KNOX Genes Were Involved in Regulating Axillary Bud Formation of Chrysanthemum × morifolium
Abstract
1. Introduction
2. Results
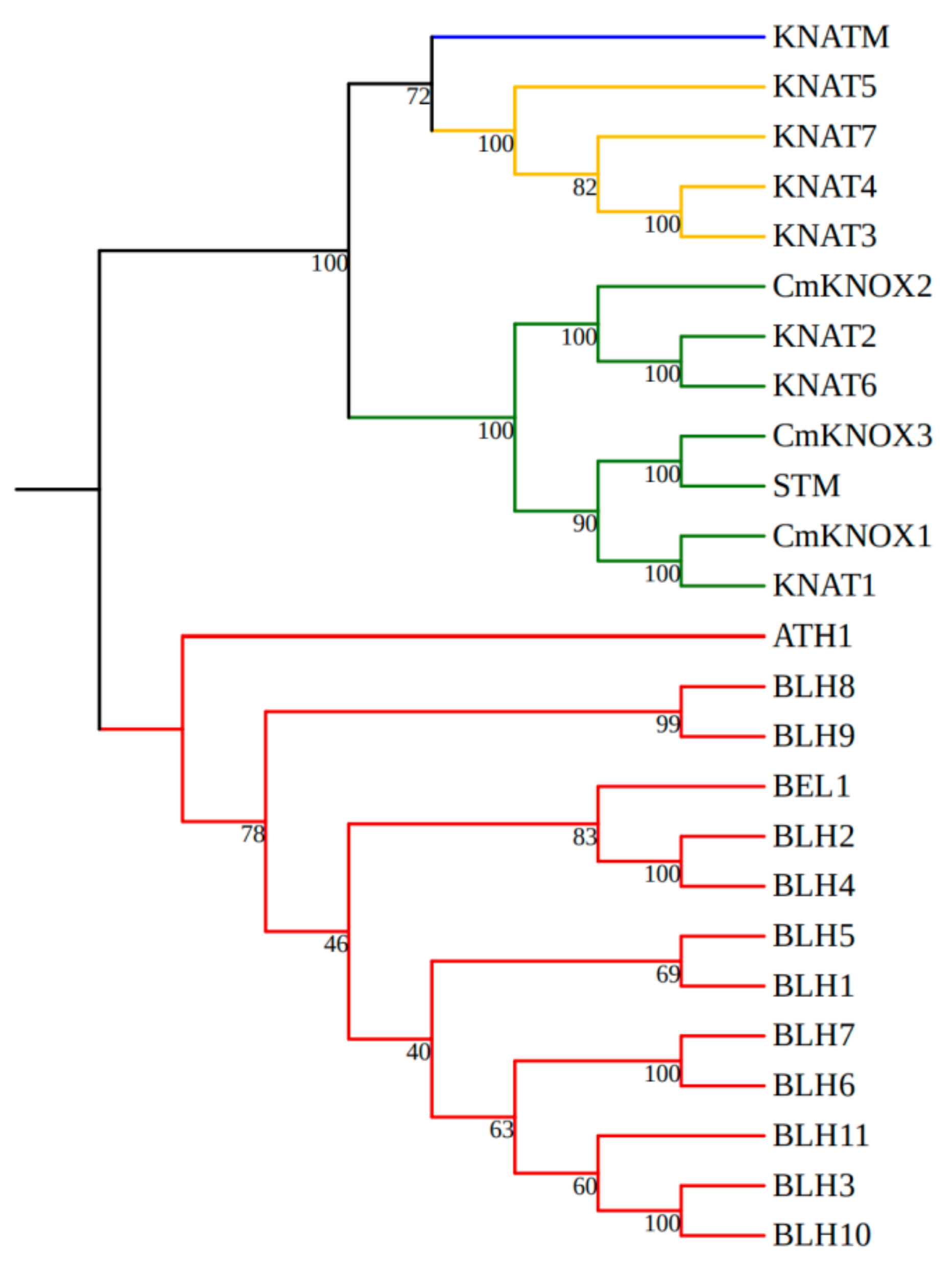
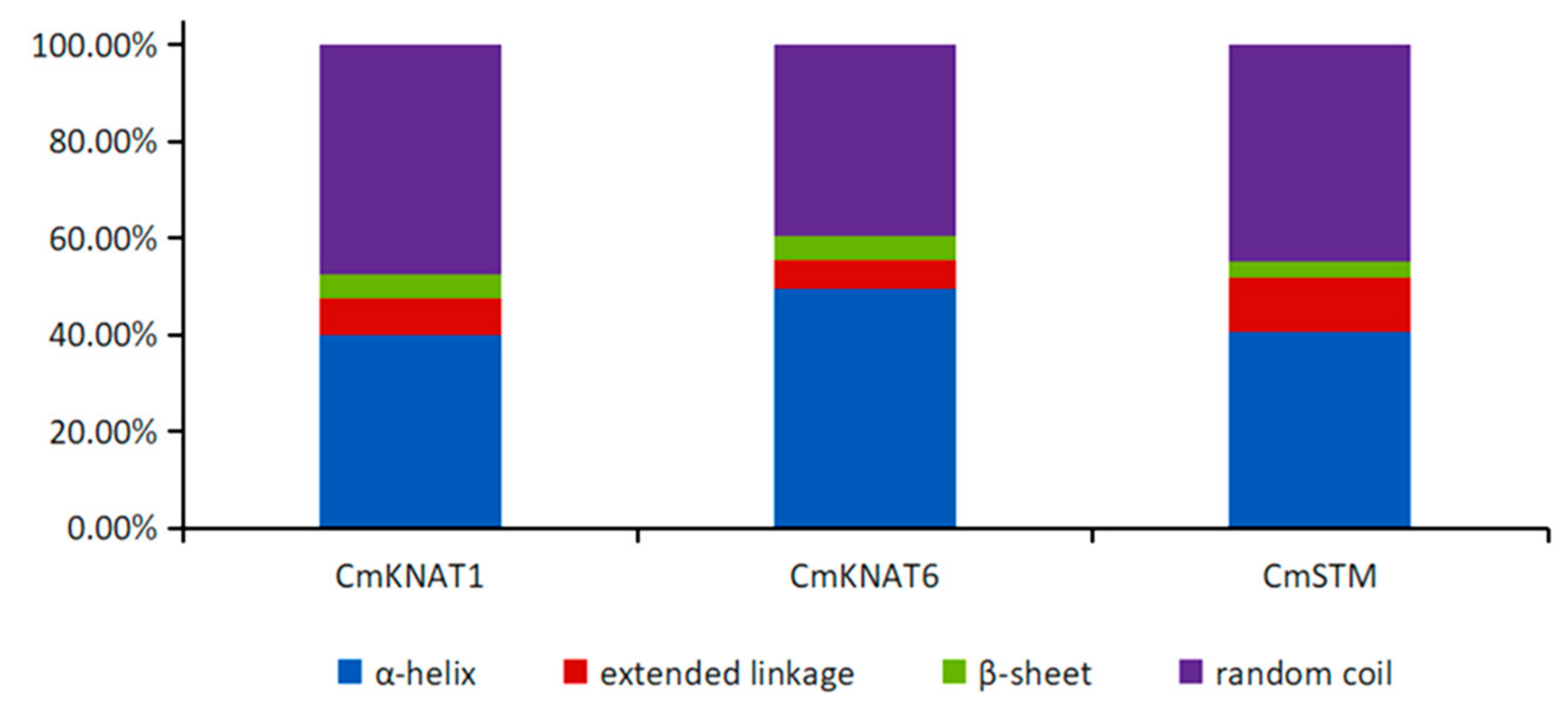
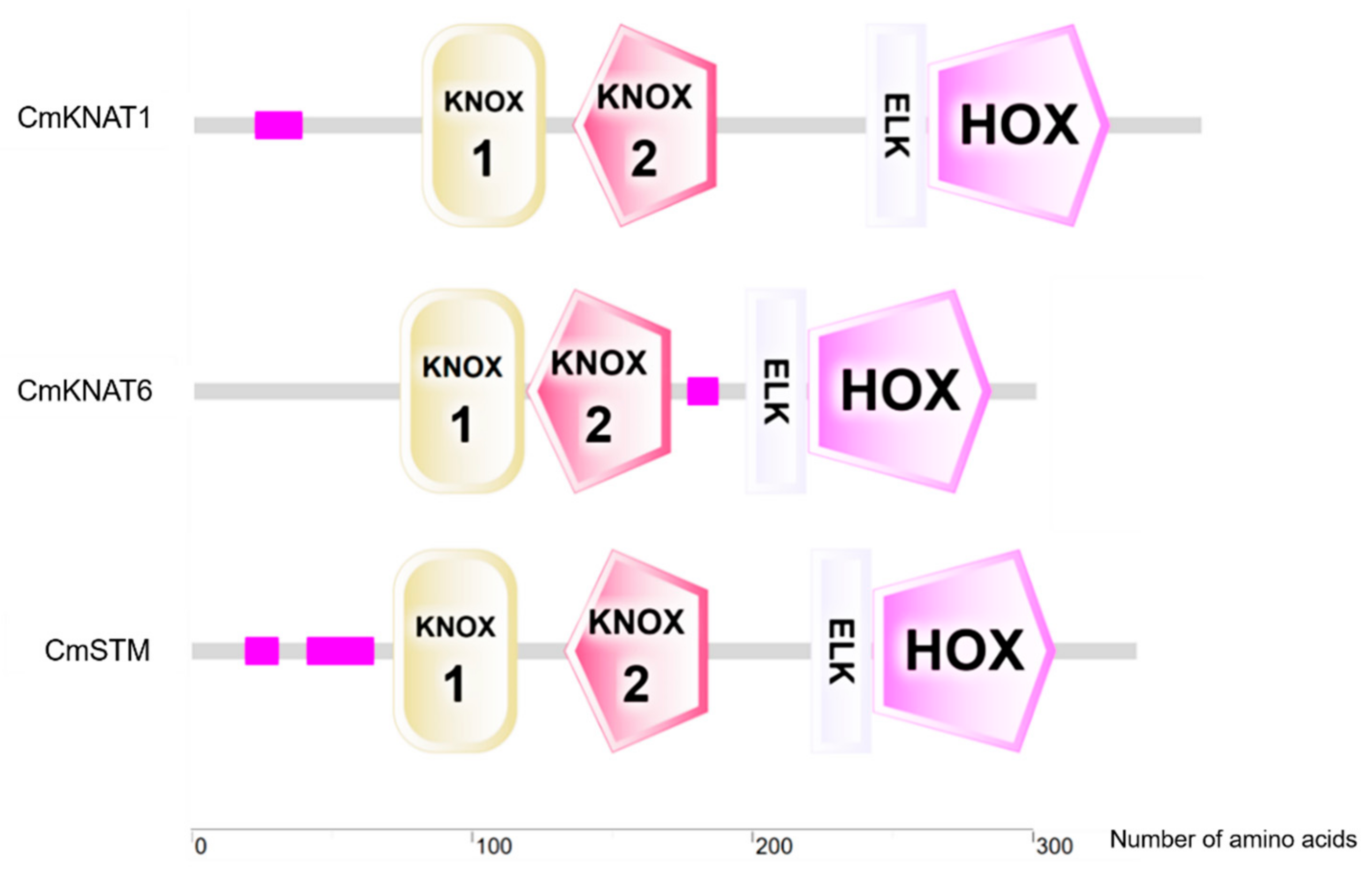
2.1. Cloning and Bioinformatics Analysis of Three KNOX Genes in Chrysanthemum
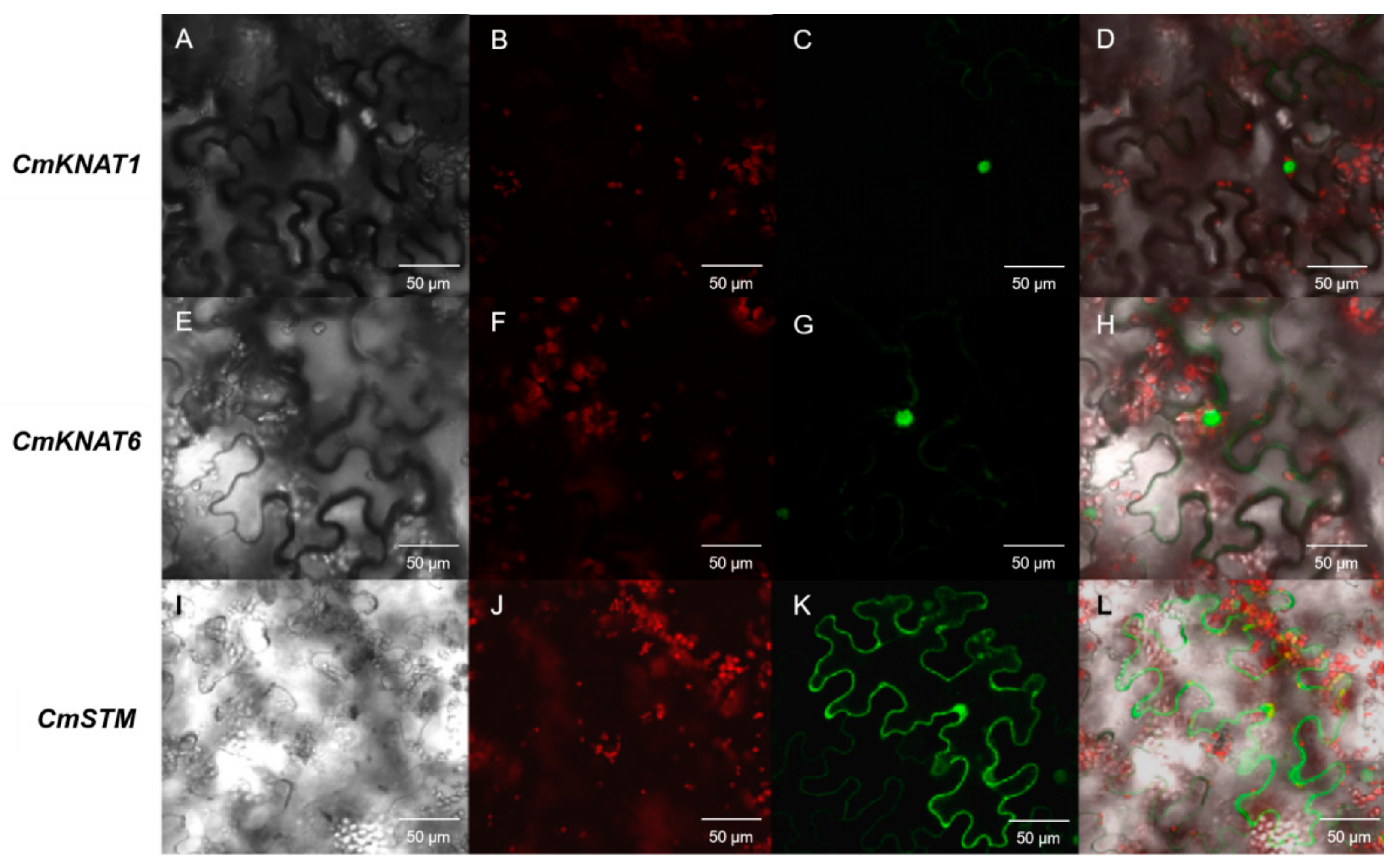
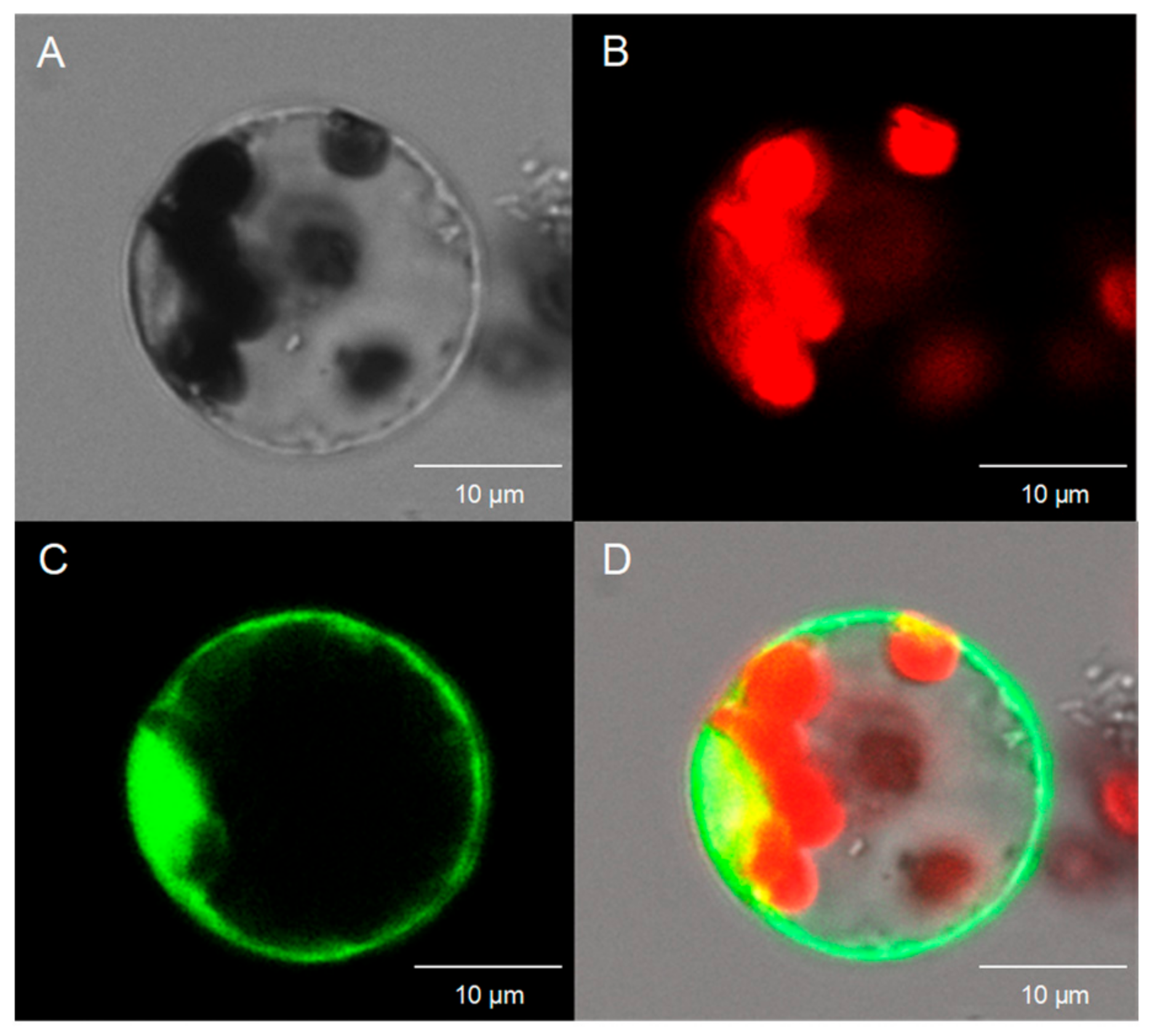
2.2. Subcellular Localization of These Three Chrysanthemum KNOX Genes
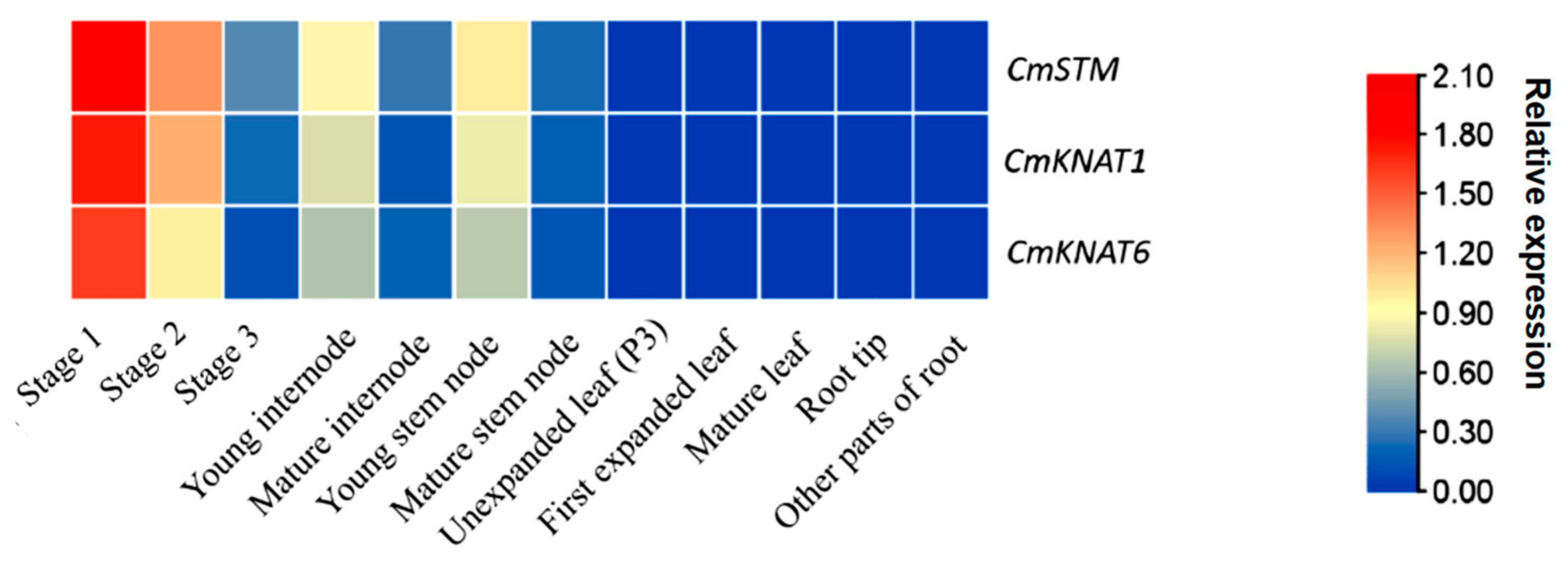
2.3. Analysis of the Relative Expression Level of These Three Chrysanthemum KNOX Genes
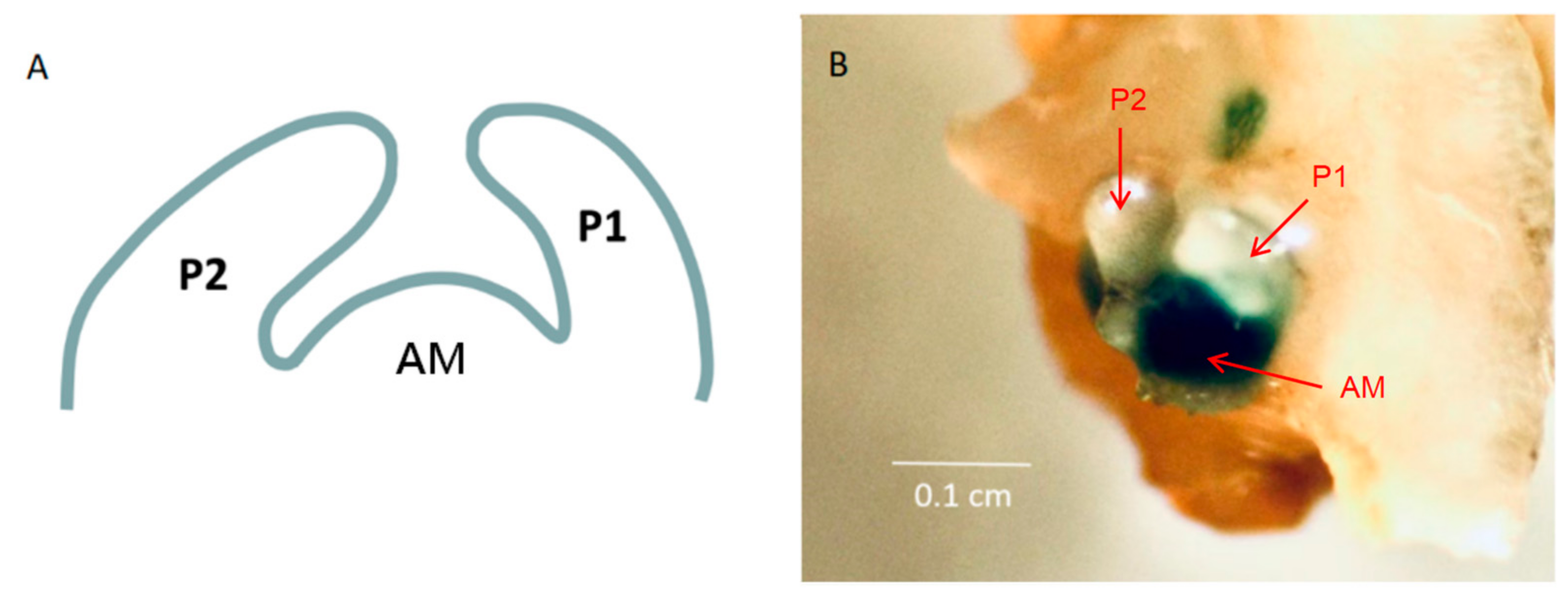
2.4. Phenotypic Observation and Regeneration Test of Model Plants Overexpressing Chrysanthemum KNOX Gene
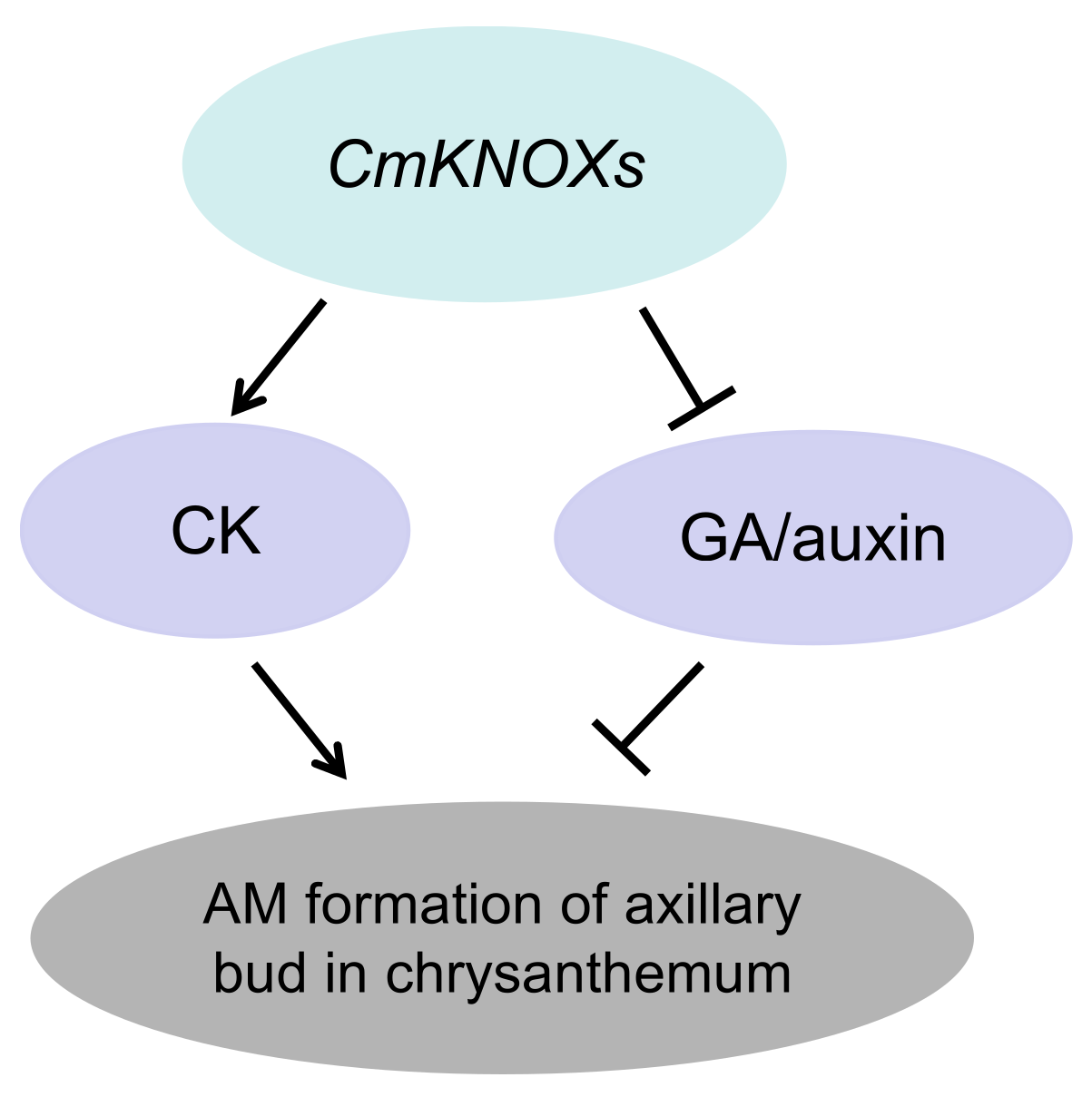
2.5. Prediction of the Effect of These Three KNOX Genes on Hormone Pathway
3. Discussion
4. Materials and Methods
4.1. Bioinformatics Analysis of Three KNOX Genes in Chrysanthemum
4.2. Subcellular Localization of Chrysanthemum KNOX Genes
4.3. Analysis of the Relative Expression Level of Chrysanthemum KNOX Genes
4.4. Acquisition and Phenotypic Observation of Model Plants Overexpressing Chrysanthemum KNOX Gene
4.5. Prediction of the Effect of KNOX Genes on Hormone Pathway
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Chan, R.L.; Gago, G.M.; Palena, C.M.; Gonzalez, D.H. Homeoboxes in Plant Development. Biochim. Biophys. Acta 1998, 1442, 1–19. [Google Scholar] [CrossRef] [PubMed]
- Billeter, M.; Qian, Y.Q.; Otting, G.; Muller, M.; Gehring, W.; Wuthrich, K. Determination of the Nuclear Magnetic Resonance Solution Structure of an Antennapedia Homeodomain-DNA Complex. J. Mol. Biol. 1993, 234, 1084–1093. [Google Scholar] [CrossRef] [PubMed]
- Gehring, W.J.; Affolter, M.; Bürglin, T. Homeodomain Proteins. Annu. Rev. Biochem. 1994, 63, 487–526. [Google Scholar] [CrossRef] [PubMed]
- Vollbrecht, E.; Veit, B.; Sinha, N.; Hake, S. The Developmental Gene Knotted-1 Is a Member of a Maize Homeobox Gene Family. Nature 1991, 350, 241–243. [Google Scholar] [CrossRef] [PubMed]
