Exome-Based Genomic Markers Could Improve Prediction of Checkpoint Inhibitor Efficacy Independently of Tumor Type
Abstract
1. Introduction
2. Results
2.1. Patient Characteristics
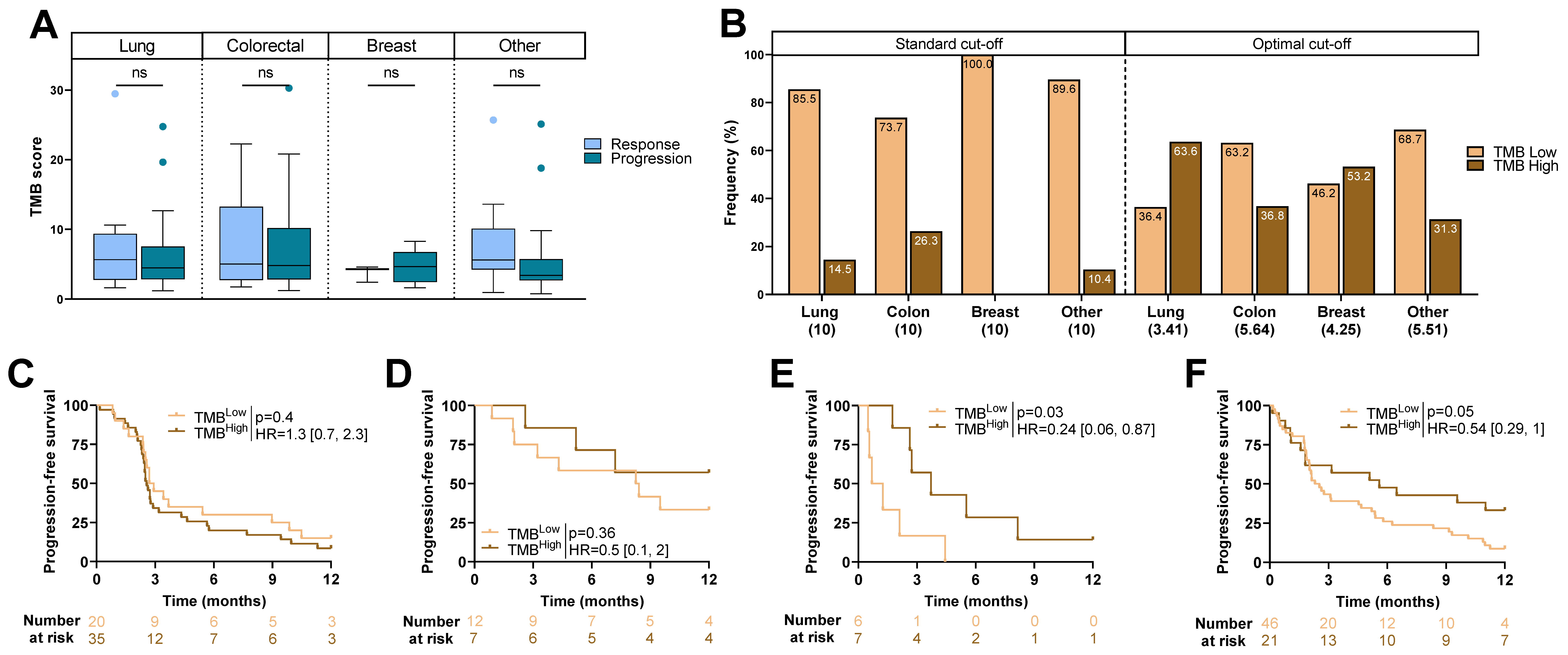
2.2. TMB Score and Type of Cancer
2.3. Association between Clinical Variables and Outcome
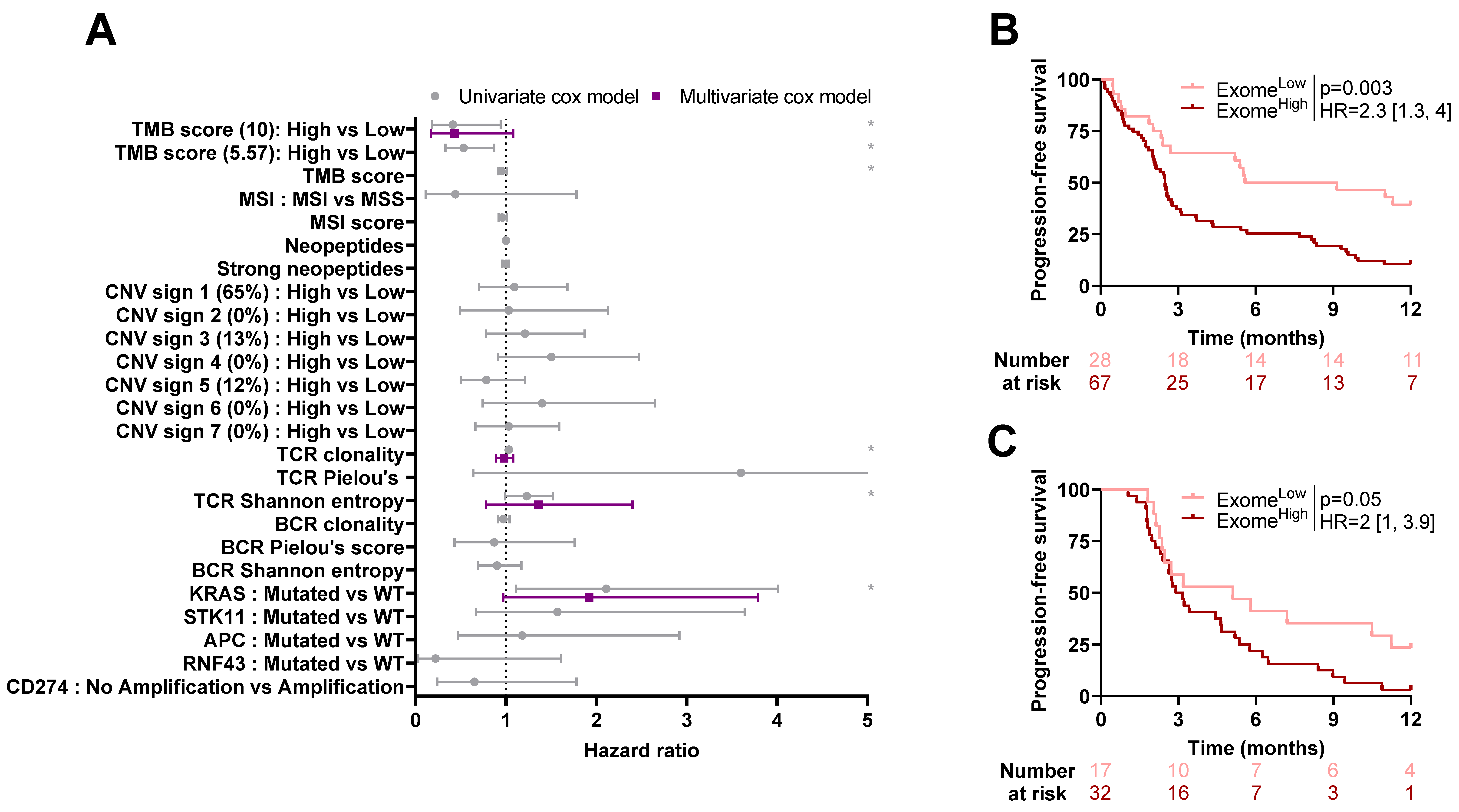
2.4. Association between Exome-Derived Variables and Outcome
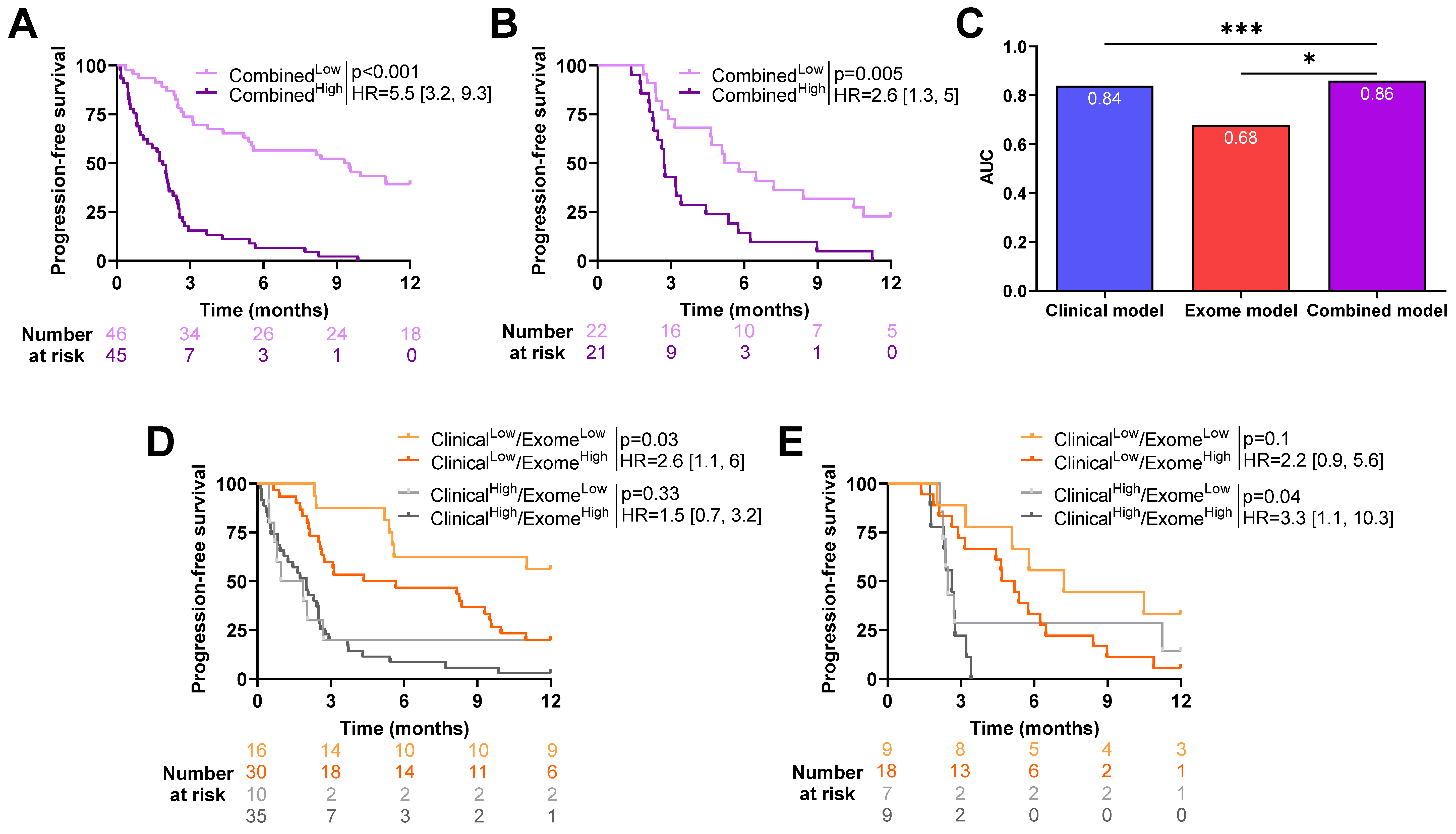
2.5. Exome-Derived Variables Add Predictive Power on Top of Clinical Variables
3. Discussion
4. Materials and Methods
4.1. Sample Selection
4.2. DNA Isolation
4.3. Whole-Exome Capture and Sequencing
4.4. Exome Analysis Pipeline
4.5. Analysis of Somatic Mutations
4.6. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| AUC | Area Under the Curve |
| BCR | B-cell receptor |
| CNV | Copy Number Variant |
| CT | Computed Tomography |
| CTLA-4 | Cytotoxic T Lymphocyte–Associated protein 4 |
| CR | Complete Response |
| FDA | Food and Drug Administration |
| FDR | False Discovery rate |
| HR | Hazard Ratio |
| ICI | Immune Checkpoint Inhibitor |
| IQR | Interquartile Range |
| LAG-3 | Lymphocyte-Activation Gene 3 |
| LRT | Likelihood Ratio Test |
| MSI | Microsatellite Instability |
| MSS | Microsatellite Stable |
| NS | Not Significant |
| NSCL | Non-Small-Cell Lung cancer |
| NR | Not Reached |
| PD | Progressive Disease |
| PD1 | Programmed cell Death protein-1 |
| PD-L1 | Programmed Death-Ligand 1 |
| PFS | Progression-Free Survival |
| PR | Partial Response |
| PS | Performance Status |
| SD | Stable Disease |
| SNV | Single-Nucleotide Variants |
| TMB | Tumor Mutational Burden |
| TCR | T-cell Receptor |
| WHO | World Health Organization |
| WT | Wild-Type |
References
- Waldman, A.D.; Fritz, J.M.; Lenardo, M.J. A guide to cancer immunotherapy: From T cell basic science to clinical practice. Nat. Rev. Immunol. 2020, 20, 651–668. [Google Scholar] [CrossRef] [PubMed]
- Zitvogel, L.; Tesniere, A.; Kroemer, G. Cancer despite immunosurveillance: Immunoselection and immunosubversion. Nat. Rev. Immunol. 2006, 6, 715–727. [Google Scholar] [CrossRef] [PubMed]
- Pardoll, D.M. The blockade of immune checkpoints in cancer immunotherapy. Nat. Rev. Cancer 2012, 12, 252–264. [Google Scholar] [CrossRef] [PubMed]
- Hodi, F.S.; O’Day, S.J.; McDermott, D.F.; Weber, R.W.; Sosman, J.A.; Haanen, J.B.; Gonzalez, R.; Robert, C.; Schadendorf, D.; Hassel, J.C.; et al. Improved Survival with Ipilimumab in Patients with Metastatic Melanoma. N. Engl. J. Med. 2010, 363, 711–723. [Google Scholar] [CrossRef] [PubMed]
- Brahmer, J.; Reckamp, K.L.; Baas, P.; Crinò, L.; Eberhardt, W.E.E.; Poddubskaya, E.; Antonia, S.; Pluzanski, A.; Vokes, E.E.; Holgado, E.; et al. Nivolumab versus Docetaxel in Advanced Squamous-Cell Non–Small-Cell Lung Cancer. N. Engl. J. Med. 2015, 373, 123–135. [Google Scholar] [CrossRef] [PubMed]
