TcbHLH14 a Jasmonate Associated MYC2-like Transcription Factor Positively Regulates Pyrethrin Biosynthesis in Tanacetum cinerariifolium
Abstract
1. Introduction
2. Results
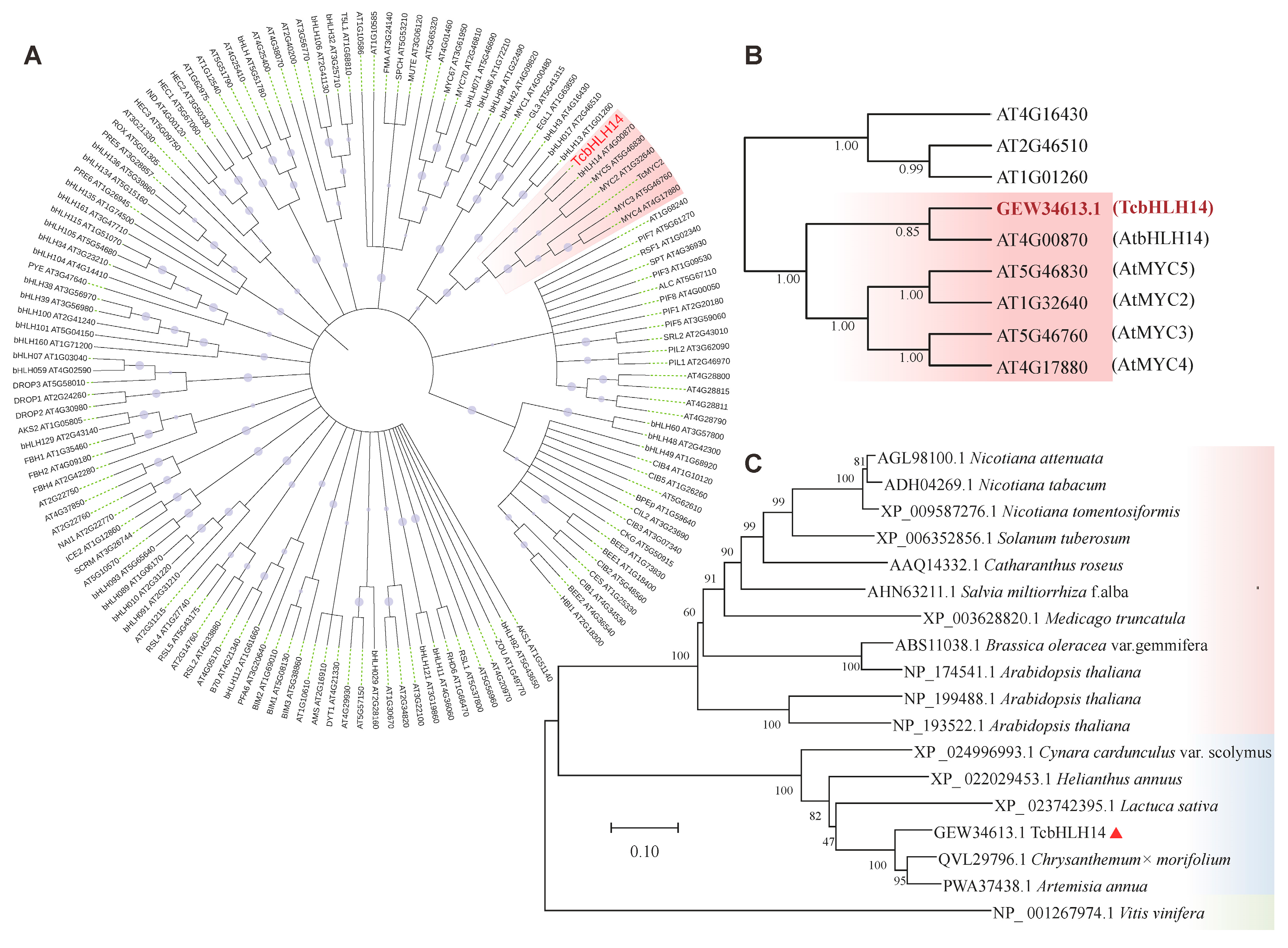
2.1. Domain and Homology Analysis of the TcbHLH14 Gene
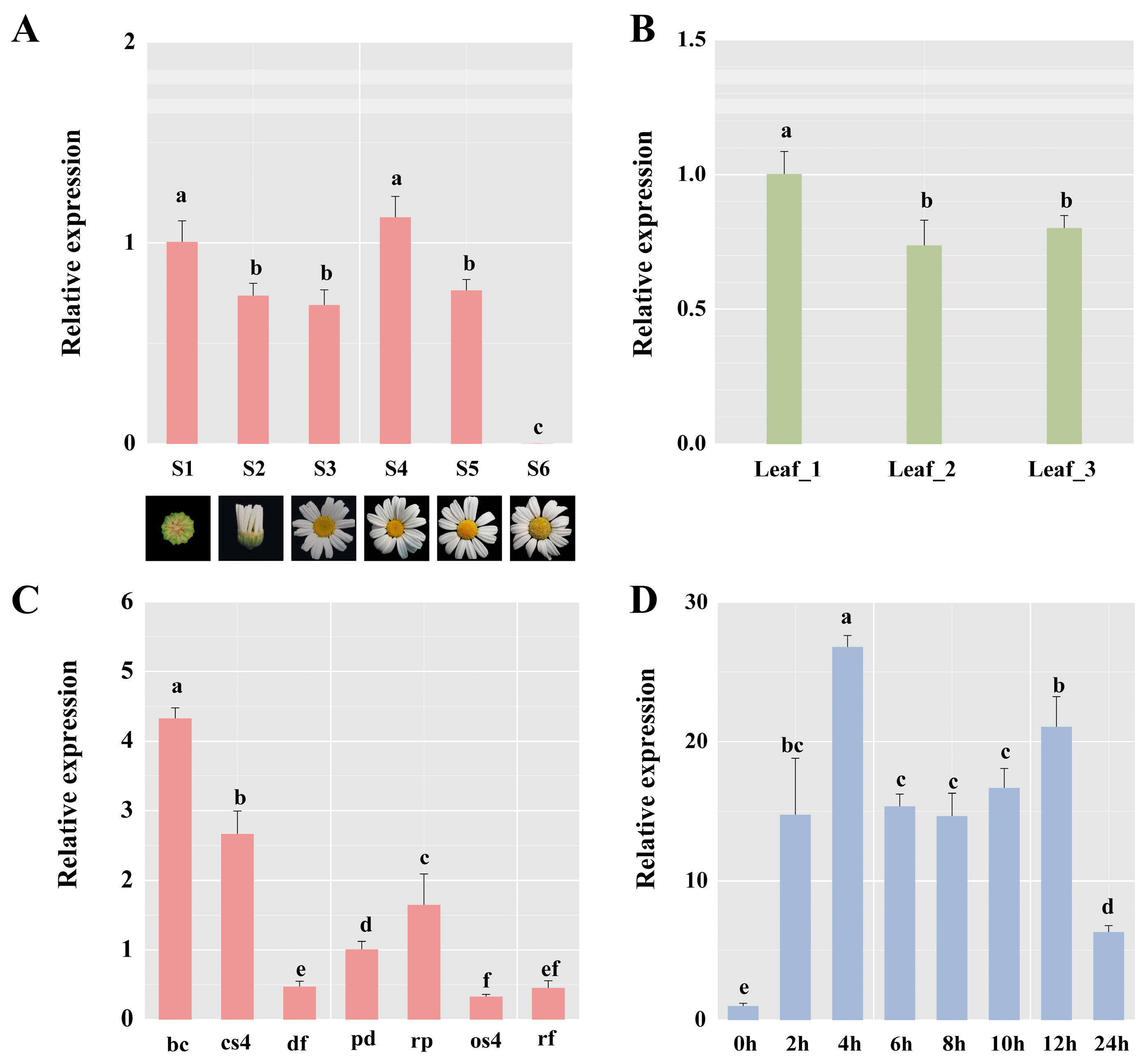
2.2. TcbHLH14 Gene Expression in Tissues
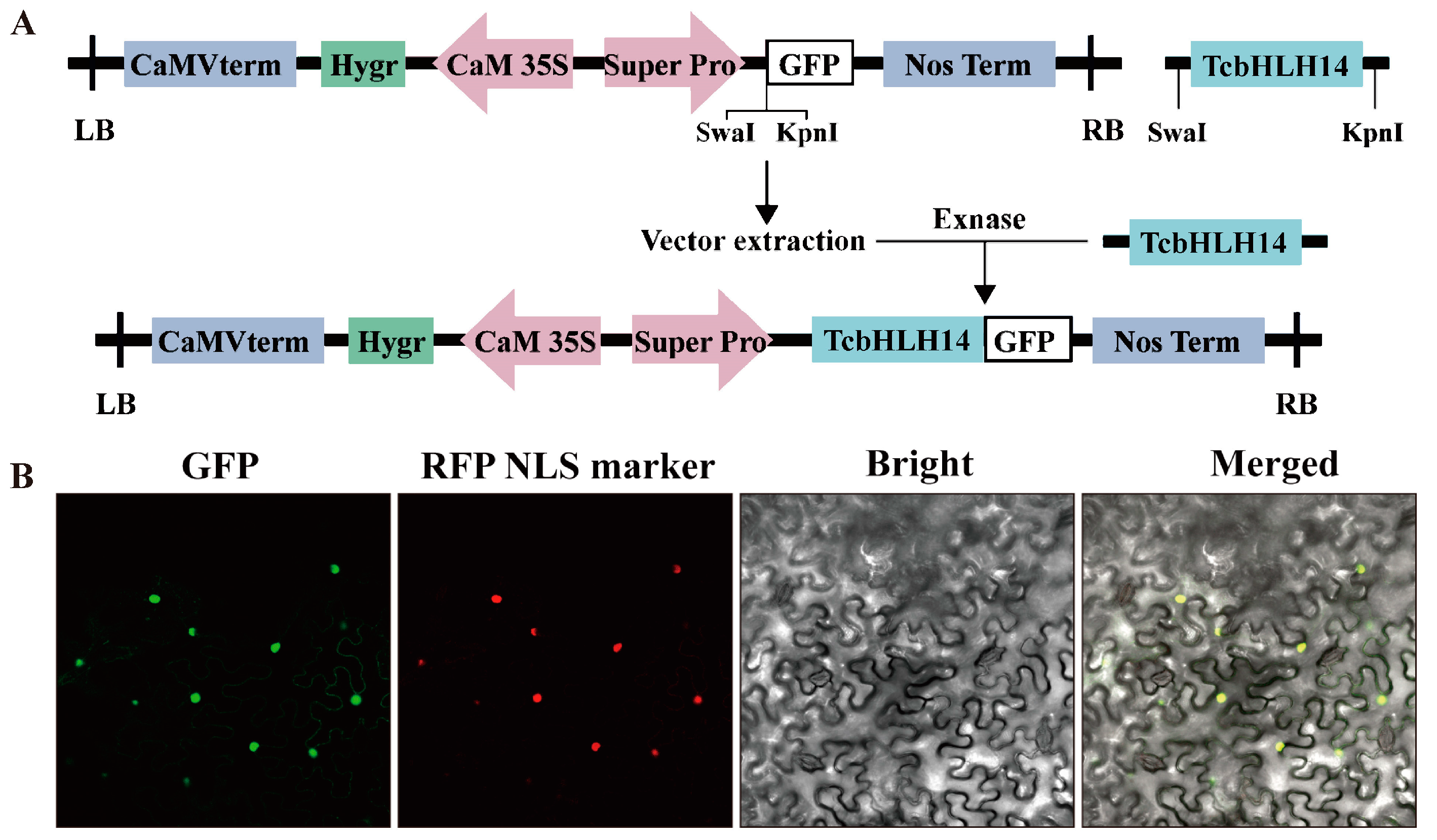
2.3. Subcellular Localization of TcbHLH14
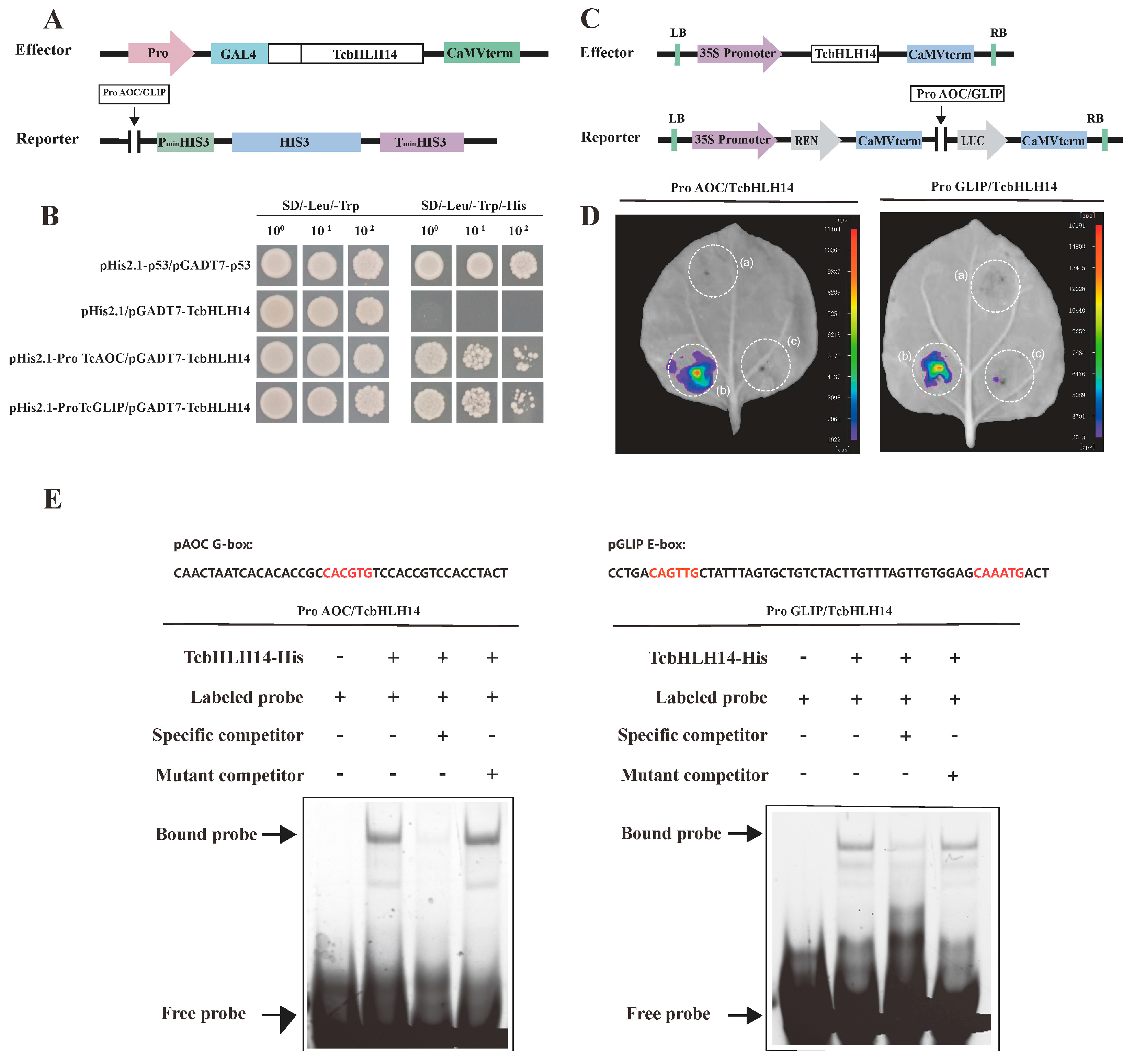
2.4. TcbHLH14 Directly Binds to the Promoters of the TcAOC and TcGLIP Genes
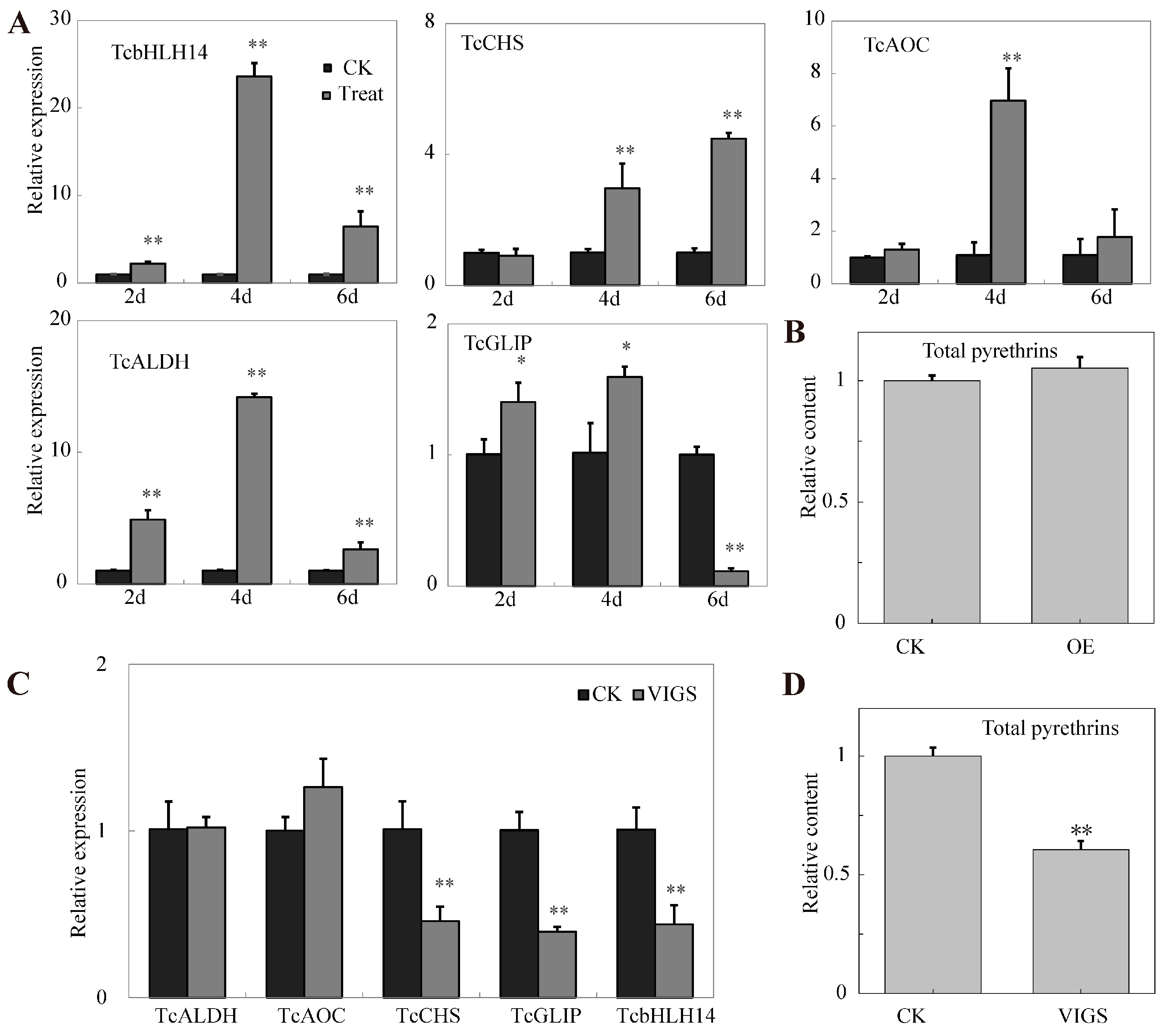
2.5. Transient Overexpression and VIGS of TcbHLH14 in T. cinerariifolium
3. Discussion
4. Materials and Methods
4.1. Sequence and Phylogenetic Analysis
4.2. Plant Materials
4.3. RNA Extraction and Reverse Transcription-Quantitative Polymerase Chain Reaction (RT-qPCR)
4.4. Subcellular Localization Analysis
4.5. Yeast One-Hybrid (Y1H) Assay
4.6. Dual-Luciferase (LUC) Transient Expression Assays
4.7. Electrophoretic Mobility Shift Assay (EMSA)
4.8. Virus-Induced Gene Silencing (VIGS)
4.9. Transient Overexpression of TcbHLH14
4.10. Pyrethrin Content Measurement by HPLC
4.11. Statistical Analysis
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Casida, J.E.; Quistad, G.B. Pyrethrum Flowers: Production, Chemistry, Toxicology, and Uses; Oxford University Press: New York, NY, USA, 1995. [Google Scholar]
- Jeran, N.; Grdiša, M.; Varga, F.; Šatović, Z.; Liber, Z.; Dabić, D.; Biošić, M. Pyrethrin from Dalmatian pyrethrum (Tanacetum cinerariifolium/Trevir./Sch. Bip.): Biosynthesis, biological activity, methods of extraction and determination. Phytochem. Rev. 2021, 20, 875–905. [Google Scholar] [CrossRef]
- Grdisa, M.; Jeran, N.; Varga, F.; Klepo, T.; Nincevic, T.; Satovic, Z. Accumulation patterns of six pyrethrin compounds across the flower developmental stages-comparative analysis in six natural dalmatian pyrethrum populations. Agronomy 2022, 12, 252. [Google Scholar] [CrossRef]