- Mukherjee, K.; Brocchieri, L.; Burglin, T.R. A Comprehensive Classification and Evolutionary Analysis of Plant Homeobox Genes. Mol. Biol. Evol. 2009, 26, 2775–2794. [Google Scholar] [CrossRef]
- Burglin, T.R.; Affolter, M. Homeodomain Protein: An Update. Chromosoma 2016, 125, 497–521. [Google Scholar] [CrossRef]
- Jin, J.P.; Tian, F.; Yang, D.C.; Meng, Y.Q.; Kong, L.; Luo, J.C.; Gao, G. PlantTFDB 4.0: Toward a Central Hub for Transcription Factors and Regulatory Interactions in Plants. Nucleic Acids Res. 2017, 45, D1040–D1045. [Google Scholar] [CrossRef]
- Lee, J.-H.; Lin, H.; Joo, S.; Goodenough, U. Early Sexual Origins of Homeoprotein Heterodimerization and Evolution of the Plant KNOX/BELL Family. Cell 2008, 133, 829–840. [Google Scholar] [CrossRef]
- Hamant, O.; Pautot, V. Plant Development: A TALE Story. Comptes Rendus Biol. 2010, 333, 371–381. [Google Scholar] [CrossRef]
- Bürglin, T.R. Analysis of TALE Superclass Homeobox Genes (MEIS, PBC, KNOX, Iroquois, TGIF) Reveals a Novel Domain Conserved between Plants and Animals. Nucleic Acids Res. 1997, 25, 4173–4180. [Google Scholar] [CrossRef]
- Smith, H.M.S.; Hake, S. The Interaction of Two Homeobox Genes, BREVIPEDICELLUS and PENNYWISE, Regulates Internode Patterning in the Arabidopsis Inflorescence. Plant Cell 2003, 15, 1717–1727. [Google Scholar] [CrossRef] [PubMed]
- Cao, X.W.; Wang, J.; Xiong, Y.Y.; Yang, H.B.; Yang, M.L.; Ye, P.; Bencivenga, S.; Sablowski, R.; Jiao, Y.L. A Self-Activation Loop Maintains Meristematic Cell Fate for Branching. Curr. Biol. 2020, 30, 1893–1904. [Google Scholar] [CrossRef] [PubMed]
- Meng, L.; Liu, X.; He, C.; Xu, B.; Li, Y.; Hu, Y. Functional Divergence and Adaptive Selection of KNOX Gene Family in Plants. Open Life Sci. 2020, 15, 346–363. [Google Scholar] [CrossRef] [PubMed]
- Gao, J.; Yang, X.; Zhao, W.; Lang, T.G.; Samuelsson, T. Evolution, Diversification, and Expression of KNOX Proteins in Plants. Front. Plant Sci. 2015, 6, 882. [Google Scholar] [CrossRef]
- Hake, S.; Smith, H.M.S.; Holtan, H.; Magnani, E.; Mele, G.; Ramirez, J. The Role of Knox Genes in Plant Development. Annu. Rev. Cell Dev. Biol. 2004, 20, 125–151. [Google Scholar] [CrossRef] [PubMed]