- Borghaei, H.; Paz-Ares, L.; Horn, L.; Spigel, D.R.; Steins, M.; Ready, N.E.; Chow, L.Q.; Vokes, E.E.; Felip, E.; Holgado, E.; et al. Nivolumab versus Docetaxel in Advanced Nonsquamous Non-Small-Cell Lung Cancer. N. Engl. J. Med. 2015, 373, 1627–1639. [Google Scholar] [CrossRef] [PubMed]
- Herbst, R.S.; Garon, E.B.; Kim, D.-W.; Cho, B.C.; Gervais, R.; Perez-Gracia, J.L.; Han, J.-Y.; Majem, M.; Forster, M.D.; Monnet, I.; et al. Five Year Survival Update From KEYNOTE-010: Pembrolizumab Versus Docetaxel for Previously Treated, Programmed Death-Ligand 1–Positive Advanced NSCLC. J. Thorac. Oncol. 2021, 16, 1718–1732. [Google Scholar] [CrossRef]
- Motzer, R.J.; Escudier, B.; McDermott, D.F.; George, S.; Hammers, H.J.; Srinivas, S.; Tykodi, S.S.; Sosman, J.A.; Procopio, G.; Plimack, E.R.; et al. Nivolumab versus Everolimus in Advanced Renal-Cell Carcinoma. N. Engl. J. Med. 2015, 373, 1803–1813. [Google Scholar] [CrossRef]
- Motzer, R.J.; Tannir, N.M.; McDermott, D.F.; Aren Frontera, O.; Melichar, B.; Choueiri, T.K.; Plimack, E.R.; Barthélémy, P.; Porta, C.; George, S.; et al. Nivolumab plus Ipilimumab versus Sunitinib in Advanced Renal-Cell Carcinoma. N. Engl. J. Med. 2018, 378, 1277–1290. [Google Scholar] [CrossRef]
- Rini, B.I.; Plimack, E.R.; Stus, V.; Gafanov, R.; Hawkins, R.; Nosov, D.; Pouliot, F.; Alekseev, B.; Soulières, D.; Melichar, B.; et al. Pembrolizumab plus Axitinib versus Sunitinib for Advanced Renal-Cell Carcinoma. N. Engl. J. Med. 2019, 380, 1116–1127. [Google Scholar] [CrossRef]
- Ferris, R.L.; Blumenschein, G.; Fayette, J.; Guigay, J.; Colevas, A.D.; Licitra, L.; Harrington, K.; Kasper, S.; Vokes, E.E.; Even, C.; et al. Nivolumab for Recurrent Squamous-Cell Carcinoma of the Head and Neck. N. Engl. J. Med. 2016, 375, 1856–1867. [Google Scholar] [CrossRef] [PubMed]
- Paz-Ares, L.; Dvorkin, M.; Chen, Y.; Reinmuth, N.; Hotta, K.; Trukhin, D.; Statsenko, G.; Hochmair, M.J.; Özgüroğlu, M.; Ji, J.H.; et al. Durvalumab plus platinum–etoposide versus platinum–etoposide in first-line treatment of extensive-stage small-cell lung cancer (CASPIAN): A randomised, controlled, open-label, phase 3 trial. Lancet 2019, 394, 1929–1939. [Google Scholar] [CrossRef] [PubMed]
- Powles, T.; Park, S.H.; Voog, E.; Caserta, C.; Valderrama, B.P.; Gurney, H.; Kalofonos, H.; Radulović, S.; Demey, W.; Ullén, A.; et al. Avelumab Maintenance Therapy for Advanced or Metastatic Urothelial Carcinoma. N. Engl. J. Med. 2020, 383, 1218–1230. [Google Scholar] [CrossRef]
- Makker, V.; Colombo, N.; Herráez, A.C.; Santin, A.D.; Colomba, E.; Miller, D.S.; Fujiwara, K.; Pignata, S.; Baron-Hay, S.; Ray-Coquard, I.; et al. Lenvatinib plus Pembrolizumab for Advanced Endometrial Cancer. N. Engl. J. Med. 2022, 386, 437–448. [Google Scholar] [CrossRef] [PubMed]
- Colombo, N.; Dubot, C.; Lorusso, D.; Caceres, M.V.; Hasegawa, K.; Shapira-Frommer, R.; Tewari, K.S.; Salman, P.; Usta, E.H.; Yañez, E.; et al. Pembrolizumab for Persistent, Recurrent, or Metastatic Cervical Cancer. N. Engl. J. Med. 2021, 385, 1856–1867. [Google Scholar] [CrossRef] [PubMed]
