Concomitant Activation of OsNAS2 and OsNAS3 Contributes to the Enhanced Accumulation of Iron and Zinc in Rice
Abstract
1. Introduction
2. Results
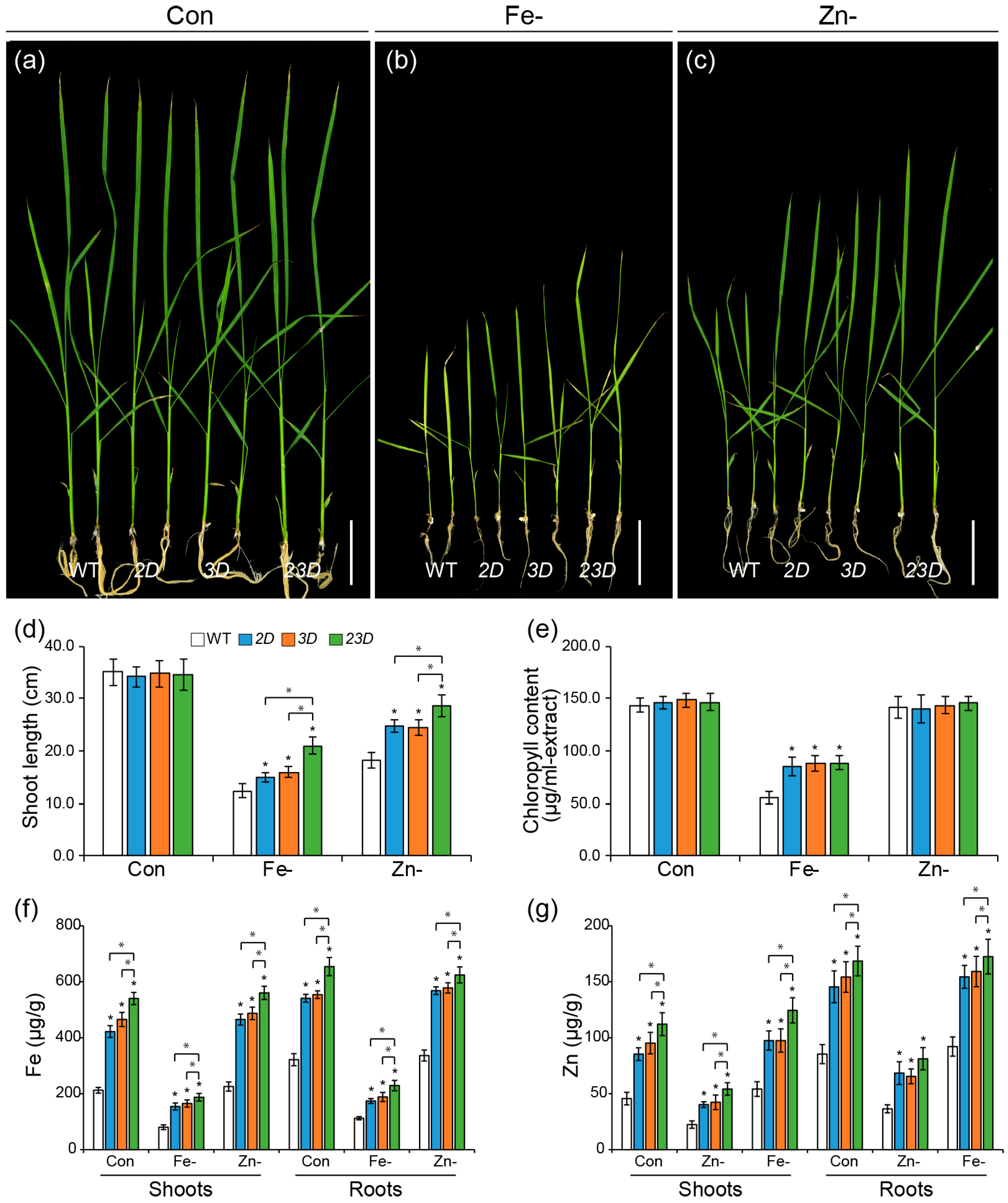
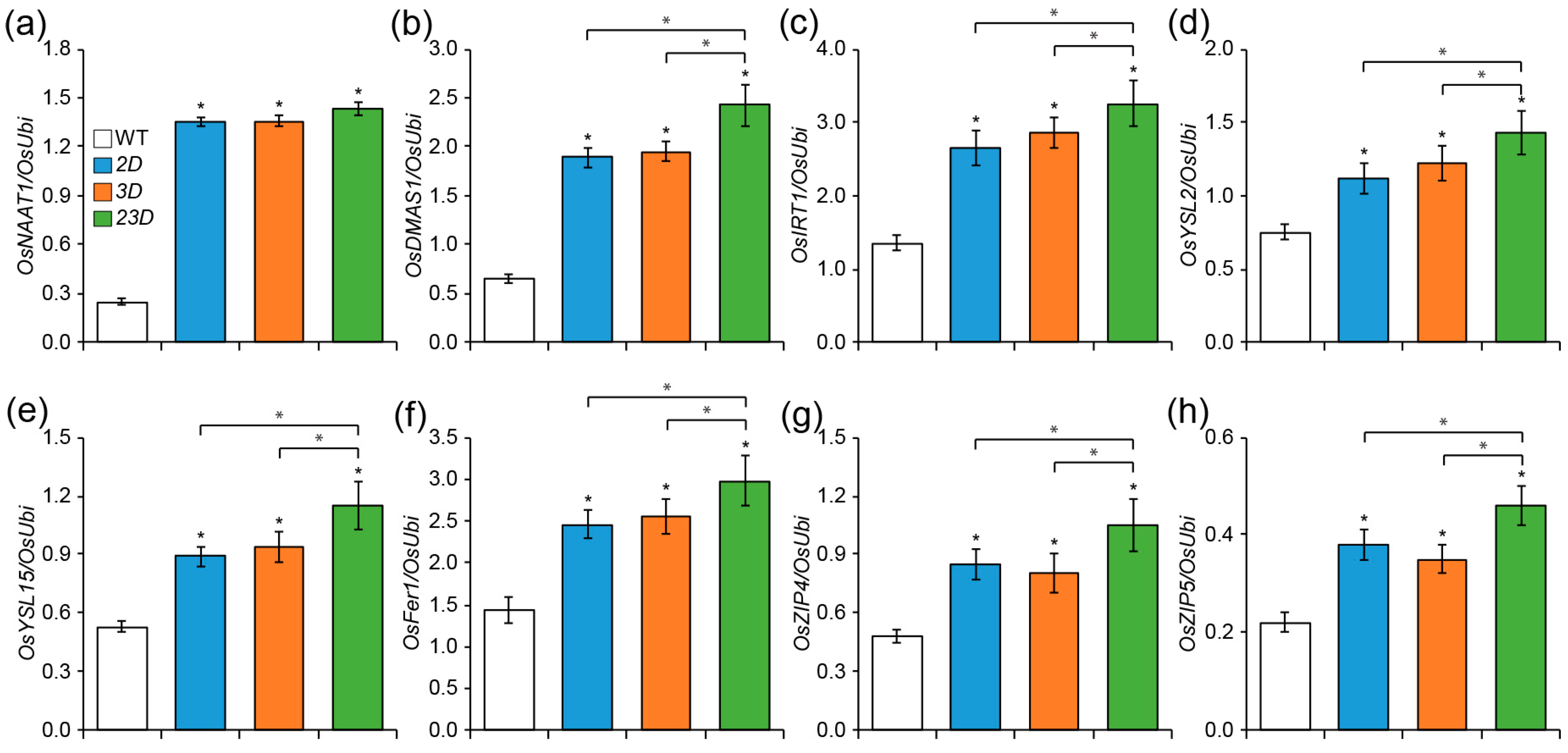
2.1. Isolation of Double-Activation-Tagged Mutant and Its Enhanced Tolerance to Fe and Zn Deficiencies
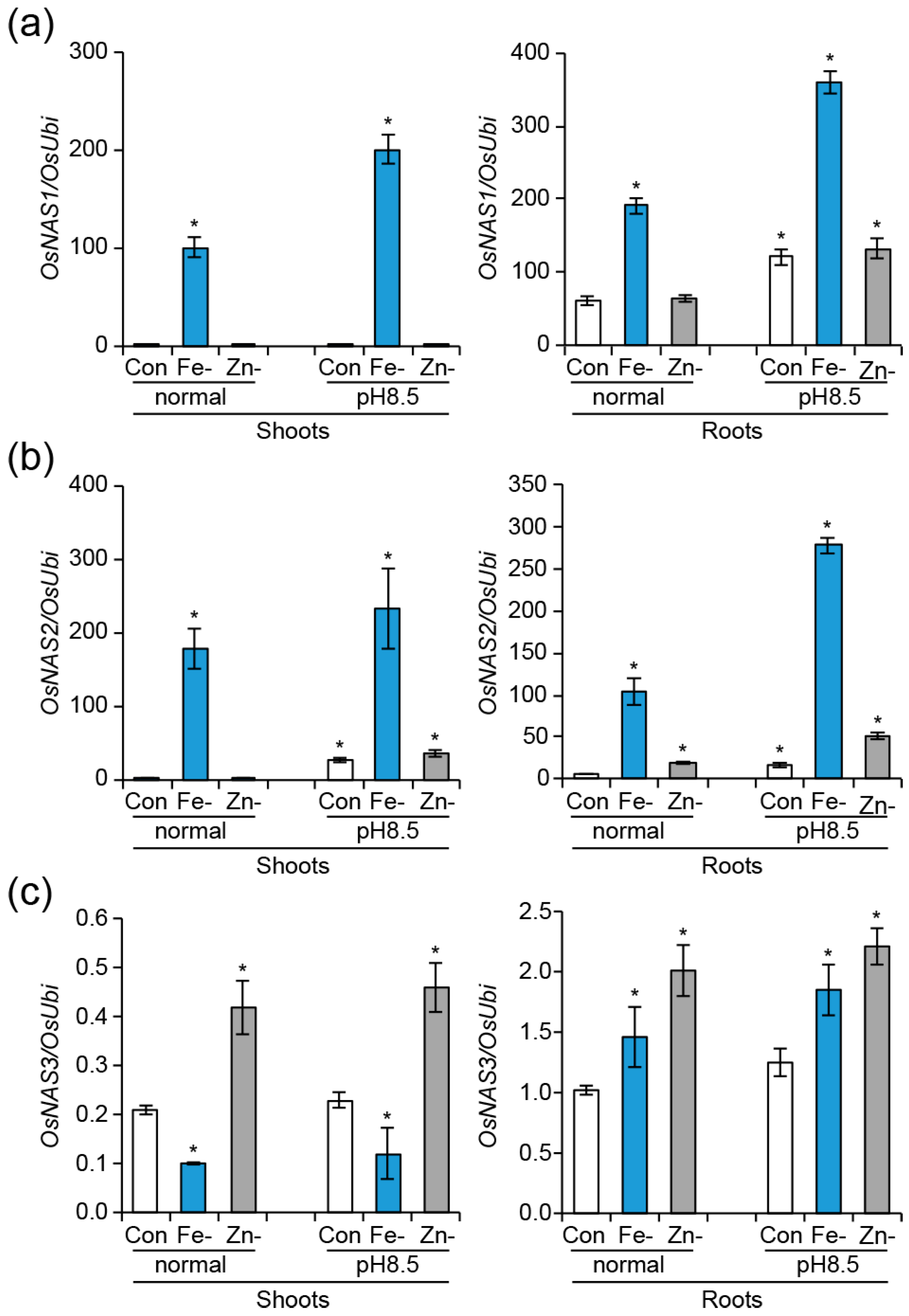
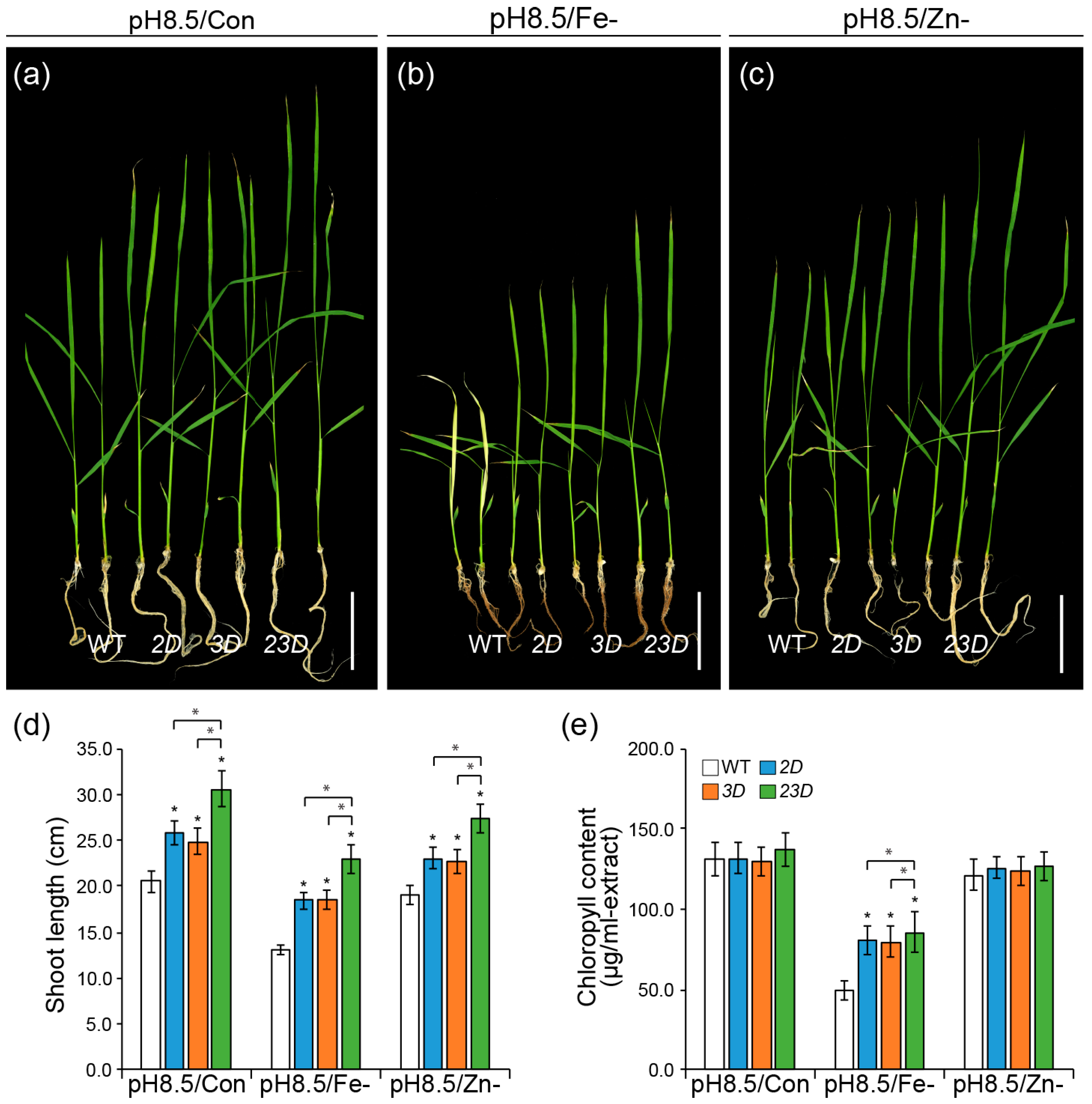
2.2. Expression Analysis of Three OsNAS Genes and the Comparison of Seedling Growth under High pH Conditions
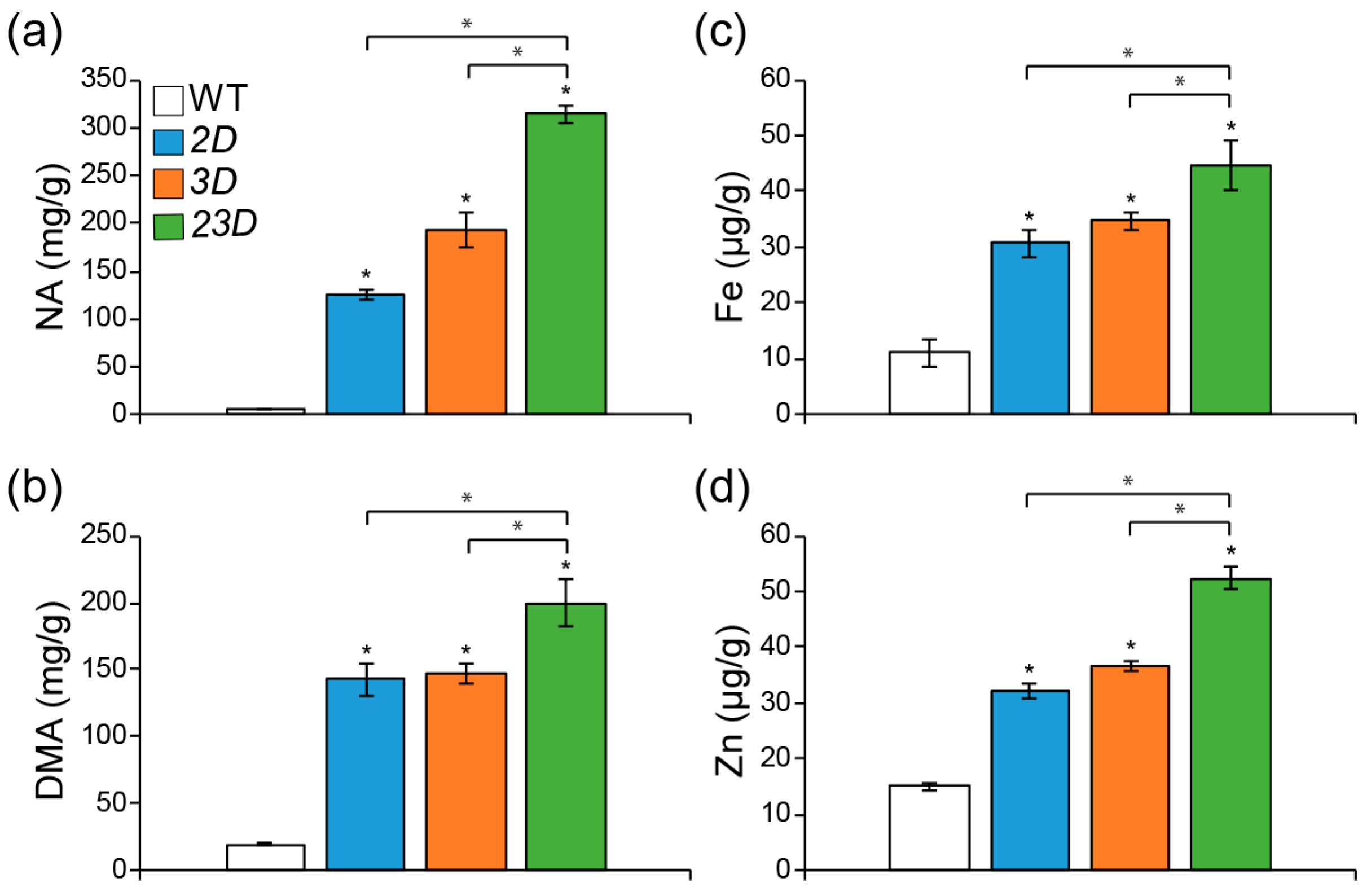
2.3. Increased Expression of Two OsNAS Genes Boosted Fe and Zn Contents in Mature Seeds
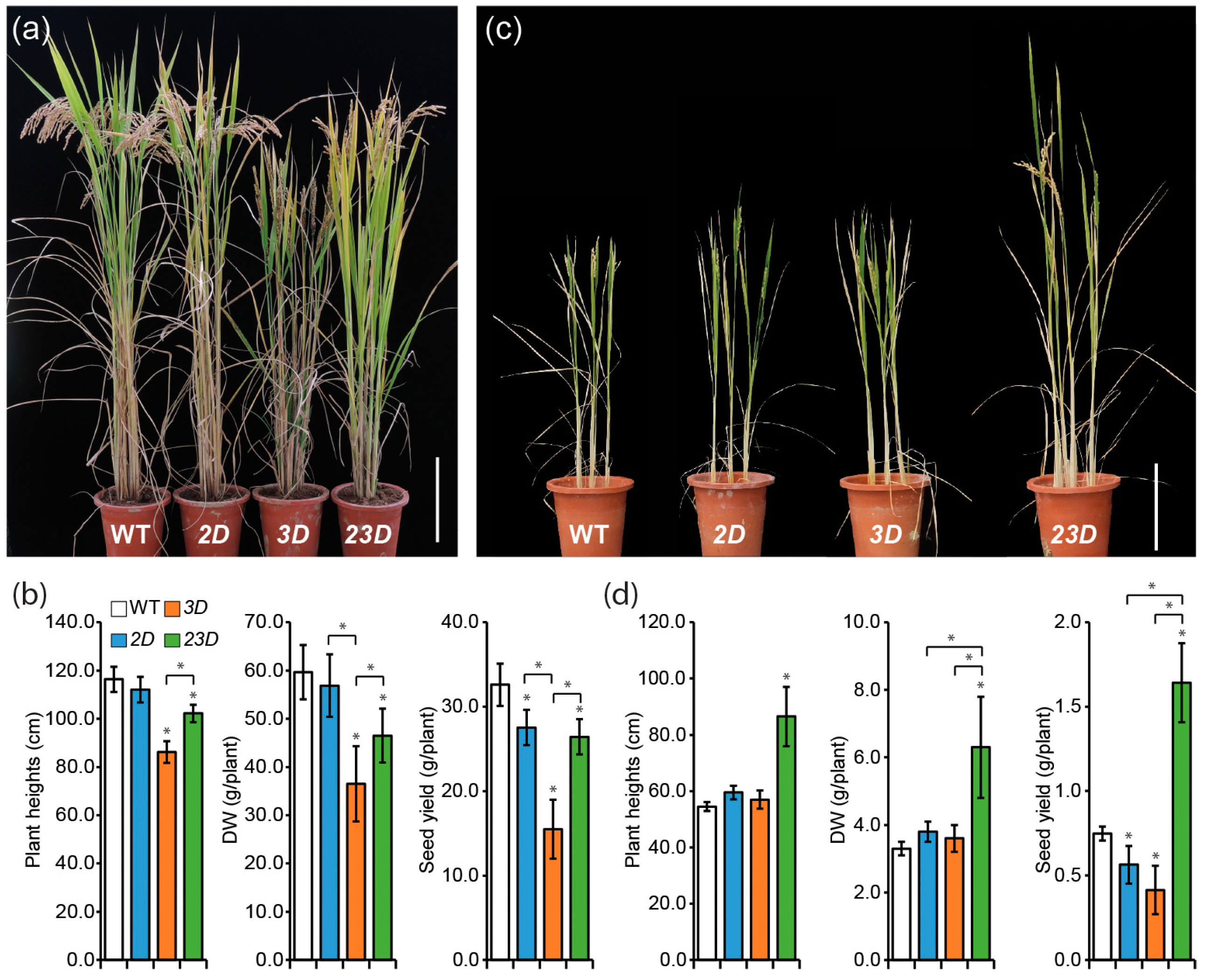
2.4. Increased Tolerance to Excess Metals during the Seedling Growth
3. Discussion
4. Materials and Methods
4.1. Plant Materials and Growth
4.2. Hydroponic Culture for the Growth Test under Fe and Zn Deficiencies
4.3. Seedling Growth on MS Solid Medium under Fe and Zn Deficiencies
4.4. RNA Isolation and Quantitative Real-Time PCR
4.5. Measurement of Chlorophyll Concentrations
4.6. Element Analysis in Plant Tissues
4.7. Perls’ Staining of the Mature Seeds
4.8. Determination of NA and DMA Levels
4.9. Excess Metal Tolerance
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Stangoulis, J.C.R.; Knez, M. Biofortification of major crop plants with iron and zinc—Achievements and future directions. Plant Soil 2022, 474, 57–76. [Google Scholar] [CrossRef]
- Wairich, A.; Ricachenevsky, F.K.; Lee, S. A tale of two metals: Biofortification of rice grains with iron and zinc. Front. Plant Sci. 2022, 13, 944624. [Google Scholar] [CrossRef] [PubMed]
- Wu, T.-Y.; Gruissem, W.; Bhullar, N.K. Facilitated citrate-dependent iron translocation increases rice endosperm iron and zinc concentrations. Plant Sci. 2018, 270, 13–22. [Google Scholar] [CrossRef] [PubMed]
- Clemens, S. The cell biology of zinc. J. Exp. Bot. 2021, 73, 1688–1698. [Google Scholar] [CrossRef] [PubMed]