- Magnani, E.; Hake, S. KNOX Lost the OX: The Arabidopsis KNATM Gene Defines a Novel Class of KNOX Transcriptional Regulators Missing the Homeodomain. Plant Cell 2008, 20, 875–887. [Google Scholar] [CrossRef] [PubMed]
- Hay, A.; Tsiantis, M. KNOX Genes: Versatile Regulators of Plant Development and Diversity. Development 2010, 137, 3153–3165. [Google Scholar] [CrossRef]
- Scofield, S.; Dewitte, W.; Murray, J.A. A Model for Arabidopsis Class-1 KNOX Gene Function. Plant Signal. Behav. 2008, 3, 257–259. [Google Scholar] [CrossRef]
- Shani, E.; Burko, Y.; Ben-Yaakov, L.; Berger, Y.; Amsellem, Z.; Goldshmidt, A.; Sharon, E.; Ori, N. Stage-Specific Regulation of Solanum Lycopersicum Leaf Maturation by Class 1 KNOTTED1-LIKE HOMEOBOX Proteins. Plant Cell 2009, 21, 3078–3092. [Google Scholar] [CrossRef]
- Byrne, M.E.; Simorowski, J.; Martienssen, R.A. ASYMMETRIC LEAVES1 Reveals Knox Gene Redundancy in Arabidopsis. Development 2002, 129, 1957–1965. [Google Scholar] [CrossRef]
- Belles-Boix, E.; Hamant, O.; Witiak, S.M.; Morin, H.; Traas, J.; Pautot, V. KNAT6: An Arabidopsis Homeobox Gene Involved in Meristem Activity and Organ Separation. Plant Cell 2006, 18, 1900–1907. [Google Scholar] [CrossRef] [PubMed]
- Bharathan, G.; Goliber, T.E.; Moore, C.; Kessler, S.; Pham, T.; Sinha, N.R. Homologies in Leaf Form Inferred from KNOXI Gene Expression during Development. Science 2002, 296, 1858–1860. [Google Scholar] [CrossRef] [PubMed]
- Ragni, L.; Belles-Boix, E.; Gunl, M.; Pautot, V. Interaction of KNAT6 and KNAT2 with BREVIPEDICELLUS and PENNYWISE in Arabidopsis Inflorescences. Plant Cell 2008, 20, 888–900. [Google Scholar] [CrossRef] [PubMed]
- Ma, Q.; Wang, N.H.; Hao, P.B.; Sun, H.R.; Wang, C.C.; Ma, L.; Wang, H.T.; Zhang, X.L.; Wei, H.L.; Yu, S.X. Genome-Wide Identification and Characterization of TALE Superfamily Genes in Cotton Reveals Their Functions in Regulating Secondary Cell Wall Biosynthesis. BMC Plant Biol. 2019, 19, 432. [Google Scholar] [CrossRef]
