Energy Metabolism Is Altered in Radioresistant Rectal Cancer
Abstract
1. Introduction
2. Results
2.1. Identification of an In Vitro Model of Radioresistant Rectal Cancer
Clonogenic Survival of SW837 and HCT116 Cells Following X-ray Radiation and 5-Fluorouracil (5-FU) Chemotherapy
2.2. Characterization of an In Vitro Model of Inherent Radioresistant/Radiosensitive CRC
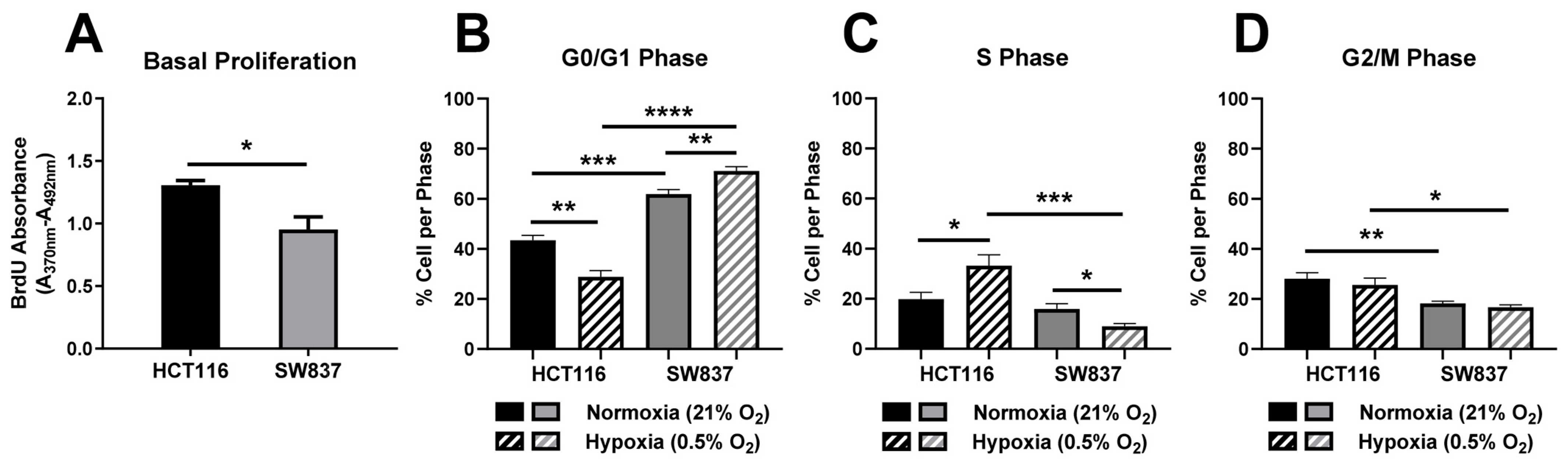
2.2.1. Basal Proliferation and Cell Cycle Distribution Is Altered in Radioresistant SW837 Cells
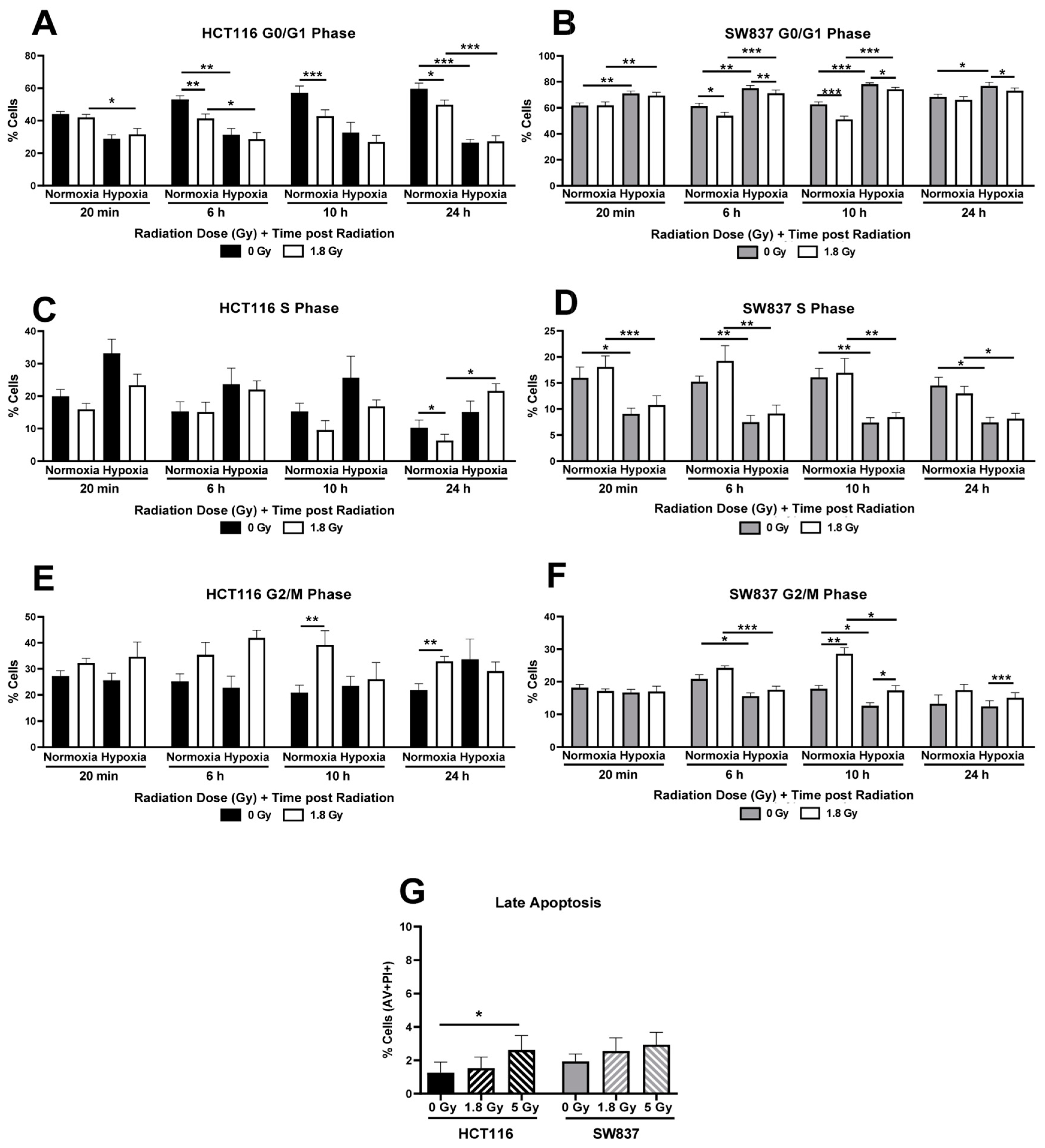
2.2.2. Cell Cycle Progression Following Radiation Exposure Is Altered in SW837 and HCT116 Cells under Normoxic and Hypoxic Conditions
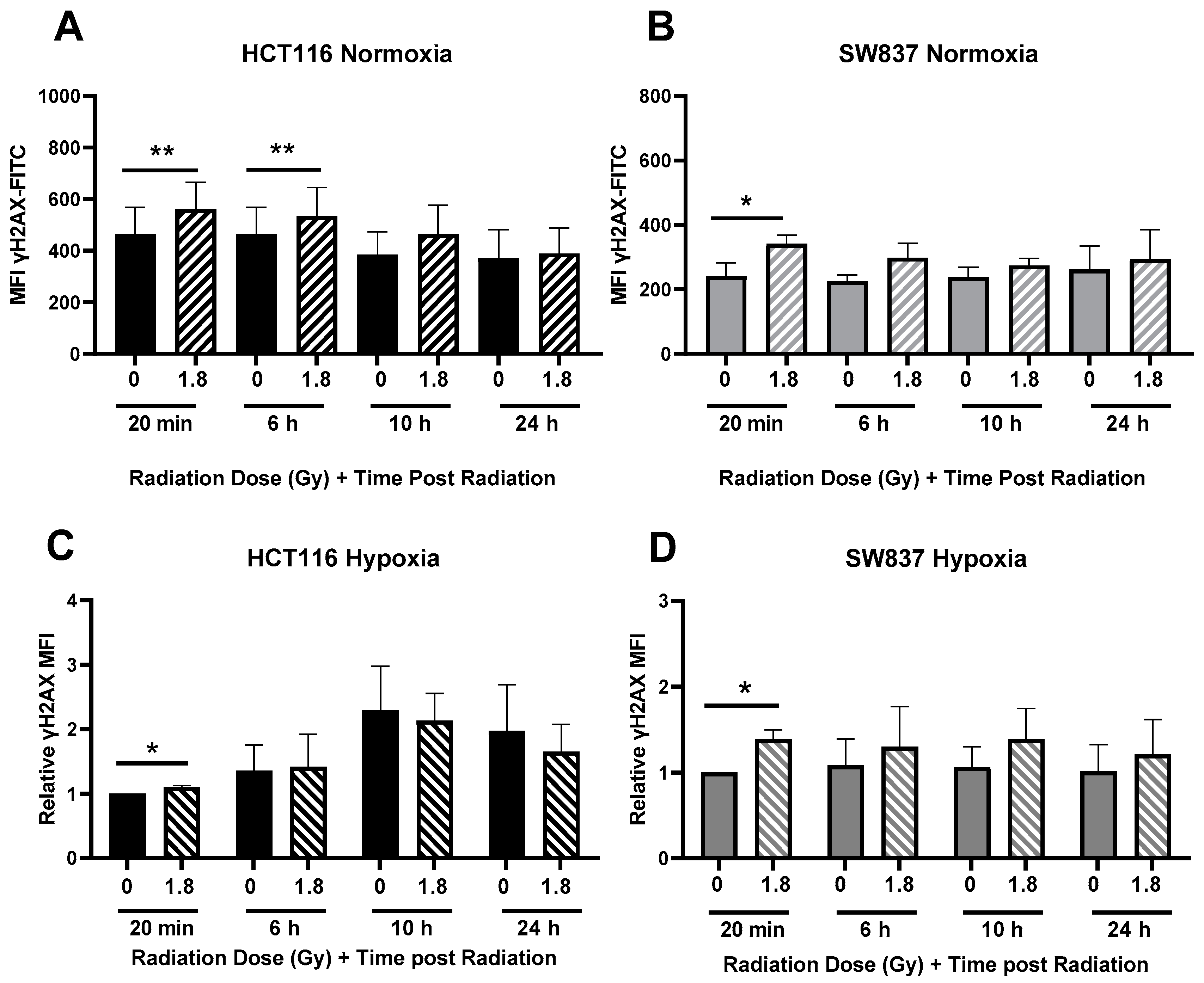
2.2.3. Radiation-Induced DNA Damage Is More Efficiently Repaired in Radioresistant SW837 Cells
2.3. Metabolic Reprograming in Radioresistant CRC Cells
2.3.1. SW837 Cells Display a Significantly Altered Basal Transcriptome When Compared to HCT116 Cells
2.3.2. Radioresistant SW837 Cells Display Decreased Reliance on Glycolysis
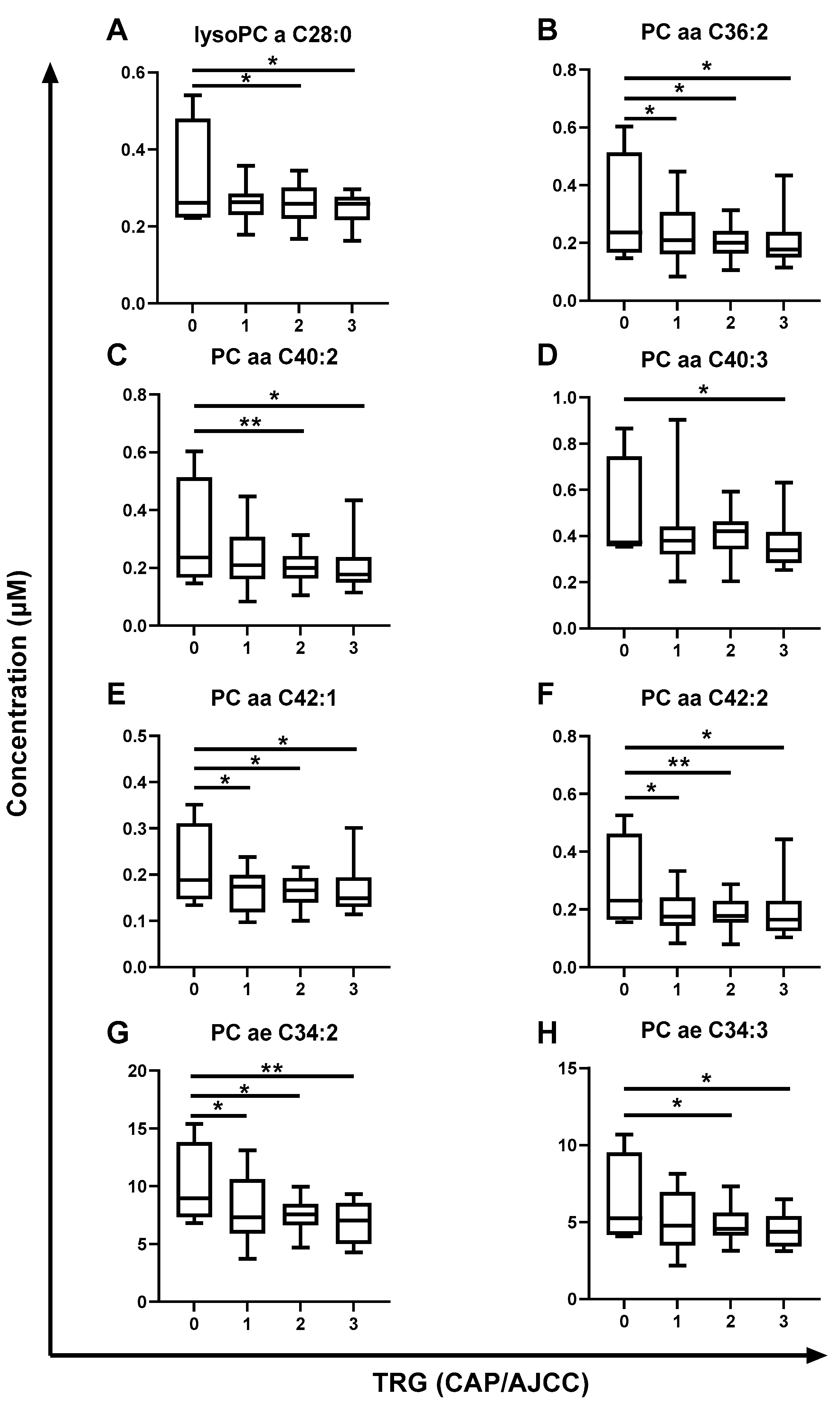
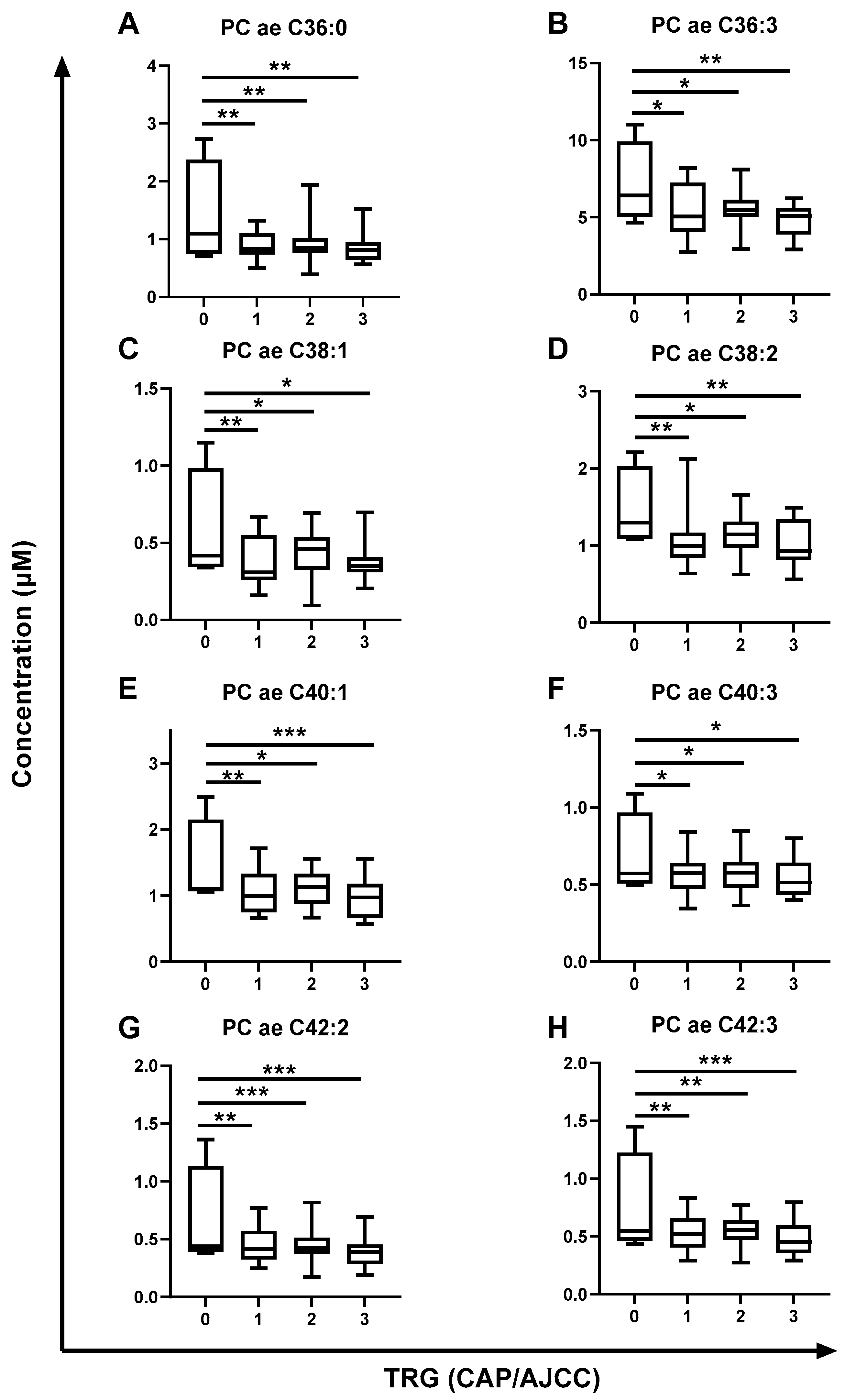
2.4. The Metabolome of Pre-Treatment Serum Is Significantly Altered in Rectal Cancer Patients Having a Poor Response to neoCRT and Poorer Outcomes
3. Discussion
4. Materials and Methods
4.1. Colorectal Adenocarcinoma Cell Lines
4.2. X-ray Radiation
4.3. Clonogenic Assessment of Radiosensitivity under Hypoxia (0.5% O2) or Normoxia
4.4. Clonogenic Assessment of 5-FU Sensitivity under Hypoxia (0.5% O2) or Normoxia
4.5. Fixation, Staining and Counting of Clonogenic Assay
4.6. BrdU ELISA
4.7. Assessment of Cell Cycle and DNA Damage under Hypoxia (0.5% O2) and Normoxia
4.8. Assessment of Apoptosis by Flow Cytometry
4.9. Crystal Violet Assay
4.10. RNA Isolation and Quantification
4.11. Transcriptomic Profiling
4.12. Transcriptomic Data Analaysis
4.13. IPA Analysis
4.14. Live-Cell Metabolic Profiling by Seahorse XFtechnology
4.15. 2-DG Clonogenic Assay
4.16. Patient Ethics, Treatment and Sample Collection
4.17. Preparation of Patient Samples
4.18. Metabolomic Profiling by AbsoluteIDQ®® p180 Assay
4.19. Data Processing and Metabolite Quantification
4.20. Statistical Analysis
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Sung, H.; Ferlay, J.; Siegel, R.L.; Laversanne, M.; Soerjomataram, I.; Jemal, A.; Bray, F. Global Cancer Statistics 2020: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA Cancer J. Clin. 2021, 71, 209–249. [Google Scholar] [CrossRef] [PubMed]
