A Novel Strategy to Identify Endolysins with Lytic Activity against Methicillin-Resistant Staphylococcus aureus
Abstract
1. Introduction
2. Results
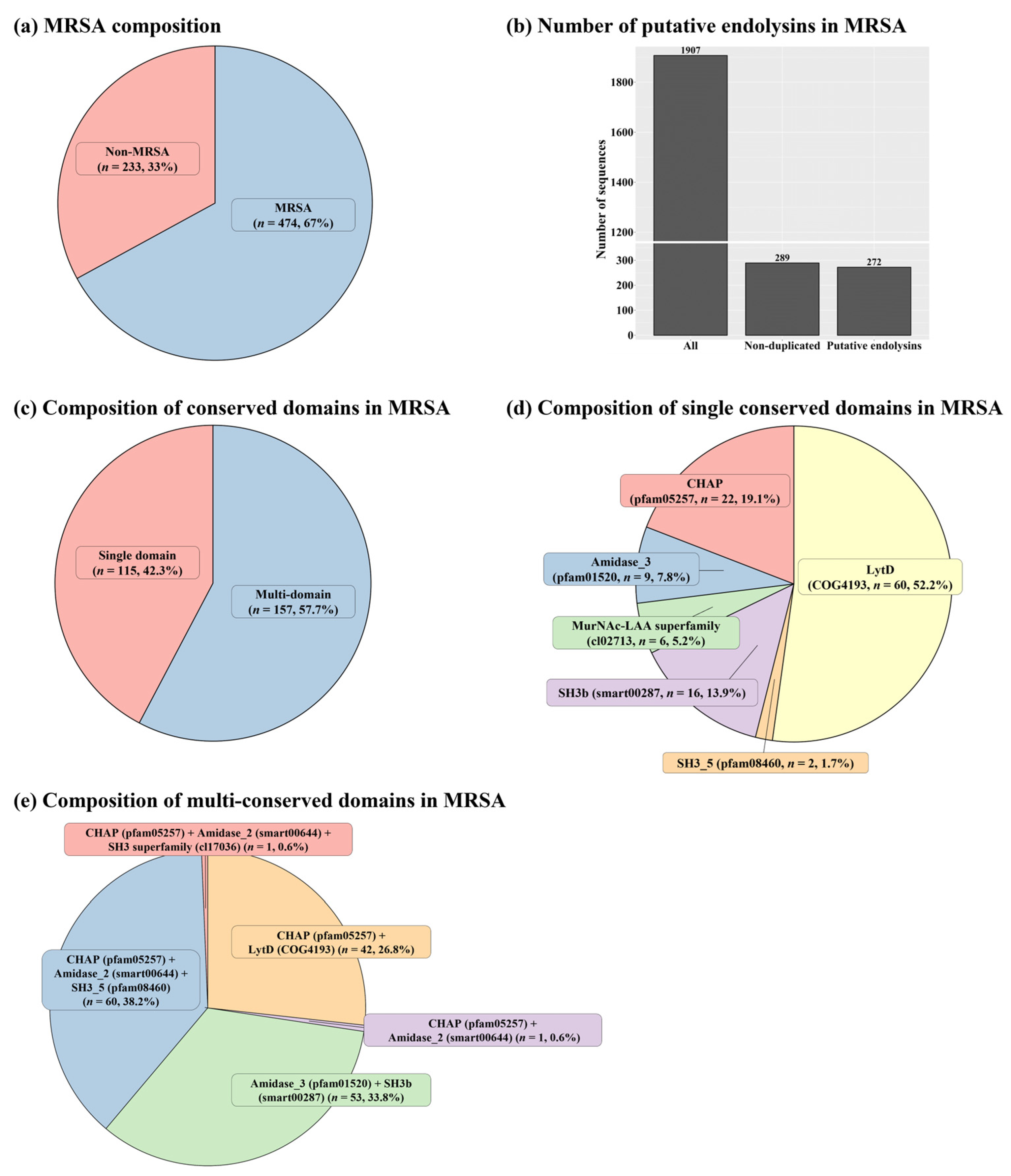
2.1. Identification of Putative Endolysins against MRSA
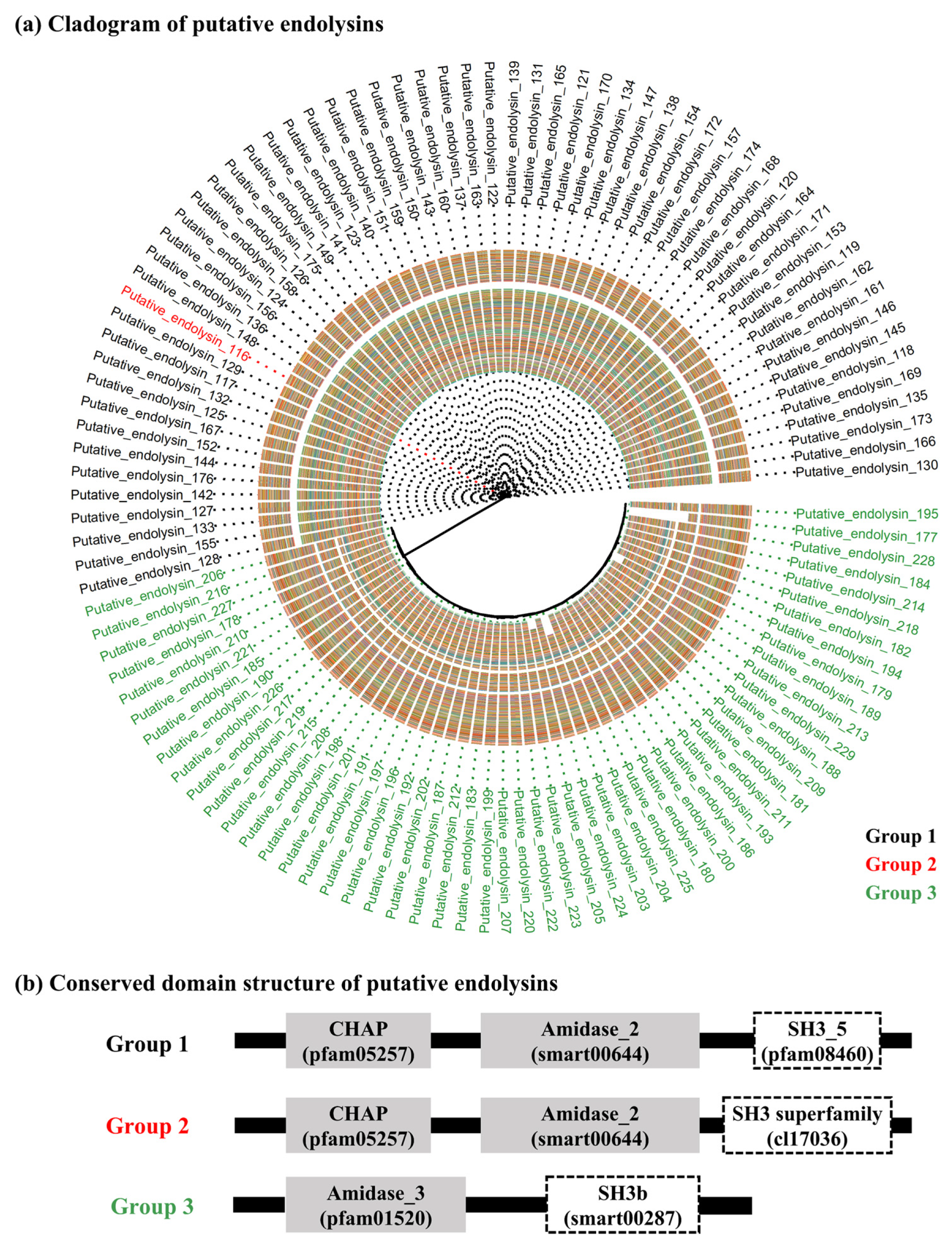
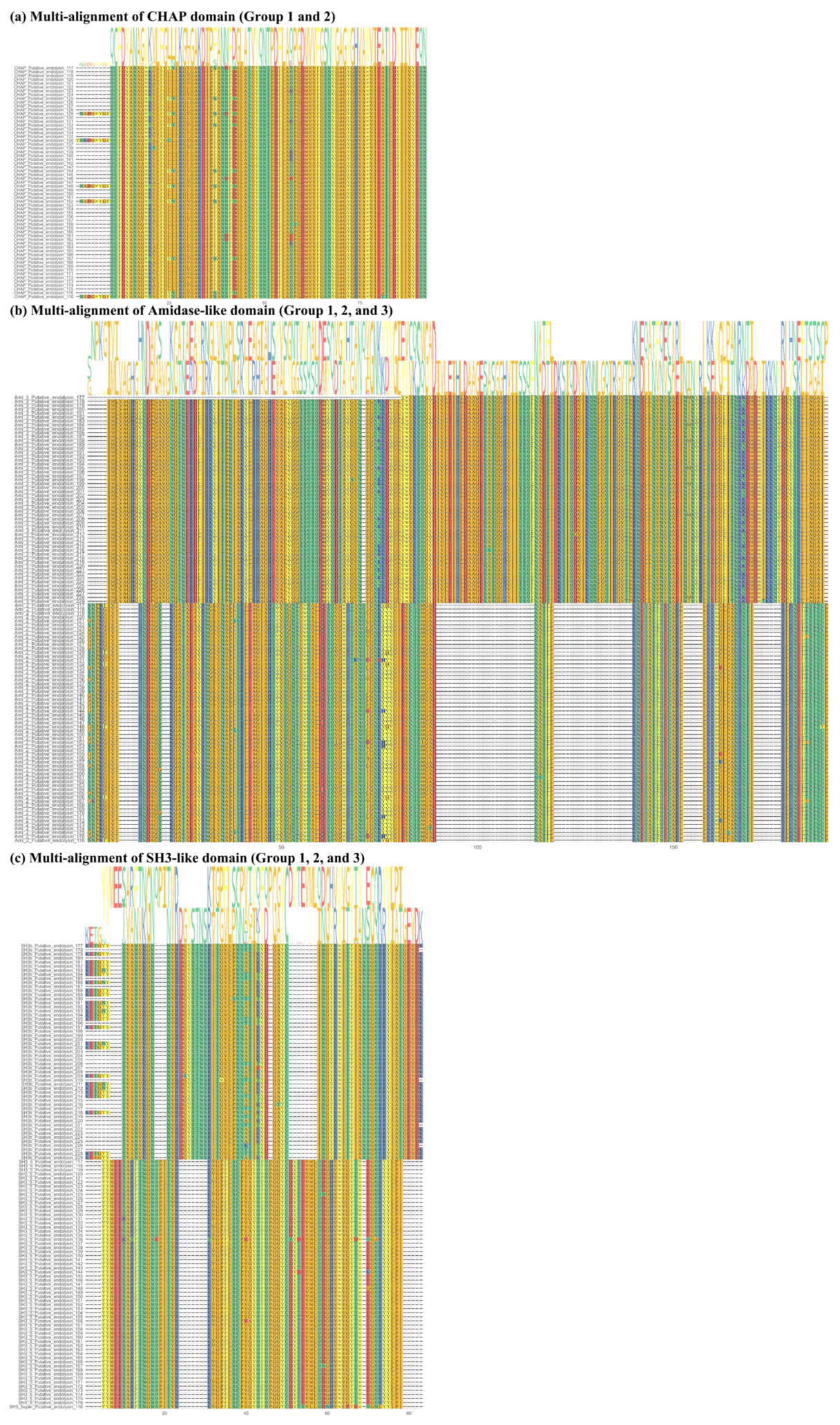