- Colvin, R.A.; Holmes, W.R.; Fontaine, C.P.; Maret, W. Cytosolic zinc buffering and muffling: Their role in intracellular zinc homeostasis. Metallomics 2010, 2, 306–317. [Google Scholar] [CrossRef] [PubMed]
- Muñoz, M.; Acheson, A.G.; Auerbach, M.; Besser, M.; Habler, O.; Kehlet, H.; Liumbruno, G.M.; Lasocki, S.; Meybohm, P.; Baikady, R.R.; et al. International consensus statement on the peri-operative management of anaemia and iron deficiency. Anaesthesia 2017, 72, 233–247. [Google Scholar] [CrossRef]
- Piskin, E.; Cianciosi, D.; Gulec, S.; Tomas, M.; Capanoglu, E. Iron Absorption: Factors, Limitations, and Improvement Methods. ACS Omega 2022, 7, 20441–20456. [Google Scholar] [CrossRef]
- Ozyildirim, S.; Baltaci, S.B. Cardiovascular Diseases and Zinc. Biol. Trace Element Res. 2022, 201, 1615–1626. [Google Scholar] [CrossRef]
- Anusha, G.; Rao, D.S.; Jaldhani, V.; Beulah, P.; Neeraja, C.N.; Gireesh, C.; Anantha, M.S.; Suneetha, K.; Santhosha, R.; Prasad, A.S.H.; et al. Grain Fe and Zn content, heterosis, combining ability and its association with grain yield in irrigated and aerobic rice. Sci. Rep. 2021, 11, 10579. [Google Scholar] [CrossRef]
- Assunção, A.G.; Cakmak, I.; Clemens, S.; González-Guerrero, M.; Nawrocki, A.; Thomine, S. Micronutrient homeostasis in plants for more sustainable agriculture and healthier human nutrition. J. Exp. Bot. 2022, 73, 1789–1799. [Google Scholar] [CrossRef]
- Garg, M.; Sharma, N.; Sharma, S.; Kapoor, P.; Kumar, A.; Chunduri, V.; Arora, P. Biofortified Crops Generated by Breeding, Agronomy, and Transgenic Approaches Are Improving Lives of Millions of People around the World. Front. Nutr. 2018, 5, 12. [Google Scholar] [CrossRef] [PubMed]
- Laffont, C.; Arnoux, P. The ancient roots of nicotianamine: Diversity, role, regulation and evolution of nicotianamine-like metallophores. Metallomics 2020, 12, 1480–1493. [Google Scholar] [CrossRef] [PubMed]