- Li, E.Y.; Bhargava, A.; Qiang, W.Y.; Friedmann, M.C.; Forneris, N.; Savidge, R.A.; Johnson, L.A.; Mansfield, S.D.; Ellis, B.E.; Douglas, C.J. The Class II KNOX Gene KNAT7 Negatively Regulates Secondary Wall Formation in Arabidopsis and Is Functionally Conserved in Populus. New Phytol. 2012, 194, 102–115. [Google Scholar] [CrossRef]
- Wang, S.G.; Yang, H.L.; Mei, J.S.; Liu, X.L.; Wen, Z.; Zhang, L.J.; Xu, Z.P.; Zhang, B.C.; Zhou, Y.H. Rice Homeobox Protein KNAT7 Integrates the Pathways Regulating Cell Expansion and Wall Stiffness. Plant Physiol. 2019, 181, 669–682. [Google Scholar] [CrossRef]
- Truernit, E.; Haseloff, J. A Role for KNAT Class II Genes in Root Development. Plant Signal. Behav. 2007, 2, 10–12. [Google Scholar] [CrossRef]
- Di Giacomo, E.; Laffont, C.; Sciarra, F.; Iannelli, M.A.; Frugier, F.; Frugis, G. KNAT3/4/5-like Class 2 KNOX Transcription Factors Are Involved in Medicago Truncatula Symbiotic Nodule Organ Development. New Phytol. 2017, 213, 822–837. [Google Scholar] [CrossRef]
- Wang, Q.; Hasson, A.; Rossmann, S.; Theres, K. Divide et Impera: Boundaries Shape the Plant Body and Initiate New Meristems. New Phytol. 2016, 209, 485–498. [Google Scholar] [CrossRef]
- Scofield, S.; Murray, J.A.H. KNOX Gene Function in Plant Stem Cell Niches. Plant Mol. Biol. 2006, 60, 929–946. [Google Scholar] [CrossRef]
- Wang, Y.; Jiao, Y.L. Axillary Meristem Initiation—A Way to Branch Out. Curr. Opin. Plant Biol. 2018, 41, 61–66. [Google Scholar] [CrossRef] [PubMed]
- Scofield, S.; Dewitte, W.; Murray, J.A. STM Sustains Stem Cell Function in the Arabidopsis Shoot Apical Meristem and Controls KNOX Gene Expression Independently of the Transcriptional Repressor AS1. Plant Signal. Behav. 2014, 9, e28934. [Google Scholar] [CrossRef] [PubMed]
- Shi, B.H.; Zhang, C.; Tian, C.H.; Wang, J.; Wang, Q.; Xu, T.F.; Xu, Y.; Ohno, C.; Sablowski, R.; Heisler, M.G.; et al. Two-Step Regulation of a Meristematic Cell Population Acting in Shoot Branching in Arabidopsis. PLoS Genet. 2016, 12, e1006168. [Google Scholar] [CrossRef] [PubMed]
- Steeves, T.A.; Sussex, I.M. Patterns in Plant Development (2nd Edn). J. Agric. Sci. 1990, 114, 354. [Google Scholar] [CrossRef]