- Baas, P.; Scherpereel, A.; Nowak, A.K.; Fujimoto, N.; Peters, S.; Tsao, A.S.; Mansfield, A.S.; Popat, S.; Jahan, T.; Antonia, S.; et al. First-line nivolumab plus ipilimumab in unresectable malignant pleural mesothelioma (CheckMate 743): A multicentre, randomised, open-label, phase 3 trial. Lancet 2021, 397, 375–386. [Google Scholar] [CrossRef]
- Finn, R.S.; Qin, S.; Ikeda, M.; Galle, P.R.; Ducreux, M.; Kim, T.-Y.; Kudo, M.; Breder, V.; Merle, P.; Kaseb, A.O.; et al. Atezolizumab plus Bevacizumab in Unresectable Hepatocellular Carcinoma. N. Engl. J. Med. 2020, 382, 1894–1905. [Google Scholar] [CrossRef]
- Sun, J.-M.; Shen, L.; Shah, M.A.; Enzinger, P.; Adenis, A.; Doi, T.; Kojima, T.; Metges, J.-P.; Li, Z.; Kim, S.-B.; et al. Pembrolizumab plus chemotherapy versus chemotherapy alone for first-line treatment of advanced oesophageal cancer (KEYNOTE-590): A randomised, placebo-controlled, phase 3 study. Lancet 2021, 398, 759–771. [Google Scholar] [CrossRef]
- Janjigian, Y.Y.; Shitara, K.; Moehler, M.; Garrido, M.; Salman, P.; Shen, L.; Wyrwicz, L.; Yamaguchi, K.; Skoczylas, T.; Bragagnoli, A.C.; et al. First-line nivolumab plus chemotherapy versus chemotherapy alone for advanced gastric, gastro-oesophageal junction, and oesophageal adenocarcinoma (CheckMate 649): A randomised, open-label, phase 3 trial. Lancet 2021, 398, 27–40. [Google Scholar] [CrossRef]
- Marabelle, A.; Le, D.T.; Ascierto, P.A.; Di Giacomo, A.M.; De Jesus-Acosta, A.; Delord, J.-P.; Geva, R.; Gottfried, M.; Penel, N.; Hansen, A.R.; et al. Efficacy of Pembrolizumab in Patients with Noncolorectal High Microsatellite Instability/Mismatch Repair–Deficient Cancer: Results From the Phase II KEYNOTE-158 Study. J. Clin. Oncol. 2020, 38, 1–10. [Google Scholar] [CrossRef]
- Borcoman, E.; Kanjanapan, Y.; Champiat, S.; Kato, S.; Servois, V.; Kurzrock, R.; Goel, S.; Bedard, P.; Le Tourneau, C. Novel patterns of response under immunotherapy. Ann. Oncol. 2019, 30, 385–396. [Google Scholar] [CrossRef]
- Khunger, M.; Hernandez, A.V.; Pasupuleti, V.; Rakshit, S.; Pennell, N.A.; Stevenson, J.; Mukhopadhyay, S.; Schalper, K.; Velcheti, V. Programmed Cell Death 1 (PD-1) Ligand (PD-L1) Expression in Solid Tumors as a Predictive Biomarker of Benefit from PD-1/PD-L1 Axis Inhibitors: A Systematic Review and Meta-Analysis. JCO Precis. Oncol. 2017, 1, 1–15. [Google Scholar] [CrossRef] [PubMed]
- Marabelle, A.; Fakih, M.; Lopez, J.; Shah, M.; Shapira-Frommer, R.; Nakagawa, K.; Chung, H.C.; Kindler, H.L.; Lopez-Martin, J.A.; Miller, W.H., Jr.; et al. Association of tumour mutational burden with outcomes in patients with advanced solid tumours treated with pembrolizumab: Prospective biomarker analysis of the multicohort, open-label, phase 2 KEYNOTE-158 study. Lancet Oncol. 2020, 21, 1353–1365. [Google Scholar] [CrossRef] [PubMed]