- Lybrand, D.B.; Xu, H.; Last, R.L.; Pichersky, E. How plants synthesize pyrethrins: Safe and biodegradable insecticides. Trends Plant Sci. 2020, 25, 1240–1251. [Google Scholar] [CrossRef] [PubMed]
- Matsuda, K.; Kikuta, Y.; Haba, A.; Nakayama, K.; Katsuda, Y.; Hatanaka, A.; Komai, K. Biosynthesis of pyrethrin I in seedlings of Chrysanthemum cinerariaefolium. Phytochemistry 2005, 66, 1529–1535. [Google Scholar] [CrossRef] [PubMed]
- Hu, H.; Li, J.J.; Delatte, T.; Vervoort, J.; Gao, L.; Verstappen, F.; Xiong, W.; Gan, J.; Jongsma, M.A.; Wang, C.Y. Modification of chrysanthemum odour and taste with chrysanthemol synthase induces strong dual resistance against cotton aphids. Plant Biotechnol. J. 2018, 16, 1434–1445. [Google Scholar] [CrossRef] [PubMed]
- Xu, H.; Moghe, G.D.; Wiegert-Rininger, K.; Schilmiller, A.L.; Barry, C.S.; Last, R.L.; Pichersky, E. Coexpression analysis identifies two oxidoreductases involved in the biosynthesis of the monoterpene acid moiety of natural pyrethrin insecticides in Tanacetum cinerariifolium. Plant Physiol. 2018, 176, 524–537. [Google Scholar] [CrossRef]
- Xu, H.; Li, W.; Schilmiller, A.L.; van Eekelen, H.; de Vos, R.C.H.; Jongsma, M.A.; Pichersky, E. Pyrethric acid of natural pyrethrin insecticide: Complete pathway elucidation and reconstitution in Nicotiana benthamiana. New Phytol. 2019, 223, 751–765. [Google Scholar] [CrossRef] [PubMed]
- Matsui, R.; Takiguchi, K.; Kuwata, N.; Oki, K.; Takahashi, K.; Matsuda, K.; Matsuura, H. Jasmonic acid is not a biosynthetic intermediate to produce the pyrethrolone moiety in pyrethrin II. Sci. Rep. 2020, 10, 6366. [Google Scholar] [CrossRef]
- Wasternack, C.; Strnad, M. Jasmonates are signals in the biosynthesis of secondary metabolites- Pathways, transcription factors and applied aspects-A brief review. New Biotechnol. 2019, 48, 1–11. [Google Scholar] [CrossRef]