- Nozoye, T. The Nicotianamine Synthase Gene Is a Useful Candidate for Improving the Nutritional Qualities and Fe-Deficiency Tolerance of Various Crops. Front. Plant Sci. 2018, 9, 340. [Google Scholar] [CrossRef] [PubMed]
- Shojima, S.; Nishizawa, N.-K.; Mori, S. Establishment of a Cell-Free System for the Biosynthesis of Nicotianamine. Plant Cell Physiol. 1989, 30, 673–677. [Google Scholar] [CrossRef]
- Inoue, H.; Higuchi, K.; Takahashi, M.; Nakanishi, H.; Mori, S.; Nishizawa, N.K. Three rice nicotianamine synthase genes, OsNAS1, OsNAS2, and OsNAS3 are expressed in cells involved in long-distance transport of iron and differentially regulated by iron. Plant J. 2003, 36, 366–381. [Google Scholar] [CrossRef]
- Johnson, A.A.T.; Kyriacou, B.; Callahan, D.L.; Carruthers, L.; Stangoulis, J.; Lombi, E.; Tester, M. Constitutive Overexpression of the OsNAS Gene Family Reveals Single-Gene Strategies for Effective Iron- and Zinc-Biofortification of Rice Endosperm. PLoS ONE 2011, 6, e24476. [Google Scholar] [CrossRef]
- Lee, S.; Jeon, U.S.; Lee, S.J.; Kim, Y.-K.; Persson, D.P.; Husted, S.; Schjørring, J.K.; Kakei, Y.; Masuda, H.; Nishizawa, N.K.; et al. Iron fortification of rice seeds through activation of the nicotianamine synthase gene. Proc. Natl. Acad. Sci. USA 2009, 106, 22014–22019. [Google Scholar] [CrossRef]
- Lee, S.; Persson, D.P.; Hansen, T.H.; Husted, S.; Schjoerring, J.K.; Kim, Y.-S.; Jeon, U.S.; Kim, Y.-K.; Kakei, Y.; Masuda, H.; et al. Bio-available zinc in rice seeds is increased by activation tagging of nicotianamine synthase. Plant Biotechnol. J. 2011, 9, 865–873. [Google Scholar] [CrossRef]
- Lee, S.; Kim, Y.-S.; Jeon, U.S.; Kim, Y.-K.; Schjoerring, J.K.; An, G. Activation of Rice nicotianamine synthase 2 (OsNAS2) enhances iron availability for biofortification. Mol. Cells 2012, 33, 269–275. [Google Scholar] [CrossRef]