- Rousseau, B.; Foote, M.B.; Maron, S.B.; Diplas, B.H.; Lu, S.; Argilés, G.; Cercek, A.; Diaz, L.A. The Spectrum of Benefit from Checkpoint Blockade in Hypermutated Tumors. N. Engl. J. Med. 2021, 384, 1168–1170. [Google Scholar] [CrossRef] [PubMed]
- Wang, S.; Xie, K.; Liu, T. Cancer Immunotherapies: From Efficacy to Resistance Mechanisms—Not Only Checkpoint Matters. Front. Immunol. 2021, 12, 690112. [Google Scholar] [CrossRef] [PubMed]
- Réda, M.; Richard, C.; Bertaut, A.; Niogret, J.; Collot, T.; Fumet, J.D.; Blanc, J.; Truntzer, C.; Desmoulins, I.; Ladoire, S.; et al. Implementation and use of whole exome sequencing for metastatic solid cancer. Ebiomedicine 2020, 51, 102624. [Google Scholar] [CrossRef] [PubMed]
- Litchfield, K.; Reading, J.L.; Puttick, C.; Thakkar, K.; Abbosh, C.; Bentham, R.; Watkins, T.B.K.; Rosenthal, R.; Biswas, D.; Rowan, A.; et al. Meta-analysis of tumor- and T cell-intrinsic mechanisms of sensitization to checkpoint inhibition. Cell 2021, 184, 596–614.e14. [Google Scholar] [CrossRef]
- Johansen, A.F.B.; Kassentoft, C.G.; Knudsen, M.; Laursen, M.B.; Madsen, A.H.; Iversen, L.H.; Sunesen, K.G.; Rasmussen, M.H.; Andersen, C.L. Validation of computational determination of microsatellite status using whole exome sequencing data from colorectal cancer patients. BMC Cancer 2019, 19, 971. [Google Scholar] [CrossRef]
- Jeanson, A.; Tomasini, P.; Souquet-Bressand, M.; Brandone, N.; Boucekine, M.; Grangeon, M.; Chaleat, S.; Khobta, N.; Milia, J.; Mhanna, L.; et al. Efficacy of Immune Checkpoint Inhibitors in KRAS-Mutant Non-Small Cell Lung Cancer (NSCLC). J. Thorac. Oncol. 2019, 14, 1095–1101. [Google Scholar] [CrossRef]
- Zhu, G.; Pei, L.; Xia, H.; Tang, Q.; Bi, F. Role of oncogenic KRAS in the prognosis, diagnosis and treatment of colorectal cancer. Mol. Cancer 2021, 20, 143. [Google Scholar] [CrossRef]
- Goulding, R.E.; Chenoweth, M.; Carter, G.C.; Boye, M.E.; Sheffield, K.M.; John, W.J.; Leusch, M.S.; Muehlenbein, C.E.; Li, L.; Jen, M.-H.; et al. KRAS mutation as a prognostic factor and predictive factor in advanced/metastatic non-small cell lung cancer: A systematic literature review and meta-analysis. Cancer Treat. Res. Commun. 2020, 24, 100200. [Google Scholar] [CrossRef] [PubMed]
- Valpione, S.; Mundra, P.A.; Galvani, E.; Campana, L.G.; Lorigan, P.; De Rosa, F.; Gupta, A.; Weightman, J.; Mills, S.; Dhomen, N.; et al. The T cell receptor repertoire of tumor infiltrating T cells is predictive and prognostic for cancer survival. Nat. Commun. 2021, 12, 4098. [Google Scholar] [CrossRef] [PubMed]
- Richard, C.; Fumet, J.-D.; Chevrier, S.; Derangère, V.; Ledys, F.; Lagrange, A.; Favier, L.; Coudert, B.; Arnould, L.; Truntzer, C.; et al. Exome Analysis Reveals Genomic Markers Associated with Better Efficacy of Nivolumab in Lung Cancer Patients. Clin. Cancer Res. 2019, 25, 957–966. [Google Scholar] [CrossRef] [PubMed]
