24097850
Vidovic T, Dakhovnik A, Hrabovskyi O, MacArthur MR, Ewald CY. AI-Predicted mTOR Inhibitor Reduces Cancer Cell Proliferation and Extends the Lifespan of C. elegans. International Journal of Molecular Sciences. 2023; 24(9):7850. https://doi.org/10.3390/ijms24097850
Chicago/Turabian StyleVidovic, Tinka, Alexander Dakhovnik, Oleksii Hrabovskyi, Michael R. MacArthur, and Collin Y. Ewald. 2023. "AI-Predicted mTOR Inhibitor Reduces Cancer Cell Proliferation and Extends the Lifespan of C. elegans" International Journal of Molecular Sciences 24, no. 9: 7850. https://doi.org/10.3390/ijms24097850
APA StyleVidovic, T., Dakhovnik, A., Hrabovskyi, O., MacArthur, M. R., & Ewald, C. Y. (2023). AI-Predicted mTOR Inhibitor Reduces Cancer Cell Proliferation and Extends the Lifespan of C. elegans. International Journal of Molecular Sciences, 24(9), 7850. https://doi.org/10.3390/ijms24097850