- Masuda, H.; Usuda, K.; Kobayashi, T.; Ishimaru, Y.; Kakei, Y.; Takahashi, M.; Higuchi, K.; Nakanishi, H.; Mori, S.; Nishizawa, N.K. Overexpression of the Barley Nicotianamine Synthase Gene HvNAS1 Increases Iron and Zinc Concentrations in Rice Grains. Rice 2009, 2, 155–166. [Google Scholar] [CrossRef]
- Usuda, K.; Wada, Y.; Ishimaru, Y.; Kobayashi, T.; Takahashi, M.; Nakanishi, H.; Nagato, Y.; Mori, S.; Nishizawa, N.K. Genetically engineered rice containing larger amounts of nicotianamine to enhance the antihypertensive effect. Plant Biotechnol. J. 2009, 7, 87–95. [Google Scholar] [CrossRef] [PubMed]
- Moreno-Moyano, L.T.; Bonneau, J.P.; Sánchez-Palacios, J.T.; Tohme, J.; Johnson, A.A.T. Association of Increased Grain Iron and Zinc Concentrations with Agro-morphological Traits of Biofortified Rice. Front. Plant Sci. 2016, 7, 1463. [Google Scholar] [CrossRef] [PubMed]
- Zheng, L.; Cheng, Z.; Ai, C.; Jiang, X.; Bei, X.; Zheng, Y.; Glahn, R.P.; Welch, R.M.; Miller, D.D.; Lei, X.G.; et al. Nicotianamine, a Novel Enhancer of Rice Iron Bioavailability to Humans. PLoS ONE 2010, 5, e10190. [Google Scholar] [CrossRef] [PubMed]
- Wirth, J.; Poletti, S.; Aeschlimann, B.; Yakandawala, N.; Drosse, B.; Osorio, S.; Tohge, T.; Fernie, A.R.; Günther, D.; Gruissem, W.; et al. Rice endosperm iron biofortification by targeted and synergistic action of nicotianamine synthase and ferritin. Plant Biotechnol. J. 2009, 7, 631–644. [Google Scholar] [CrossRef] [PubMed]
- Banakar, R.; Fernández, A.A.; Díaz-Benito, P.; Abadia, J.; Capell, T.; Christou, P. Phytosiderophores determine thresholds for iron and zinc accumulation in biofortified rice endosperm while inhibiting the accumulation of cadmium. J. Exp. Bot. 2017, 68, 4983–4995. [Google Scholar] [CrossRef] [PubMed]
- Díaz-Benito, P.; Banakar, R.; Rodríguez-Menéndez, S.; Capell, T.; Pereiro, R.; Christou, P.; Abadía, J.; Fernández, B.; Álvarez-Fernández, A. Iron and zinc in the embryo and endosperm of rice (Oryza sativa L.) seeds in contrasting 2′-deoxymugineic acid/nicotianamine scenarios. Front. Plant Sci. 2018, 9, 1190. [Google Scholar] [CrossRef]