- Scofield, S.; Murison, A.; Jones, A.; Fozard, J.; Aida, M.; Band, L.R.; Bennett, M.; Murray, J.A.H. Coordination of Meristem and Boundary Functions by Transcription Factors in the SHOOT MERISTEMLESS Regulatory Network. Development 2018, 145, dev157081. [Google Scholar] [CrossRef]
- Zadnikova, P.; Simon, R. How Boundaries Control Plant Development. Curr. Opin. Plant Biol. 2014, 17, 116–125. [Google Scholar] [CrossRef] [PubMed]
- Tanaka, W.; Tsuda, K.; Hirano, H.Y. Class I KNOX Gene OSH1 Is Indispensable for Axillary Meristem Development in Rice. Cytologia 2019, 84, 343–346. [Google Scholar] [CrossRef]
- Yang, D.H.; Yun, P.-Y.; Park, S.Y.; Plaha, P.; Lee, D.S.; Lee, I.S.; Hwang, Y.S.; Kim, Y.A.; Lee, J.S.; Han, B.H.; et al. Cloning, Characterization and Expression of a Lateral Suppressor-like Gene from Chrysanthemum (Dendranthema grandiflorum Kitamura). Plant Physiol. Biochem. PPB 2005, 43, 1044–1051. [Google Scholar] [CrossRef]
- Han, B.H.; Suh, E.J.; Lee, S.Y.; Shin, H.K.; Lim, Y.P. Selection of Non-Branching Lines Induced by Introducing Ls-like CDNA into Chrysanthemum (Dendranthema × grandiflorum (Ramat.) Kitamura) “Shuho-No-Chikara”. Sci. Hortic. 2007, 115, 70–75. [Google Scholar] [CrossRef]
- Jiang, B.; Miao, H.; Chen, S.; Zhang, S.; Chen, F.; Fang, W. The Lateral Suppressor-like Gene, DgLsL, Alternated the Axillary Branching in Transgenic Chrysanthemum (Chrysanthemum × morifolium) by Modulating IAA and GA Content. Plant Mol. Biol. Report. 2010, 28, 144–151. [Google Scholar] [CrossRef]
- Huh, E.J.; Lee, Y.R.; Choi, S.Y.; Do, K.R.; Pak, C.H. Genotypic Differences of Axillary Budding in Response to Temperature and Ethephon Treatment in Non-Branching Chrysanthemums. Hortic. Environ. Biotechnol. 2007, 48, 370–375. [Google Scholar]
- Domagalska, M.A.; Leyser, O. Signal Integration in the Control of Shoot Branching. Nat. Rev. Mol. Cell Biol. 2011, 12, 211–221. [Google Scholar] [CrossRef] [PubMed]
- Grbic, V.; Bleecker, A.B. Axillary Meristem Development in Arabidopsis thaliana. Plant J. 2000, 21, 215–223. [Google Scholar] [CrossRef] [PubMed]