- Yoeun, S.; Cho, K.; Han, O. Structural evidence for the substrate channeling of rice allene oxide cyclase in biologically analogous Nazarov reaction. Front. Chem. 2018, 6, 500. [Google Scholar] [CrossRef]
- Li, W.; Zhou, F.; Pichersky, E. Jasmone hydroxylase, a key enzyme in the synthesis of the alcohol moiety of pyrethrin insecticides. Plant Physiol. 2018, 177, 1498–1509. [Google Scholar] [CrossRef] [PubMed]
- Li, W.; Lybrand, D.B.; Zhou, F.; Last, R.L.; Pichersky, E. Pyrethrin biosynthesis: The cytochrome P450 oxidoreductase CYP82Q3 converts jasmolone to pyrethrolone. Plant Physiol. 2019, 181, 934–944. [Google Scholar] [CrossRef]
- Kikuta, Y.; Ueda, H.; Takahashi, M.; Mitsumori, T.; Yamada, G.; Sakamori, K.; Takeda, K.; Furutani, S.; Nakayama, K.; Katsuda, Y.; et al. Identification and characterization of a GDSL lipase-like protein that catalyzes the ester-forming reaction for pyrethrin biosynthesis in Tanacetum cinerariifolium- a new target for plant protection. Plant J. 2012, 71, 183–193. [Google Scholar] [CrossRef] [PubMed]
- Carretero-Paulet, L.; Galstyan, A.; Roig-Villanova, I.; Martinez-Garcia, J.F.; Bilbao-Castro, J.R.; Robertson, D.L. Genome-wide classification and evolutionary analysis of the bHLH family of transcription factors in Arabidopsis, poplar, rice, moss, and algae. Plant Physiol. 2010, 153, 1398–1412. [Google Scholar] [CrossRef] [PubMed]
- Heim, M.A.; Jakoby, M.; Werber, M.; Martin, C.; Weisshaar, B.; Bailey, P.C. The basic helix-loop-helix transcription factor family in plants: A genome-wide study of protein structure and functional diversity. Mol. Biol. Evol. 2003, 20, 735–747. [Google Scholar] [CrossRef]
- Fernández-Calvo, P.; Chini, A.; Fernández-Barbero, G.; Chico, J.-M.; Gimenez-Ibanez, S.; Geerinck, J.; Eeckhout, D.; Schweizer, F.; Godoy, M.; Franco-Zorrilla, J.M. The Arabidopsis bHLH transcription factors MYC3 and MYC4 are targets of JAZ repressors and act additively with MYC2 in the activation of jasmonate responses. Plant Cell 2011, 23, 701–715. [Google Scholar] [CrossRef]