- Hundal, J.; Kiwala, S.; McMichael, J.; Miller, C.A.; Xia, H.; Wollam, A.T.; Liu, C.J.; Zhao, S.; Feng, Y.-Y.; Graubert, A.P.; et al. pVACtools: A Computational Toolkit to Identify and Visualize Cancer Neoantigens. Cancer Immunol. Res. 2020, 8, 409–420. [Google Scholar] [CrossRef] [PubMed]
- Warren, R.L.; Choe, G.; Freeman, D.J.; Castellarin, M.; Munro, S.; Moore, R.; Holt, R.A. Derivation of HLA types from shotgun sequence datasets. Genome Med. 2012, 4, 95. [Google Scholar] [CrossRef] [PubMed]
- Middha, S.; Zhang, L.; Nafa, K.; Jayakumaran, G.; Wong, D.; Kim, H.R.; Sadowska, J.; Berger, M.F.; Delair, D.F.; Shia, J.; et al. Reliable Pan-Cancer Microsatellite Instability Assessment by Using Targeted Next-Generation Sequencing Data. JCO Precis. Oncol. 2017, 2017, 1–17. [Google Scholar] [CrossRef]
- Flensburg, C.; Sargeant, T.; Oshlack, A.; Majewski, I.J. SuperFreq: Integrated mutation detection and clonal tracking in cancer. PLoS Comput. Biol. 2020, 16, e1007603. [Google Scholar] [CrossRef]
- MacIntyre, G.; Goranova, T.E.; De Silva, D.; Ennis, D.; Piskorz, A.M.; Eldridge, M.; Sie, D.; Lewsley, L.-A.; Hanif, A.; Wilson, C.; et al. Copy-number signatures and mutational processes in ovarian carcinoma. Nat. Genet. 2018, 50, 1262–1270. [Google Scholar] [CrossRef]
- Bolotin, D.; Poslavsky, S.; Mitrophanov, I.; Shugay, M.; Mamedov, I.Z.; Putintseva, E.; Chudakov, D.M. MiXCR: Software for comprehensive adaptive immunity profiling. Nat. Methods 2015, 12, 380–381. [Google Scholar] [CrossRef]
- Schneider-Hohendorf, T.; Mohan, H.; Bien, C.G.; Breuer, J.; Becker, A.; Görlich, D.; Kuhlmann, T.; Widman, G.; Herich, S.; Elpers, C.; et al. CD8+ T-cell pathogenicity in Rasmussen encephalitis elucidated by large-scale T-cell receptor sequencing. Nat. Commun. 2016, 7, 11153. [Google Scholar] [CrossRef]
- Chiffelle, J.; Genolet, R.; Perez, M.A.; Coukos, G.; Zoete, V.; Harari, A. T-cell repertoire analysis and metrics of diversity and clonality. Curr. Opin. Biotechnol. 2020, 65, 284–295. [Google Scholar] [CrossRef] [PubMed]
- Ricciuti, B.; Arbour, K.C.; Lin, J.J.; Vajdi, A.; Vokes, N.; Hong, L.; Zhang, J.; Tolstorukov, M.Y.; Li, Y.Y.; Spurr, L.F.; et al. Diminished Efficacy of Programmed Death-(Ligand)1 Inhibition in STK11- and KEAP1-Mutant Lung Adenocarcinoma Is Affected by KRAS Mutation Status. J. Thorac. Oncol. 2022, 17, 399–410. [Google Scholar] [CrossRef] [PubMed]
- Bachelot, T.; Filleron, T.; Bieche, I.; Arnedos, M.; Campone, M.; Dalenc, F.; Coussy, F.; Sablin, M.-P.; Debled, M.; Lefeuvre-Plesse, C.; et al. Durvalumab compared to maintenance chemotherapy in metastatic breast cancer: The randomized phase II SAFIR02-BREAST IMMUNO trial. Nat. Med. 2021, 27, 250–255. [Google Scholar] [CrossRef] [PubMed]
- Lin, A.; Zhang, H.; Hu, X.; Chen, X.; Wu, G.; Luo, P.; Zhang, J. Age, sex, and specific gene mutations affect the effects of immune checkpoint inhibitors in colorectal cancer. Pharmacol. Res. 2020, 159, 105028. [Google Scholar] [CrossRef] [PubMed]
- Liu, D.; Jenkins, R.W.; Sullivan, R.J. Mechanisms of Resistance to Immune Checkpoint Blockade. Am. J. Clin. Dermatol. 2019, 20, 41–54. [Google Scholar] [CrossRef]