- Wang, Q.; Kohlen, W.; Rossmann, S.; Vernoux, T.; Theres, K. Correction: Auxin Depletion from the Leaf Axil Conditions Competence for Axillary Meristem Formation in Arabidopsis and Tomato. Plant Cell 2015, 27, 946. [Google Scholar] [CrossRef]
- Tanaka, W.; Ohmori, Y.; Ushijima, T.; Matsusaka, H.; Matsushita, T.; Kumamaru, T.; Kawano, S.; Hirano, H.Y. Axillary Meristem Formation in Rice Requires the WUSCHEL Ortholog TILLERS ABSENT1. Plant Cell 2015, 27, 1173–1184. [Google Scholar] [CrossRef]
- Hibara, K.; Karim, M.R.; Takada, S.; Taoka, K.I.; Furutani, M.; Aida, M.; Tasaka, M. Arabidopsis CUP-SHAPED COTYLEDON3 Regulates Postembryonic Shoot Meristem and Organ Boundary Formation. Plant Cell 2006, 18, 2946–2957. [Google Scholar] [CrossRef]
- Raman, S.; Greb, T.; Peaucelle, A.; Blein, T.; Laufs, P.; Theres, K. Interplay of MiR164, CUP-SHAPED COTYLEDON Genes and LATERAL SUPPRESSOR Controls Axillary Meristem Formation in Arabidopsis Thaliana. Plant J. 2008, 55, 65–76. [Google Scholar] [CrossRef]
- Keller, T.; Abbott, J.; Moritz, T.; Doerner, P. Arabidopsis REGULATOR OF AXILLARY MERISTEMS1 Controls a Leaf Axil Stem Cell Niche and Modulates Vegetative Development. Plant Cell 2006, 18, 598–611. [Google Scholar] [CrossRef]
- Yang, F.; Wang, Q.; Schmitz, G.; Muller, D.; Theres, K. The BHLH Protein ROX Acts in Concert with RAX1 and LAS to Modulate Axillary Meristem Formation in Arabidopsis. Plant J. 2012, 71, 61–70. [Google Scholar] [CrossRef]
- Reiser, L.; Sánchez-Baracaldo, P.; Hake, S. Knots in the Family Tree: Evolutionary Relationships and Functions of Knox Homeobox Genes. Plant Mol. Biol. 2000, 42, 151–166. [Google Scholar] [CrossRef]
- Su, Y.H.; Zhou, C.; Li, Y.J.; Yu, Y.; Tang, L.P.; Zhang, W.J.; Yao, W.J.; Huang, R.F.; Laux, T.; Zhang, X.S. Integration of Pluripotency Pathways Regulates Stem Cell Maintenance in the Arabidopsis Shoot Meristem. Proc. Natl. Acad. Sci. USA 2020, 117, 22561–22571. [Google Scholar] [CrossRef] [PubMed]
- Ikezaki, M.; Kojima, M.; Sakakibara, H.; Kojima, S.; Ueno, Y.; Machida, C.; Machida, Y. Genetic Networks Regulated by ASYMMETRIC LEAVES1 (AS1) and AS2 in Leaf Development in Arabidopsis Thaliana: KNOX Genes Control Five Morphological Events. Plant J. 2010, 61, 70–82. [Google Scholar] [CrossRef] [PubMed]