- Pérez-Alonso, M.-M.; Sánchez-Parra, B.; Ortiz-García, P.; Santamaría, M.E.; Díaz, I.; Pollmann, S. Jasmonic acid-dependent myc transcription factors bind to a tandem g-box motif in the yucca8 and yucca9 promoters to regulate biotic stress responses. Int. J. Mol. Sci. 2021, 22, 9768. [Google Scholar] [CrossRef]
- Goklany, S.; Rizvi, N.F.; Loring, R.H.; Cram, E.J.; Lee-Parsons, C.W.T. Jasmonate-Dependent Alkaloid Biosynthesis in Catharanthus roseus Hairy Root Cultures Is Correlated with the Relative Expression of Orca and Zct Transcription Factors. Biotechnol. Prog. 2013, 29, 1367–1376. [Google Scholar] [CrossRef]
- Alfieri, M.; Vaccaro, M.C.; Cappetta, E.; Ambrosone, A.; De Tommasi, N.; Leone, A. Coactivation of MEP-biosynthetic genes and accumulation of abietane diterpenes in Salvia sclarea by heterologous expression of WRKY and MYC2 transcription factors. Sci. Rep. 2018, 8, 11009. [Google Scholar] [CrossRef]
- Xiang, L.; Jian, D.; Zhang, F.; Yang, C.; Bai, G.; Lan, X.; Chen, M.; Tang, K.; Liao, Z. The cold-induced bHLH transcription factor AabHLH112 promotes artemisinin biosynthesis in Artemisia annua. J. Exp. Bot. 2019, 70, 4835–4848. [Google Scholar] [CrossRef]
- Ji, Y.P.; Xiao, J.W.; Shen, Y.L.; Ma, D.M.; Li, Z.Q.; Pu, G.B.; Li, X.; Huang, L.L.; Liu, B.Y.; Ye, H.C.; et al. Cloning and Characterization of AabHLH1, a bHLH Transcription Factor that Positively Regulates Artemisinin Biosynthesis in Artemisia annua. Plant Cell Physiol. 2014, 55, 1592–1604. [Google Scholar] [CrossRef]
- Yang, N.; Zhou, W.; Su, J.; Wang, X.; Li, L.; Wang, L.; Cao, X.; Wang, Z. Overexpression of SmMYC2 increases the production of phenolic acids in Salvia miltiorrhiza. Front. Plant Sci. 2017, 8, 1804. [Google Scholar] [CrossRef] [PubMed]