- National Cancer Registry Ireland (NCRI). Cancer Incidence Projections for Ireland 2020–2045; National Cancer Registry: Cork, Ireland, 2019. [Google Scholar]
- McCarthy, K.; Pearson, K.; Fulton, R.; Hewitt, J. Pre-operative chemoradiation for non-metastatic locally advanced rectal cancer. Cochrane Database Syst. Rev. 2012, 12, Cd008368. [Google Scholar] [CrossRef] [PubMed]
- Glynne-Jones, R.; Wyrwicz, L.; Tiret, E.; Brown, G.; Rödel, C.D.; Cervantes, A.; Arnold, D. Rectal cancer: ESMO Clinical Practice Guidelines for diagnosis, treatment and follow-up. Ann. Oncol. 2017, 28 (Suppl. S4), iv22–iv40. [Google Scholar] [CrossRef] [PubMed]
- Ferrari, L.; Fichera, A. Neoadjuvant chemoradiation therapy and pathological complete response in rectal cancer. Gastroenterol. Rep. 2015, 3, 277–288. [Google Scholar] [CrossRef] [PubMed]
- Roeder, F.; Meldolesi, E.; Gerum, S.; Valentini, V.; Rödel, C. Recent advances in (chemo-)radiation therapy for rectal cancer: A comprehensive review. Radiat. Oncol. 2020, 15, 262. [Google Scholar] [CrossRef]
- Pawlik, T.M.; Keyomarsi, K. Role of cell cycle in mediating sensitivity to radiotherapy. Int. J. Radiat. Oncol. Biol. Phys. 2004, 59, 928–942. [Google Scholar] [CrossRef]
- Huerta, S.; Gao, X.; Saha, D. Mechanisms of resistance to ionizing radiation in rectal cancer. Expert Rev. Mol. Diagn. 2009, 9, 469–480. [Google Scholar] [CrossRef]
- Ganesh, K.; Wu, C.; O’Rourke, K.P.; Adileh, M.; Szeglin, B.C.; Wasserman, I.; Marco, M.R.; Shady, M.; Zheng, Y.; Karthaus, W.R. A rectal cancer model establishes a platform to study individual responses to chemoradiation. bioRxiv 2019, 640193. [Google Scholar]
- Janakiraman, H.; Zhu, Y.; Becker, S.A.; Wang, C.; Cross, A.; Curl, E.; Lewin, D.; Hoffman, B.J.; Warren, G.W.; Hill, E.G. Modeling rectal cancer to advance neoadjuvant precision therapy. Int. J. Cancer 2020, 147, 1405–1418. [Google Scholar] [CrossRef]
- Gollins, S.; Moran, B.; Adams, R.; Cunningham, C.; Bach, S.; Myint, A.S.; Renehan, A.; Karandikar, S.; Goh, V.; Prezzi, D. Association of Coloproctology of Great Britain & Ireland (ACPGBI): Guidelines for the Management of Cancer of the Colon, Rectum and Anus (2017)–Multidisciplinary Management. Colorectal Dis. 2017, 19 (Suppl. S1), 37–66. [Google Scholar]