- Ha, C.M.; Jun, J.H.; Fletcher, J.C. Control of Arabidopsis Leaf Morphogenesis through Regulation of the YABBY and KNOX Families of Transcription Factors. Genetics 2010, 186, 197–206. [Google Scholar] [CrossRef] [PubMed]
- Khan, M.; Tabb, P.; Hepworth, S.R. BLADE-ON-PETIOLE1 and 2 Regulate Arabidopsis Inflorescence Architecture in Conjunction with Homeobox Genes KNAT6 and ATH1. Plant Signal. Behav. 2012, 7, 788–792. [Google Scholar] [CrossRef]
- Khan, M.; Xu, M.; Murmu, J.; Tabb, P.; Liu, Y.; Storey, K.; McKim, S.M.; Douglas, C.J.; Hepworth, S.R. Antagonistic Interaction of BLADE-ON-PETIOLE1 and 2 with BREVIPEDICELLUS and PENNYWISE Regulates Arabidopsis Inflorescence Architecture. Plant Physiol. 2012, 158, 946–960. [Google Scholar] [CrossRef]
- Lodha, M.; Marco, C.F.; Timmermans, M.C.P. The ASYMMETRIC LEAVES Complex Maintains Repression of KNOX Homeobox Genes via Direct Recruitment of Polycomb-Repressive Complex2. Genes Dev. 2013, 27, 596–601. [Google Scholar] [CrossRef]
- Sakamoto, T.; Kamiya, N.; Ueguchi-Tanaka, M.; Iwahori, S.; Matsuoka, M. KNOX Homeodomain Protein Directly Suppresses the Expression of a Gibberellin Biosynthetic Gene in the Tobacco Shoot Apical Meristem. Genes Dev. 2001, 15, 581–590. [Google Scholar] [CrossRef]
- Hay, A.; Kaur, H.; Phillips, A.; Hedden, P.; Hake, S.; Tsiantis, M. The Gibberellin Pathway Mediates KNOTTED1-Type Homeobox Function in Plants with Different Body Plans. Curr. Biol. 2002, 12, 1557–1565. [Google Scholar] [CrossRef]
- Bolduc, N.; Hake, S. The Maize Transcription Factor KNOTTED1 Directly Regulates the Gibberellin Catabolism Gene Ga2ox1. Plant Cell 2009, 21, 1647–1658. [Google Scholar] [CrossRef]
- Jasinski, S.; Piazza, P.; Craft, J.; Hay, A.; Woolley, L.; Rieu, I.; Phillips, A.; Hedden, P.; Tsiantis, M. KNOX Action in Arabidopsis Is Mediated by Coordinate Regulation of Cytokinin and Gibberellin Activities. Curr. Biol. 2005, 15, 1560–1565. [Google Scholar] [CrossRef]
- Yanai, O.; Shani, E.; Dolezal, K.; Tarkowski, P.; Sablowski, R.; Sandberg, G.; Samach, A.; Ori, N. Arabidopsis KNOXI Proteins Activate Cytokinin Biosynthesis. Curr. Biol. 2005, 15, 1566–1571. [Google Scholar] [CrossRef] [PubMed]