- Trijatmiko, K.R.; Dueñas, C.; Tsakirpaloglou, N.; Torrizo, L.; Arines, F.M.; Adeva, C.; Balindong, J.; Oliva, N.; Sapasap, M.V.; Borrero, J.; et al. Biofortified indica rice attains iron and zinc nutrition dietary targets in the field. Sci. Rep. 2016, 6, 19792. [Google Scholar] [CrossRef]
- Boonyaves, K.; Wu, T.-Y.; Gruissem, W.; Bhullar, N.K. Enhanced grain iron levels in rice expressing an iron-regulated metal transporter, nicotianamine synthase, and ferritin gene cassette. Front. Plant Sci. 2017, 8, 130. [Google Scholar] [CrossRef]
- Kawakami, Y.; Gruissem, W.; Bhullar, N.K. Novel rice iron biofortification approaches using expression of ZmYS1 and OsTOM1 controlled by tissue-specific promoters. J. Exp. Bot. 2022, 73, 5440–5459. [Google Scholar] [CrossRef]
- Bashir, K.; Inoue, H.; Nagasaka, S.; Takahashi, M.; Nakanishi, H.; Mori, S.; Nishizawa, N.K. Cloning and Characterization of Deoxymugineic Acid Synthase Genes from Graminaceous Plants. J. Biol. Chem. 2006, 281, 32395–32402. [Google Scholar] [CrossRef]
- Suzuki, M.; Tsukamoto, T.; Inoue, H.; Watanabe, S.; Matsuhashi, S.; Takahashi, M.; Nakanishi, H.; Mori, S.; Nishizawa, N.K. Deoxymugineic acid increases Zn translocation in Zn-deficient rice plants. Plant Mol. Biol. 2008, 66, 609–617. [Google Scholar] [CrossRef] [PubMed]
- Inoue, H.; Kobayashi, T.; Nozoye, T.; Takahashi, M.; Kakei, Y.; Suzuki, K.; Nakazono, M.; Nakanishi, H.; Mori, S.; Nishizawa, N.K. Rice OsYSL15 Is an Iron-regulated Iron(III)-Deoxymugineic Acid Transporter Expressed in the Roots and Is Essential for Iron Uptake in Early Growth of the Seedlings. J. Biol. Chem. 2009, 284, 3470–3479. [Google Scholar] [CrossRef] [PubMed]
- Ishimaru, Y.; Suzuki, M.; Tsukamoto, T.; Suzuki, K.; Nakazono, M.; Kobayashi, T.; Wada, Y.; Watanabe, S.; Matsuhashi, S.; Takahashi, M.; et al. Rice plants take up iron as an Fe3+-phytosiderophore and as Fe2+. Plant J. 2006, 45, 335–346. [Google Scholar] [CrossRef] [PubMed]