- Lee, G.H.; Malietzis, G.; Askari, A.; Bernardo, D.; Al-Hassi, H.O.; Clark, S.K. Is right-sided colon cancer different to left-sided colorectal cancer?–a systematic review. Eur. J. Surg. Oncol. 2015, 41, 300–308. [Google Scholar] [CrossRef] [PubMed]
- Stintzing, S.; Tejpar, S.; Gibbs, P.; Thiebach, L.; Lenz, H.J. Understanding the role of primary tumour localisation in colorectal cancer treatment and outcomes. Eur. J. Cancer 2017, 84, 69–80. [Google Scholar] [CrossRef] [PubMed]
- Yang, S.Y.; Cho, M.S.; Kim, N.K. Difference between right-sided and left-sided colorectal cancers: From embryology to molecular subtype. Expert Rev. Anticancer. Ther. 2018, 18, 351–358. [Google Scholar] [CrossRef] [PubMed]
- Ishibashi, N.; Maebayashi, T.; Aizawa, T.; Sakaguchi, M.; Nishimaki, H.; Masuda, S. Correlation between the Ki-67 proliferation index and response to radiation therapy in small cell lung cancer. Radiat. Oncol. 2017, 12, 16. [Google Scholar] [CrossRef]
- Freudlsperger, C.; Freier, K.; Hoffmann, J.; Engel, M. Ki-67 expression predicts radiosensitivity in oral squamous cell carcinoma. Int. J. Oral Maxillofac. Surg. 2012, 41, 965–969. [Google Scholar] [CrossRef]
- Sinclair, W.K.; Morton, R.A. X-Ray Sensitivity during the Cell Generation Cycle of Cultured Chinese Hamster Cells. Radiat. Res. 1966, 29, 450–474. [Google Scholar] [CrossRef]
- Brown, K.G.M.; Solomon, M.J.; Mahon, K.; O’Shannassy, S. Management of colorectal cancer. BMJ 2019, 366, l4561. [Google Scholar] [CrossRef]
- Kendziorra, E.; Ahlborn, K.; Spitzner, M.; Rave-Fränk, M.; Emons, G.; Gaedcke, J.; Kramer, F.; Wolff, H.A.; Becker, H.; Beissbarth, T. Silencing of the Wnt transcription factor TCF4 sensitizes colorectal cancer cells to (chemo-) radiotherapy. Carcinogenesis 2011, 32, 1824–1831. [Google Scholar] [CrossRef]
- Koerdel, K.; Spitzner, M.; Meyer, T.; Engels, N.; Krause, F.; Gaedcke, J.; Conradi, L.-C.; Haubrock, M.; Beißbarth, T.; Leha, A. NOTCH Activation via gp130/STAT3 Signaling Confers Resistance to Chemoradiotherapy. Cancers 2021, 13, 455. [Google Scholar] [CrossRef]
- Jing, X.; Yang, F.; Shao, C.; Wei, K.; Xie, M.; Shen, H.; Shu, Y. Role of hypoxia in cancer therapy by regulating the tumor microenvironment. Mol. Cancer 2019, 18, 157. [Google Scholar] [CrossRef]
- Wilson, W.R.; Hay, M.P. Targeting hypoxia in cancer therapy. Nat. Rev. Cancer 2011, 11, 393–410. [Google Scholar] [CrossRef] [PubMed]