- Hay, A.; Barkoulas, M.; Tsiantis, M. ASYMMETRIC LEAVES1 and Auxin Activities Converge to Repress BREVIPEDICELLUS Expression and Promote Leaf Development in Arabidopsis. Development 2006, 133, 3955–3961. [Google Scholar] [CrossRef] [PubMed]




| Protein Name | Protein Length | MW | pI | GRAVY | Coefficient of Instability | Subcellular Localization Prediction | Homolog in Arabidopsis |
|---|---|---|---|---|---|---|---|
| CmKNAT1 | 361 | 41,484.11 | 5.56 | −0.981 | 53.59 | Nucleus | KNAT1 |
| CmKNAT6 | 300 | 34,586.26 | 5.03 | −0.857 | 44.25 | Nucleus | KNAT6 |
| CmSTM | 337 | 37,637.28 | 5.72 | −0.654 | 48.34 | Nucleus | STM |
| Protein Name | Serine Residues (s) Number/Proportion | Lysine Residues (Y) Number/Proportion | Threonine Residues (T) Number/Proportion | Total |
|---|---|---|---|---|
| CmKNAT1 | 26 (65%) | 9 (23%) | 5 (13%) | 40 |
| CmKNAT6 | 23 (56%) | 8 (20%) | 10 (24%) | 41 |
| CmSTM | 23 (74%) | 4 (13%) | 4 (13%) | 31 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yang, Q.; Cong, T.; Yao, Y.; Cheng, T.; Yuan, C.; Zhang, Q. KNOX Genes Were Involved in Regulating Axillary Bud Formation of Chrysanthemum × morifolium. Int. J. Mol. Sci. 2023, 24, 7081. https://doi.org/10.3390/ijms24087081
Yang Q, Cong T, Yao Y, Cheng T, Yuan C, Zhang Q. KNOX Genes Were Involved in Regulating Axillary Bud Formation of Chrysanthemum × morifolium. International Journal of Molecular Sciences. 2023; 24(8):7081. https://doi.org/10.3390/ijms24087081
Chicago/Turabian StyleYang, Qingqing, Tianci Cong, Yicen Yao, Tangren Cheng, Cunquan Yuan, and Qixiang Zhang. 2023. "KNOX Genes Were Involved in Regulating Axillary Bud Formation of Chrysanthemum × morifolium" International Journal of Molecular Sciences 24, no. 8: 7081. https://doi.org/10.3390/ijms24087081
APA StyleYang, Q., Cong, T., Yao, Y., Cheng, T., Yuan, C., & Zhang, Q. (2023). KNOX Genes Were Involved in Regulating Axillary Bud Formation of Chrysanthemum × morifolium. International Journal of Molecular Sciences, 24(8), 7081. https://doi.org/10.3390/ijms24087081