- Dabiri, M.; Majdi, M.; Bahramnejad, B. Partial sequence isolation of DXS and AOS genes and gene expression analysis of terpenoids and pyrethrin biosynthetic pathway of Chrysanthemum cinerariaefolium under abiotic elicitation. Acta Physiol. Plant. 2020, 42, 30. [Google Scholar] [CrossRef]
- Xu, Y.H.; Liao, Y.C.; Lv, F.F.; Zhang, Z.; Sun, P.W.; Gao, Z.H.; Hu, K.P.; Sui, C.; Jin, Y.; Wei, J.H. Transcription factor AsMYC2 controls the jasmonate-responsive expression of ASS1 regulating sesquiterpene biosynthesis in Aquilaria sinensis (Lour.) Gilg. Plant Cell Physiol. 2017, 58, 1924–1933. [Google Scholar] [CrossRef] [PubMed]
- An, J.P.; Li, H.H.; Song, L.Q.; Su, L.; Liu, X.; You, C.X.; Wang, X.F.; Hao, Y.J. The molecular cloning and functional characterization of MdMYC2, a bHLH transcription factor in apple. Plant Physiol. Biochem. 2016, 108, 24–31. [Google Scholar] [CrossRef]
- Huo, Y.B.; Zhang, J.; Zhang, B.; Chen, L.; Zhang, X.; Zhu, C.A.S. MYC2 Transcription Factors TwMYC2a and TwMYC2b Negatively Regulate Triptolide Biosynthesis in Tripterygium wilfordii Hairy Roots. Plants 2021, 10, 679. [Google Scholar] [CrossRef]
- Zhang, C.; Xing, B.; Yang, D.; Ren, M.; Guo, H.; Yang, S.; Liang, Z. SmbHLH3 acts as a transcription repressor for both phenolic acids and tanshinone biosynthesis in Salvia miltiorrhiza hairy roots. Phytochemistry 2020, 169, 112183. [Google Scholar] [CrossRef]
- Dombrecht, B.; Xue, G.P.; Sprague, S.J.; Kirkegaard, J.A.; Ross, J.J.; Reid, J.B.; Fitt, G.P.; Sewelam, N.; Schenk, P.M.; Manners, J.M. MYC2 differentially modulates diverse jasmonate-dependent functions in Arabidopsis. Plant Cell 2007, 19, 2225–2245. [Google Scholar] [CrossRef]
- Zeng, T.; Li, J.-W.; Xu, Z.-Z.; Zhou, L.; Li, J.-J.; Yu, Q.; Luo, J.; Chan, Z.-L.; Jongsma, M.A.; Hu, H.; et al. TcMYC2 regulates Pyrethrin biosynthesis in Tanacetum cinerariifolium. Hortic. Res. 2022, 9, uhac178. [Google Scholar] [CrossRef]