- Bazzocco, S.; Dopeso, H.; Carton-Garcia, F.; Macaya, I.; Andretta, E.; Chionh, F.; Rodrigues, P.; Garrido, M.; Alazzouzi, H.; Nieto, R. Highly Expressed Genes in Rapidly Proliferating Tumor Cells as New Targets for Colorectal Cancer Treatment. Clin. Cancer Res. 2015, 21, 3695–3704. [Google Scholar] [CrossRef] [PubMed]
- Melling, N.; Kowitz, C.M.; Simon, R.; Bokemeyer, C.; Terracciano, L.; Sauter, G.; Izbicki, J.R.; Marx, A.H. High Ki67 expression is an independent good prognostic marker in colorectal cancer. J. Clin. Pathol. 2016, 69, 209. [Google Scholar] [CrossRef] [PubMed]
- Fu, C.G.; Tominaga, O.; Nagawa, H.; Eidi Nita, M.; Masaki, T.; Ishimaru, G.; Higuchi, Y.; Tsuruo, T.; Muto, T. Role of p53 and p21/WAF1 detection in patient selection for preoperative radiotherapy in rectal cancer patients. Dis. Colon Rectum. 1998, 41, 68–74. [Google Scholar] [CrossRef]
- Carcereri de Prati, A.; Butturini, E.; Rigo, A.; Oppici, E.; Rossin, M.; Boriero, D.; Mariotto, S. Metastatic Breast Cancer Cells Enter Into Dormant State and Express Cancer Stem Cells Phenotype Under Chronic Hypoxia. J. Cell Biochem. 2017, 118, 3237–3248. [Google Scholar] [CrossRef]
- Yao, K.; Gietema, J.A.; Shida, S.; Selvakumaran, M.; Fonrose, X.; Haas, N.B.; O’Dwyer, P.J. In vitro hypoxia-conditioned colon cancer cell lines derived from HCT116 and HT29 exhibit altered apoptosis susceptibility and a more angiogenic profile in vivo. Br. J. Cancer 2005, 93, 1356–1363. [Google Scholar] [CrossRef]
- Rakotomalala, A.; Escande, A.; Furlan, A.; Meignan, S.; Lartigau, E. Hypoxia in Solid Tumors: How Low Oxygenation Impacts the “Six Rs” Radiother. Front. Endocrinol. 2021, 12, 742215. [Google Scholar] [CrossRef]
- Visconti, R.; Della Monica, R.; Grieco, D. Cell cycle checkpoint in cancer: A therapeutically targetable double-edged sword. J. Exp. Clin. Cancer Res. 2016, 35, 153. [Google Scholar] [CrossRef]
- Raleigh, D.R.; Haas-Kogan, D.A. Molecular targets and mechanisms of radiosensitization using DNA damage response pathways. Future Oncol. 2013, 9, 219–233. [Google Scholar] [CrossRef]
- Bouquet, F.; Ousset, M.; Biard, D.; Fallone, F.; Dauvillier, S.; Frit, P.; Salles, B.; Muller, C. A DNA-dependent stress response involving DNA-PK occurs in hypoxic cells and contributes to cellular adaptation to hypoxia. J. Cell Sci. 2011, 124, 1943–1951. [Google Scholar] [CrossRef]