- Lee, S.; An, G. Over-expression of OsIRT1 leads to increased iron and zinc accumulations in rice. Plant. Cell Environ. 2009, 32, 408–416. [Google Scholar] [CrossRef]
- Lee, S.; Chiecko, J.C.; Kim, S.A.; Walker, E.L.; Lee, Y.; Guerinot, M.L.; An, G. Disruption of OsYSL15 Leads to Iron Inefficiency in Rice Plants. Plant Physiol. 2009, 150, 786–800. [Google Scholar] [CrossRef]
- Ishimaru, Y.; Masuda, H.; Bashir, K.; Inoue, H.; Tsukamoto, T.; Takahashi, M.; Nakanishi, H.; Aoki, N.; Hirose, T.; Ohsugi, R.; et al. Rice metal-nicotianamine transporter, OsYSL2, is required for the long-distance transport of iron and manganese. Plant J. 2010, 62, 379–390. [Google Scholar] [CrossRef]
- Koike, S.; Inoue, H.; Mizuno, D.; Takahashi, M.; Nakanishi, H.; Mori, S.; Nishizawa, N.K. OsYSL2 is a rice metal-nicotianamine transporter that is regulated by iron and expressed in the phloem. Plant J. 2004, 39, 415–424. [Google Scholar] [CrossRef]
- Harrison, P.M.; Arosio, P. The ferritins: Molecular properties, iron storage function and cellular regulation. Biochim. Biophys. Acta (BBA) Bioenerg. 1996, 1275, 161–203. [Google Scholar] [CrossRef]
- Ishimaru, Y.; Suzuki, M.; Kobayashi, T.; Takahashi, M.; Nakanishi, H.; Mori, S.; Nishizawa, N.K. OsZIP4, a novel zinc-regulated zinc transporter in rice. J. Exp. Bot. 2005, 56, 3207–3214. [Google Scholar] [CrossRef]
- Lee, S.; Jeong, H.J.; Kim, S.A.; Lee, J.; Guerinot, M.L.; An, G. OsZIP5 is a plasma membrane zinc transporter in rice. Plant Mol. Biol. 2010, 73, 507–517. [Google Scholar] [CrossRef]
- Penn, C.J.; Camberato, J.J. Critical Review on Soil Chemical Processes that Control How Soil pH Affects Phosphorus Availability to Plants. Agriculture 2019, 9, 120. [Google Scholar] [CrossRef]