- Ogawa, S.; Miyamoto, K.; Nemoto, K.; Sawasaki, T.; Yamane, H.; Nojiri, H.; Okada, K. OsMYC2, an essential factor for JA-inductive sakuranetin production in rice, interacts with MYC2-like proteins that enhance its transactivation ability. Sci. Rep. 2017, 7, 40175. [Google Scholar] [CrossRef]
- Lavy, M.; Zuker, A.; Lewinsohn, E.; Larkov, O.; Ravid, U.; Vainstein, A.; Weiss, D. Linalool and linalool oxide production in transgenic carnation flowers expressing the Clarkia breweri linalool synthase gene. Mol. Breed. 2002, 9, 103–111. [Google Scholar] [CrossRef]
- Sun, M.; Shi, M.; Wang, Y.; Huang, Q.; Yuan, T.; Wang, Q.; Wang, C.; Zhou, W.; Kai, G. The biosynthesis of phenolic acids is positively regulated by the JA-responsive transcription factor ERF115 in Salvia miltiorrhiza. J. Exp. Bot. 2019, 70, 243–254. [Google Scholar] [CrossRef] [PubMed]
- Lu, X.; Zhang, L.; Zhang, F.; Jiang, W.; Shen, Q.; Zhang, L.; Lv, Z.; Wang, G.; Tang, K. A a ORA, a trichome-specific AP2/ERF transcription factor of Artemisia annua, is a positive regulator in the artemisinin biosynthetic pathway and in disease resistance to Botrytis cinerea. New Phytol. 2013, 198, 1191–1202. [Google Scholar] [CrossRef]
- Yang, R.; Wang, S.; Zou, H.; Li, L.; Li, Y.; Wang, D.; Xu, H.; Cao, X. R2R3-MYB transcription factor SmMYB52 positively regulates biosynthesis of salvianolic acid B and inhibits root growth in Salvia miltiorrhiza. Int. J. Mol. Sci. 2021, 22, 9538. [Google Scholar] [CrossRef] [PubMed]
- Lu, S.; Ye, J.; Zhu, K.; Zhang, Y.; Zhang, M.; Xu, Q.; Deng, X. A fruit ripening-associated transcription factor CsMADS5 positively regulates carotenoid biosynthesis in citrus. J. Exp. Bot. 2021, 72, 3028–3043. [Google Scholar] [CrossRef]
- Kumar, S.; Stecher, G.; Li, M.; Knyaz, C.; Tamura, K. MEGA X: Molecular evolutionary genetics analysis across computing platforms. Mol. Biol. Evol. 2018, 35, 1547–1549. [Google Scholar] [CrossRef]