- Wozny, A.S.; Alphonse, G.; Cassard, A.; Malésys, C.; Louati, S.; Beuve, M.; Lalle, P.; Ardail, D.; Nakajima, T.; Rodriguez-Lafrasse, C. Impact of hypoxia on the double-strand break repair after photon and carbon ion irradiation of radioresistant HNSCC cells. Sci. Rep. 2020, 10, 21357. [Google Scholar] [CrossRef] [PubMed]
- Macedo-Silva, C.; Miranda-Gonçalves, V.; Lameirinhas, A.; Lencart, J.; Pereira, A.; Lobo, J.; Guimarães, R.; Martins, A.T.; Henrique, R.; Bravo, I. JmjC-KDMs KDM3A and KDM6B modulate radioresistance under hypoxic conditions in esophageal squamous cell carcinoma. Cell Death Dis. 2020, 11, 1068. [Google Scholar] [CrossRef] [PubMed]
- Lynam-Lennon, N.; Maher, S.G.; Maguire, A.; Phelan, J.; Muldoon, C.; Reynolds, J.V.; O’Sullivan, J. Altered mitochondrial function and energy metabolism is associated with a radioresistant phenotype in oesophageal adenocarcinoma. PLoS ONE 2014, 9, e100738. [Google Scholar] [CrossRef] [PubMed]
- Buckley, A.M.; Dunne, M.R.; Morrissey, M.E.; Kennedy, S.A.; Nolan, A.; Davern, M.; Foley, E.K.; Clarke, N.; Lysaght, J.; Ravi, N. Real-time metabolic profiling of oesophageal tumours reveals an altered metabolic phenotype to different oxygen tensions and to treatment with Pyrazinib. Sci. Rep. 2020, 10, 12105. [Google Scholar] [CrossRef]
- Buckley, A.M.; Dunne, M.R.; Lynam-Lennon, N.; Kennedy, S.A.; Cannon, A.; Reynolds, A.L.; Maher, S.G.; Reynolds, J.V.; Kennedy, B.N.; O’Sullivan, J. Pyrazinib (P3), [(E)-2-(2-Pyrazin-2-yl-vinyl)-phenol], a small molecule pyrazine compound enhances radiosensitivity in oesophageal adenocarcinoma. Cancer Lett. 2019, 447, 115–129. [Google Scholar] [CrossRef]
- McCann, E.; O’Sullivan, J.; Marcone, S. Targeting cancer-cell mitochondria and metabolism to improve radiotherapy response. Transl. Oncol. 2021, 14, 100905. [Google Scholar] [CrossRef]
- Chekulayev, V.; Mado, K.; Shevchuk, I.; Koit, A.; Kaldma, A.; Klepinin, A.; Timohhina, N.; Tepp, K.; Kandashvili, M.; Ounpuu, L.; et al. Metabolic remodeling in human colorectal cancer and surrounding tissues: Alterations in regulation of mitochondrial respiration and metabolic fluxes. Biochem. Biophys. Rep. 2015, 4, 111–125. [Google Scholar] [CrossRef]
- Sulciner, M.L.; Gartung, A.; Gilligan, M.M.; Serhan, C.N.; Panigrahy, D. Targeting lipid mediators in cancer biology. Cancer Metastasis Rev. 2018, 37, 557–572. [Google Scholar] [CrossRef]