- Mateo, J.; Chakravarty, D.; Dienstmann, R.; Jezdic, S.; Gonzalez-Perez, A.; Lopez-Bigas, N.; Ng, C.; Bedard, P.; Tortora, G.; Douillard, J.-Y.; et al. A framework to rank genomic alterations as targets for cancer precision medicine: The ESMO Scale for Clinical Actionability of molecular Targets (ESCAT). Ann. Oncol. 2018, 29, 1895–1902. [Google Scholar] [CrossRef]
- Benjamini, Y.; Hochberg, Y. Controlling the False Discovery Rate: A Practical and Powerful Approach to Multiple Testing. J. R. Stat. Soc. Ser. B Methodol. 1995, 57, 289–300. [Google Scholar] [CrossRef]
- Hothorn, T.; Lausen, B. On the exact distribution of maximally selected rank statistics. Comput. Stat. Data Anal. 2003, 43, 121–137. [Google Scholar] [CrossRef]


| Variables | Whole Cohort N = 154 | Training Cohort N = 101 | Validation Cohort N = 53 | p-Value | Adjusted p-Value |
|---|---|---|---|---|---|
| Sex | 0.9 | >0.9 | |||
| Male | 80 (52%) | 53 (52%) | 27 (51%) | ||
| Female | 74 (48%) | 48 (48%) | 26 (49%) | ||
| Age, years | 62 (54, 68) | 62 (55, 68) | 61 (54, 67) | 0.6 | 0.9 |
| ≤60 | 66 (43%) | 42 (42%) | 25 (47%) | 0.5 | 0.8 |
| >60 | 87 (57%) | 59 (58%) | 28 (53%) | ||
| NA | 1 | 1 | 0 | ||
| Stage at diagnosis | 0.044 | 0.4 | |||
| Local | 70 (45%) | 40 (40%) | 30 (57%) | ||
| Metastatic | 84 (55%) | 61 (60%) | 23 (43%) | ||
| PDL1 status | >0.9 | >0.9 | |||
| Positive | 18 (30%) | 13 (30%) | 5 (29%) | ||
| Negative | 43 (70%) | 31 (70%) | 12 (71%) | ||
| NA | 93 | 57 | 36 | ||
| Surgery | 0.2 | 0.7 | |||
| No | 87 (56%) | 61 (60%) | 26 (49%) | ||
| Yes | 67 (44%) | 40 (40%) | 27 (51%) | ||
| Type of surgery | 0.2 | 0.7 | |||
| Curative | 60 (90%) | 34 (85%) | 26 (96%) | ||
| Palliative | 7 (10%) | 6 (15%) | 1 (3.7%) | ||
| Histology | >0.9 | >0.9 | |||
| Adenocarcinoma | 81 (53%) | 54 (53%) | 27 (51%) | ||
| Carcinoma | 55 (36%) | 35 (35%) | 20 (38%) | ||
| Other | 18 (12%) | 12 (12%) | 6 (11%) | ||
| Line of ICI | 0.5 | 0.8 | |||
| ≤2 | 101 (66%) | 68 (67%) | 33 (62%) | ||
| >2 | 53 (34%) | 33 (33%) | 20 (38%) | ||
| Number of cycles | 0.4 | 0.8 | |||
| ≤2 | 59 (38%) | 41 (41%) | 18 (34%) | ||
| >2 | 95 (62%) | 60 (59%) | 35 (66%) | ||
| Type of ICI | 0.5 | 0.8 | |||
| PD-1 | 93 (60%) | 61 (60%) | 32 (60%) | ||
| PD-L1 | 26 (17%) | 20 (20%) | 6 (11%) | ||
| PD-1/CTLA-4 | 4 (2.6%) | 3 (3.0%) | 1 (1.9%) | ||
| PD-L1/CTLA-4 | 27 (18%) | 15 (15%) | 12 (23%) | ||
| Other | 4 (2.6%) | 2 (2.0%) | 2 (3.8%) | ||
| WHO status at ICI | >0.9 | >0.9 | |||
| 0 | 55 (36%) | 36 (36%) | 19 (36%) | ||
| >0 | 98 (64%) | 64 (64%) | 34 (64%) | ||
| NA | 1 | 1 | 0 | ||
| Smoking status | 0.9 | >0.9 | |||
| Never smoker | 35 (29%) | 22 (28%) | 13 (30%) | ||
| Smoker | 87 (71%) | 56 (72%) | 31 (70%) | ||
| NA | 32 | 23 | 9 | ||
| Cerebral metastasis | 25 (16%) | 17 (17%) | 8 (15%) | 0.8 | >0.9 |
| NA | 1 | 0 | 1 | ||
| Liver metastasis | 47 (31%) | 35 (35%) | 12 (23%) | 0.14 | 0.7 |