- Letunic, I.; Bork, P. Interactive Tree Of Life (iTOL) v5: An online tool for phylogenetic tree display and annotation. Nucleic Acids Res. 2021, 49, W293–W296. [Google Scholar] [CrossRef] [PubMed]
- Ramirez, A.M. Pyrethrum Secondary Metabolism: Biosynthesis, Localization and Ecology of Defence Compounds. Ph.D. Thesis, Wageningen University, Wageningen, The Netherlands, 2013. [Google Scholar]
- Zito, S.W.; Zieg, R.G.; Staba, E.J. Distribution of pyrethrins in oil glands and leaf tissue of Chrysanthemum cinerariaefolium. Planta Med. 1983, 47, 205–207. [Google Scholar] [CrossRef]
- Zeng, T.; Li, J.-W.; Zhou, L.; Xu, Z.-Z.; Li, J.-J.; Hu, H.; Luo, J.; Zheng, R.-R.; Wang, Y.-Y.; Wang, C.-Y. Transcriptional responses and GCMS analysis for the biosynthesis of pyrethrins and volatile terpenes in Tanacetum coccineum. Int. J. Mol. Sci. 2021, 22, 13005. [Google Scholar] [CrossRef]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods 2001, 408, 402–408. [Google Scholar] [CrossRef]
- Xu, H.; Luo, D.; Zhang, F. DcWRKY75 promotes ethylene induced petal senescence in carnation (Dianthus caryophyllus L.). Plant J. 2021, 108, 1473–1492. [Google Scholar] [CrossRef] [PubMed]
- Lück, S.; Kreszies, T.; Strickert, M.; Schweizer, P.; Kuhlmann, M.; Douchkov, D. siRNA-Finder (si-Fi) Software for RNAi-Target Design and Off-Target Prediction. Front. Plant Sci. 2019, 10, 1023. [Google Scholar] [CrossRef] [PubMed]
- Senthil-Kumar, M.; Mysore, K.S. Tobacco rattle virus-based virus-induced gene silencing in Nicotiana benthamiana. Nat. Protoc. 2014, 9, 1549–1562. [Google Scholar] [CrossRef] [PubMed]
- Zhou, L.; Li, J.; Zeng, T.; Xu, Z.; Luo, J.; Zheng, R.; Wang, Y.; Wang, C. TcMYB8, a R3-MYB transcription factor, positively regulates pyrethrin biosynthesis in Tanacetum Cinerariifolium. Int. J. Mol. Sci. 2022, 23, 12186. [Google Scholar] [CrossRef]