- Saito, R.d.F.; Andrade, L.N.d.S.; Bustos, S.O.; Chammas, R. Phosphatidylcholine-Derived Lipid Mediators: The Crosstalk Between Cancer Cells and Immune Cells. Front. Immunol. 2022, 13, 215. [Google Scholar] [CrossRef]
- Geijsen, A.J.; van Roekel, E.H.; van Duijnhoven, F.J.; Achaintre, D.; Bachleitner-Hofmann, T.; Baierl, A.; Bergmann, M.M.; Boehm, J.; Bours, M.J.L.; Brenneret, H.; et al. Plasma metabolites associated with colorectal cancer stage: Findings from an international consortium. Int. J. Cancer 2020, 146, 3256–3266. [Google Scholar] [CrossRef]
- van der Veen, J.N.; Kennelly, J.P.; Wan, S.; Vance, J.E.; Vance, D.E.; Jacobs, R.L. The critical role of phosphatidylcholine and phosphatidylethanolamine metabolism in health and disease. Biochim. Biophys. Acta Biomembr. 2017, 1859, 1558–1572. [Google Scholar] [CrossRef] [PubMed]
- Del Boccio, P.; Perrotti, F.; Rossi, C.; Cicalini, I.; Di Santo, S.; Zucchelli, M.; Sacchetta, P.; Genovesi, D.; Pieragostino, D. Serum lipidomic study reveals potential early biomarkers for predicting response to chemoradiation therapy in advanced rectal cancer: A pilot study. Adv. Radiat. Oncol. 2017, 2, 118–124. [Google Scholar] [CrossRef] [PubMed]
- Jia, H.; Shen, X.; Guan, Y.; Xu, M.; Tu, J.; Mo, M.; Xie, L.; Yuan, J.; Zhang, Z.; Cai, S.; et al. Predicting the pathological response to neoadjuvant chemoradiation using untargeted metabolomics in locally advanced rectal cancer. Radiother. Oncol. J. Eur. Soc. Ther. Radiol. Oncol. 2018, 128, 548–556. [Google Scholar] [CrossRef]
- Schmidt, D.R.; Patel, R.; Kirsch, D.G.; Lewis, C.A.; Vander Heiden, M.G.; Locasale, J.W. Metabolomics in cancer research and emerging applications in clinical oncology. CA Cancer J. Clin. 2021, 71, 333–358. [Google Scholar] [CrossRef]
- Tolstikov, V.; Moser, A.J.; Sarangarajan, R.; Narain, N.R.; Kiebish, M.A. Current Status of Metabolomic Biomarker Discovery: Impact of Study Design and Demographic Characteristics. Metabolites 2020, 10, 224. [Google Scholar] [CrossRef] [PubMed]
- Franken, N.A.; Rodermond, H.M.; Stap, J.; Haveman, J.; van Bree, C. Clonogenic assay of cells in vitro. Nat. Protoc. 2006, 1, 2315–2319. [Google Scholar] [CrossRef]
- Zhang, J.; Nuebel, E.; Wisidagama, D.R.R.; Setoguchi, K.; Hong, J.S.; Van Horn, C.M.; Imam, S.S.; Vergnes, L.; Malone, C.S.; Koehler, C.M.; et al. Measuring energy metabolism in cultured cells, including human pluripotent stem cells and differentiated cells. Nat. Protoc. 2012, 7, 1068–1085. [Google Scholar] [CrossRef]