| NA | 1 | 0 | 1 | ||
| Bone metastasis | 51 (33%) | 35 (35%) | 16 (31%) | 0.6 | 0.9 |
| NA | 1 | 0 | 1 | ||
| Lymph node metastasis | 106 (70%) | 65 (64%) | 41 (80%) | 0.042 | 0.4 |
| NA | 2 | 0 | 2 | ||
| Lung metastasis | 60 (42%) | 38 (39%) | 22 (47%) | 0.4 | 0.8 |
| NA | 10 | 4 | 6 | ||
| Pleuro-peritoneal metastasis | 58 (38%) | 42 (42%) | 16 (31%) | 0.2 | 0.7 |
| NA | 1 | 0 | 1 | ||
| Toxicity | 66 (43%) | 41 (41%) | 25 (47%) | 0.4 | 0.8 |
| Use of corticosteroids | 51 (33%) | 39 (39%) | 12 (23%) | 0.045 | 0.4 |
| RECIST | 0.3 | 0.8 | |||
| Progression | 115 (78%) | 71 (76%) | 44 (83%) | ||
| Response | 32 (22%) | 23 (24%) | 9 (17%) | ||
| NA | 7 | 7 | 0 | ||
| Cancer type | 0.2 | 0.7 | |||
| Breast | 13 (8%) | 12 (12%) | 1 (1.9%) | ||
| Colon | 19 (12%) | 12 (12%) | 7 (13%) | ||
| Lung | 55 (36%) | 36 (36%) | 19 (36%) | ||
| Other | 67 (44%) | 41 (41%) | 26 (49%) |
| Variables | Whole Cohort N = 154 | Training Cohort N = 101 | Validation Cohort N = 53 | p-Value | Adjusted p-Value |
|---|---|---|---|---|---|
| TMB status | 0.3 | >0.9 | |||
| Low | 134 (87%) | 90 (89%) | 44 (83%) | ||
| High | 20 (13%) | 11 (11%) | 9 (17%) | ||
| TMB score | 4.5 (2.8, 7.1) | 4.5 (3.1, 6.1) | 4.3 (2.6, 8.0) | >0.9 | >0.9 |
| MSI status | 0.4 | >0.9 | |||
| MSS | 146 (95%) | 97 (96%) | 49 (92%) | ||
| MSI | 8 (5.2%) | 4 (4.0%) | 4 (7.5%) | ||
| MSI score | 1.3 (1.0, 1.9) | 1.4 (1.0, 1.9) | 1.2 (0.9, 1.7) | 0.4 | >0.9 |
| TCR clonality | 7 (4, 12) | 7 (4, 12) | 7 (4, 10) | 0.5 | >0.9 |
| BCR clonality | 0.00 (0.00, 2.00) | 0.00 (0.00, 2.00) | 0.00 (0.00, 2.00) | >0.9 | >0.9 |
| NA | 2 | 2 | 0 | ||
| Neopeptides | 12 (7, 28) | 13 (6, 36) | 11 (7, 23) | 0.6 | >0.9 |
| NA | 3 | 2 | 1 | ||
| Strong neopeptides | 2.0 (1.0, 5.0) | 2.0 (1.0, 5.0) | 2.0 (1.0, 3.2) | 0.4 | >0.9 |
| NA | 3 | 2 | 1 | ||
| CNV signature 1 | 68 (53, 79) | 65 (51, 76) | 72 (61, 81) | 0.2 | >0.9 |
| Cut-off = 65.3%, 71.7% Low | 50 (50%) | 26 (50%) | >0.9 | >0.9 | |
| High | 50 (50%) | 26 (50%) | |||
| NA | 2 | 1 | 1 | ||
| CNV signature 2 | 0.00 (0.00, 0.00) | 0.00 (0.00, 0.00) | 0.00 (0.00, 0.00) | 0.4 | >0.9 |
| Cut-off = 0%, 0%, Low | 90 (90%) | 49 (94%) | 0.5 | >0.9 | |
| High | 10 (10%) | 3 (5.8%) | |||
| NA | 2 | 1 | 1 | ||
| CNV signature 3 | 13 (10, 19) | 13 (10, 19) | 13 (10, 20) | >0.9 | >0.9 |
| Cut-off = 13.2%, 13%, Low | 50 (50%) | 26 (50%) | >0.9 | >0.9 | |
| High | 50 (50%) | 26 (50%) | |||
| NA | 2 | 1 | 1 | ||
| CNV signature 4 | 0.00 (0.00, 0.00) | 0.00 (0.00, 0.00) | 0.00 (0.00, 0.00) | 0.7 | >0.9 |
| Cut-off = 0%, 0%, Low | 77 (77%) | 42 (81%) | 0.6 | >0.9 | |
| High | 23 (23%) | 10 (19%) | |||
| NA | 2 | 1 | 1 | ||
| CNV signature 5 | 11 (5, 18) | 0.12 (0.06, 0.19) | 10 (4, 16) | 0.12 | >0.9 |
| Cut-off = 11.8%, 9.8%, Low | 50 (50%) | 26 (50%) | >0.9 | >0.9 | |
| High | 50 (50%) | 26 (50%) | |||
| NA | 2 | 1 | 1 | ||
| CNV signature 6 | 0.00 (0.00, 0.00) | 0.00 (0.00, 0.00) | 0.00 (0.00, 0.00) | >0.9 | >0.9 |