- Desta, M.; Broadley, M.; McGrath, S.; Hernandez-Allica, J.; Hassall, K.; Gameda, S.; Amede, T.; Haefele, S. Plant Available Zinc Is Influenced by Landscape Position in the Amhara Region, Ethiopia. Plants 2021, 10, 254. [Google Scholar] [CrossRef] [PubMed]
- Higuchi, K.; Suzuki, K.; Nakanishi, H.; Yamaguchi, H.; Nishizawa, N.-K.; Mori, S. Cloning of Nicotianamine Synthase Genes, Novel Genes Involved in the Biosynthesis of Phytosiderophores. Plant Physiol. 1999, 119, 471–480. [Google Scholar] [CrossRef]
- Stein, R.J.; Ricachenevsky, F.K.; Fett, J.P. Differential regulation of the two rice ferritin genes (OsFER1 and OsFER2). Plant Sci. 2009, 177, 563–569. [Google Scholar] [CrossRef]
- Aung, M.S.; Masuda, H.; Nozoye, T.; Kobayashi, T.; Jeon, J.S.; An, G.; Nishizawa, N.K. Nicotianamine Synthesis by OsNAS3 Is Important for Mitigating Iron Excess Stress in Rice. Front. Plant Sci. 2019, 10, 660. [Google Scholar] [CrossRef]
- Gentili, R.; Ambrosini, R.; Montagnani, C.; Caronni, S.; Citterio, S. Effect of soil ph on the growth, reproductive investment and pollen allergenicity of Ambrosia artemisiifolia L. Front. Plant Sci. 2018, 9, 1335. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.; Barak, P. Iron nutrition of plants in calcareous soils. Adv. Agron. 1982, 35, 217–240. [Google Scholar] [CrossRef]
- George, E.; Horst, W.J.; Neumann, E. Adaptation of Plants to Adverse Chemical Soil Conditions. In Marschner’s Mineral Nutrition of Higher Plants, 3rd ed.; Marschner, P., Ed.; Academic Press: London, UK, 2012; pp. 409–472. [Google Scholar] [CrossRef]
- Yoshida, S.; Forno, D.A.; Cock, J.H.; Gomez, K.A. Laboratory Manual for Physiological Studies of Rice; IRRI: Laguna, Philippines, 1976; p. 83. [Google Scholar]
- Murashige, T.; Skoog, F. A Revised Medium for Rapid Growth and Bioassays with Tobacco Tissue Cultures. Physiol. Plant. 1962, 15, 473–497. [Google Scholar] [CrossRef]
- Arnon, D.I. Copper enzymes in isolated chloroplasts. Polyphenoloxidase in Beta vulgaris. Plant Physiol. 1949, 24, 1–15. [Google Scholar] [CrossRef]