- Bateman, A.C.; Jaynes, E.; Bateman, A.R. Rectal cancer staging post neoadjuvant therapy--how should the changes be assessed? Histopathology 2009, 54, 713–721. [Google Scholar] [CrossRef]
- Zukunft, S.; Prehn, C.; Röhring, C.; Möller, G.; Hrabě de Angelis, M.; Adamski, J.; Tokarz, J. High-throughput extraction and quantification method for targeted metabolomics in murine tissues. Metabolomics 2018, 14, 18. [Google Scholar] [CrossRef]


| Ingenuity Canonical Pathways | p-Value | z-Score |
|---|---|---|
| Oxidative Phosphorylation | 1.95 × 10−9 | 5.145 |
| Sirtuin Signalling Pathway | 6.31 × 10−9 | −2.496 |
| EIF2 Signalling | 3.80 × 10−8 | 2.746 |
| Mitochondrial Dysfunction | 9.12 × 10−8 | NaN |
| Coronavirus Pathogenesis Pathway | 3.98 × 10−6 | −2.287 |
| Regulation of eIF4 and p70S6K Signalling | 8.71 × 10−6 | −0.707 |
| Axonal Guidance Signalling | 1.02 × 10−5 | NaN |
| Actin Cytoskeleton Signalling | 1.15 × 10−5 | −0.632 |
| BAG2 Signalling Pathway | 1.91 × 10−5 | −0.632 |
| mTOR Signalling | 3.39 × 10−5 | 0.894 |
| (n = 52) | ||
|---|---|---|
| Gender | Male (n) | 35 |
| Female (n) | 17 | |
| Age | Mean (y) | 62 |
| Range (y) | 41–79 | |
| Histology | Adenocarcinoma (n) | 52 |
| Clinical T Stage | 2 (n) | 3 |
| 3 (n) | 25 | |
| 4 (n) | 24 | |
| Pathological Nodal Involvement | Yes (n) | 25 |
| No (n) | 27 | |
| Treatment | NeoCRT (n) | 52 |
| TRG (CAP/AJCC Scale) | 0 (n) | 4 |
| 1 (n) | 14 | |
| 2 (n) | 22 | |
| 3 (n) | 12 | |
| Recurrence-Free Survival | Median | 63 |
| Minimum | 2 | |
| Maximum | 103 | |
| Overall Survival (Months) | Median | 63 |
| Minimum | 9 | |
| Maximum | 103 |
| Metabolite | TRG (CAP/AJCC Scale) p-Value (FDR-Corrected) | Lymph Node Positivity p-Value (FDR-Corrected) |
|---|---|---|
| LysoPC a C28:0 | 0.0143 | |
| PC aa C36:2 | 0.0322 | |
| PC aa C40:2 | 0.0143 | |
| PC aa C40:3 | 0.025 | |
| PC aa C42:1 | 0.0322 | |
| PC aa C42:2 | 0.0243 | |
| PC ae C34:2 | 0.0184 | |
| PC ae C34:3 | 0.0250 | |
| PC ae C36:0 | 0.0072 | |
| PC ae C36:3 | 0.0322 | |
| PC ae C38:1 | 0.0207 | 0.0473 |
| PC ae C38:2 | 0.0207 | |
| PC ae C40:1 | 0.0033 | |
| PC ae C40:3 | 0.0322 | |
| PC ae C42:2 | 0.0010 | |
| PC ae C42:3 | 0.0021 |
| Recurence-Free Survival | Overall Survival | |||
|---|---|---|---|---|
| Metabolite | HR (95% CI) | p-Value | HR (95% CI) | p-Value |