- Li, J.; Jongsma, M.A.; Wang, C.Y. Comparative analysis of pyrethrin content improvement by mass selection, family selection and polycross in pyrethrum [Tanacetum cinerariifolium (Trevir.) Sch.Bip.] populations. Ind. Crops Prod. 2014, 53, 268–273. [Google Scholar] [CrossRef]
- Xu, Z.; Zeng, T.; Li, J.; Zhou, L.; Li, J.; Luo, J.; Zheng, R.; Wang, Y.; Hu, H.; Wang, C. TcbZIP60 positively regulates pyrethrins biosynthesis in Tanacetum cinerariifolium. Front. Plant Sci. 2023, 14, 521. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zeng, T.; Yu, Q.; Shang, J.; Xu, Z.; Zhou, L.; Li, W.; Li, J.; Hu, H.; Zhu, L.; Li, J.; et al. TcbHLH14 a Jasmonate Associated MYC2-like Transcription Factor Positively Regulates Pyrethrin Biosynthesis in Tanacetum cinerariifolium. Int. J. Mol. Sci. 2023, 24, 7379. https://doi.org/10.3390/ijms24087379
Zeng T, Yu Q, Shang J, Xu Z, Zhou L, Li W, Li J, Hu H, Zhu L, Li J, et al. TcbHLH14 a Jasmonate Associated MYC2-like Transcription Factor Positively Regulates Pyrethrin Biosynthesis in Tanacetum cinerariifolium. International Journal of Molecular Sciences. 2023; 24(8):7379. https://doi.org/10.3390/ijms24087379
Chicago/Turabian StyleZeng, Tuo, Qin Yu, Junzhong Shang, Zhizhuo Xu, Li Zhou, Wei Li, Jinjin Li, Hao Hu, Liyong Zhu, Jiawen Li, and et al. 2023. "TcbHLH14 a Jasmonate Associated MYC2-like Transcription Factor Positively Regulates Pyrethrin Biosynthesis in Tanacetum cinerariifolium" International Journal of Molecular Sciences 24, no. 8: 7379. https://doi.org/10.3390/ijms24087379
APA StyleZeng, T., Yu, Q., Shang, J., Xu, Z., Zhou, L., Li, W., Li, J., Hu, H., Zhu, L., Li, J., & Wang, C. (2023). TcbHLH14 a Jasmonate Associated MYC2-like Transcription Factor Positively Regulates Pyrethrin Biosynthesis in Tanacetum cinerariifolium. International Journal of Molecular Sciences, 24(8), 7379. https://doi.org/10.3390/ijms24087379