- Vasconcelos, M.; Datta, K.; Oliva, N.; Khalekuzzaman, M.; Torrizo, L.; Krishnan, S.; Oliveira, M.M.; Goto, F.; Datta, S. Enhanced iron and zinc accumulation in transgenic rice with the ferritin gene. Plant Sci. 2003, 164, 371–378. [Google Scholar] [CrossRef]
- Kakei, Y.; Yamaguchi, I.; Kobayashi, T.; Takahashi, M.; Nakanishi, H.; Yamakawa, T.; Nishizawa, N.K. A Highly Sensitive, Quick and Simple Quantification Method for Nicotianamine and 2′-Deoxymugineic Acid from Minimum Samples Using LC/ESI-TOF-MS Achieves Functional Analysis of These Components in Plants. Plant Cell Physiol. 2009, 50, 1988–1993. [Google Scholar] [CrossRef] [PubMed]

Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lee, S.; Rahman, M.M.; Nakanishi, H.; Nishizawa, N.K.; An, G.; Nam, H.G.; Jeon, J.-S. Concomitant Activation of OsNAS2 and OsNAS3 Contributes to the Enhanced Accumulation of Iron and Zinc in Rice. Int. J. Mol. Sci. 2023, 24, 6568. https://doi.org/10.3390/ijms24076568
Lee S, Rahman MM, Nakanishi H, Nishizawa NK, An G, Nam HG, Jeon J-S. Concomitant Activation of OsNAS2 and OsNAS3 Contributes to the Enhanced Accumulation of Iron and Zinc in Rice. International Journal of Molecular Sciences. 2023; 24(7):6568. https://doi.org/10.3390/ijms24076568
Chicago/Turabian StyleLee, Sichul, Md Mizanor Rahman, Hiromi Nakanishi, Naoko K. Nishizawa, Gynheung An, Hong Gil Nam, and Jong-Seong Jeon. 2023. "Concomitant Activation of OsNAS2 and OsNAS3 Contributes to the Enhanced Accumulation of Iron and Zinc in Rice" International Journal of Molecular Sciences 24, no. 7: 6568. https://doi.org/10.3390/ijms24076568
APA StyleLee, S., Rahman, M. M., Nakanishi, H., Nishizawa, N. K., An, G., Nam, H. G., & Jeon, J.-S. (2023). Concomitant Activation of OsNAS2 and OsNAS3 Contributes to the Enhanced Accumulation of Iron and Zinc in Rice. International Journal of Molecular Sciences, 24(7), 6568. https://doi.org/10.3390/ijms24076568