| LysoPC a C28:0 | 0.145 (0.000–6103.4) | 0.722 | 0.062 (0.000–316–7) | 0.523 |
| PC aa C36:2 | 0.984 (0.966–1.001) | 0.070 | 0.992 (0.981–1.004) | 0.202 |
| PC aa C40:2 | 0.003 (0.000–161.4) | 0.300 | 0.000 (0.000–0.177) | 0.017 |
| PC aa C40:3 | 0.042 (0.000–38.58) | 0.363 | 0.008 (0.000–1.034) | 0.052 |
| PC aa C42:1 | 0.000 (0.000–1.702) | 0.057 | 0.000 (0.000–0.039) | 0.014 |
| PC aa C42:2 | 0.000 (0.000–0.009) | 0.012 | 0.000 (0.000–0.017) | 0.005 |
| PC ae C34:2 | 0.808 (0.558–1.170) | 0.258 | 0.705 (0.542–0.917) | 0.009 |
| PC ae C34:3 | 0.717 (0.403–1.275) | 0.258 | 0.645 (0.432–0.963) | 0.032 |
| PC ae C36:0 | 0.042 (0.002–0.986) | 0.042 | 0.088 (0.009–0.872) | 0.038 |
| PC ae C36:3 | 0.735 (0.446–1.212) | 0.228 | 0.688 (0.480–0.985) | 0.041 |
| PC ae C38:1 | 0.022 (0.000–3.152) | 0.132 | 0.020 (0.000–0.826) | 0.039 |
| PC ae C38:2 | 0.042 (0.002–0.821) | 0.037 | 0.087 (0.014–0.540) | 0.009 |
| PC ae C40:1 | 0.132 (0.009–1.894) | 0.136 | 0.101 (0.014–0.754) | 0.025 |
| PC ae C40:3 | 0.006 (0.000–3.848) | 0.120 | 0.003 (0.000–0.207) | 0.008 |
| PC ae C42:2 | 0.008 (0.000–0.077) | 0.008 | 0.001 (0.000-0.067) | 0.002 |
| PC ae C42:3 | 0.000 (0.000–0.155) | 0.011 | 0.001 (0.000-0.064) | <0.001 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Buckley, C.E.; Yin, X.; Meltzer, S.; Ree, A.H.; Redalen, K.R.; Brennan, L.; O’Sullivan, J.; Lynam-Lennon, N. Energy Metabolism Is Altered in Radioresistant Rectal Cancer. Int. J. Mol. Sci. 2023, 24, 7082. https://doi.org/10.3390/ijms24087082
Buckley CE, Yin X, Meltzer S, Ree AH, Redalen KR, Brennan L, O’Sullivan J, Lynam-Lennon N. Energy Metabolism Is Altered in Radioresistant Rectal Cancer. International Journal of Molecular Sciences. 2023; 24(8):7082. https://doi.org/10.3390/ijms24087082
Chicago/Turabian StyleBuckley, Croí E., Xiaofei Yin, Sebastian Meltzer, Anne Hansen Ree, Kathrine Røe Redalen, Lorraine Brennan, Jacintha O’Sullivan, and Niamh Lynam-Lennon. 2023. "Energy Metabolism Is Altered in Radioresistant Rectal Cancer" International Journal of Molecular Sciences 24, no. 8: 7082. https://doi.org/10.3390/ijms24087082
APA StyleBuckley, C. E., Yin, X., Meltzer, S., Ree, A. H., Redalen, K. R., Brennan, L., O’Sullivan, J., & Lynam-Lennon, N. (2023). Energy Metabolism Is Altered in Radioresistant Rectal Cancer. International Journal of Molecular Sciences, 24(8), 7082. https://doi.org/10.3390/ijms24087082