| Cut-off = 0%, 0%, Low | 88 (88%) | 46 (88%) | >0.9 | >0.9 | |
| High | 12 (12%) | 6 (12%) | |||
| NA | 2 | 1 | 1 | ||
| CNV signature 7 | 0.00 (0.00, 0.08) | 0.00 (0.00, 11) | 0.00 (0.00, 4) | 0.2 | >0.9 |
| Cut-off = 0%, 0%, Low | 55 (56%) | 34 (65%) | 0.2 | >0.9 | |
| High | 44 (44%) | 18 (35%) | |||
| NA | 3 | 2 | 1 | ||
| TCR Pielou’s score | 0.98 (0.96, 1.00) | 0.99 (0.96, 1.00) | 0.97 (0.95, 1.00) | 0.3 | >0.9 |
| NA | 10 | 6 | 4 | ||
| BCR Pielou’s score | 1.00 (0.72, 1.00) | 0.99 (0.00, 1.00) | 1.00 (0.96, 1.00) | 0.078 | >0.9 |
| NA | 84 | 55 | 29 | ||
| TCR Shannon entropy | 2.81 (2.00, 3.54) | 2.85 (2.00, 3.55) | 2.75 (2.00, 3.32) | 0.6 | >0.9 |
| NA | 10 | 6 | 4 | ||
| BCR Shannon entropy | 1.44 (0.72, 2.00) | 1.26 (0.00, 2.30) | 1.48 (1.00, 2.00) | 0.7 | >0.9 |
| NA | 84 | 55 | 29 | ||
| KRAS | 0.7 | >0.9 | |||
| WT | 136 (88%) | 90 (89%) | 46 (87%) | ||
| Mutated | 18 (12%) | 11 (11%) | 7 (13%) | ||
| STK11 | 0.7 | >0.9 | |||
| WT | 146 (95%) | 95 (94%) | 51 (96%) | ||
| Mutated | 8 (5.2%) | 6 (5.9%) | 2 (3.8%) | ||
| APC | >0.9 | >0.9 | |||
| WT | 147 (95%) | 96 (95%) | 51 (96%) | ||
| Mutated | 7 (4.5%) | 5 (5.0%) | 2 (3.8%) | ||
| RNF43 | 0.4 | >0.9 | |||
| WT | 148 (96%) | 98 (97%) | 50 (94%) | ||
| Mutated | 6 (3.9%) | 3 (3.0%) | 3 (5.7%) | ||
| CD274 | 0.5 | >0.9 | |||
| Amplification | 9 (5.8%) | 5 (5.0%) | 4 (7.5%) | ||
| No amplification | 145 (94%) | 96 (95%) | 49 (92%) |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Dalens, L.; Lecuelle, J.; Favier, L.; Fraisse, C.; Lagrange, A.; Kaderbhai, C.; Boidot, R.; Chevrier, S.; Mananet, H.; Derangère, V.; et al. Exome-Based Genomic Markers Could Improve Prediction of Checkpoint Inhibitor Efficacy Independently of Tumor Type. Int. J. Mol. Sci. 2023, 24, 7592. https://doi.org/10.3390/ijms24087592
Dalens L, Lecuelle J, Favier L, Fraisse C, Lagrange A, Kaderbhai C, Boidot R, Chevrier S, Mananet H, Derangère V, et al. Exome-Based Genomic Markers Could Improve Prediction of Checkpoint Inhibitor Efficacy Independently of Tumor Type. International Journal of Molecular Sciences. 2023; 24(8):7592. https://doi.org/10.3390/ijms24087592
Chicago/Turabian StyleDalens, Lorraine, Julie Lecuelle, Laure Favier, Cléa Fraisse, Aurélie Lagrange, Courèche Kaderbhai, Romain Boidot, Sandy Chevrier, Hugo Mananet, Valentin Derangère, and et al. 2023. "Exome-Based Genomic Markers Could Improve Prediction of Checkpoint Inhibitor Efficacy Independently of Tumor Type" International Journal of Molecular Sciences 24, no. 8: 7592. https://doi.org/10.3390/ijms24087592
APA StyleDalens, L., Lecuelle, J., Favier, L., Fraisse, C., Lagrange, A., Kaderbhai, C., Boidot, R., Chevrier, S., Mananet, H., Derangère, V., Truntzer, C., & Ghiringhelli, F. (2023). Exome-Based Genomic Markers Could Improve Prediction of Checkpoint Inhibitor Efficacy Independently of Tumor Type. International Journal of Molecular Sciences, 24(8), 7592. https://doi.org/10.3390/ijms24087592