2.2. Sequence Comparison among Putative Endolysins Targeting MRSA
2.3. Comparison of Predicted Recombinant Protein Solubility
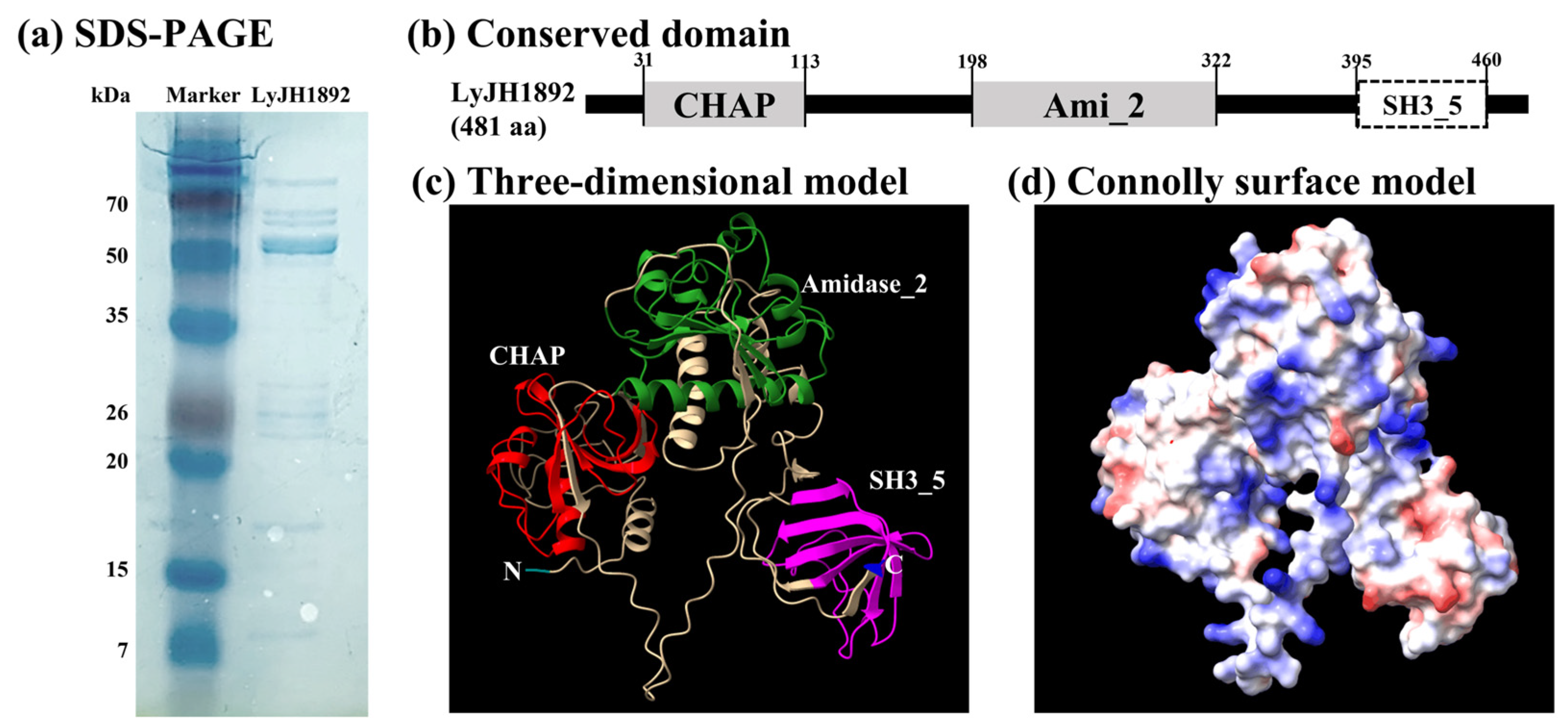
2.4. Overexpression and Structural Analysis of Selected Endolysins
2.5. Antibiotic Resistance Profiles of S. aureus Strains
2.6. Characterization of Recombinant LyJH1892
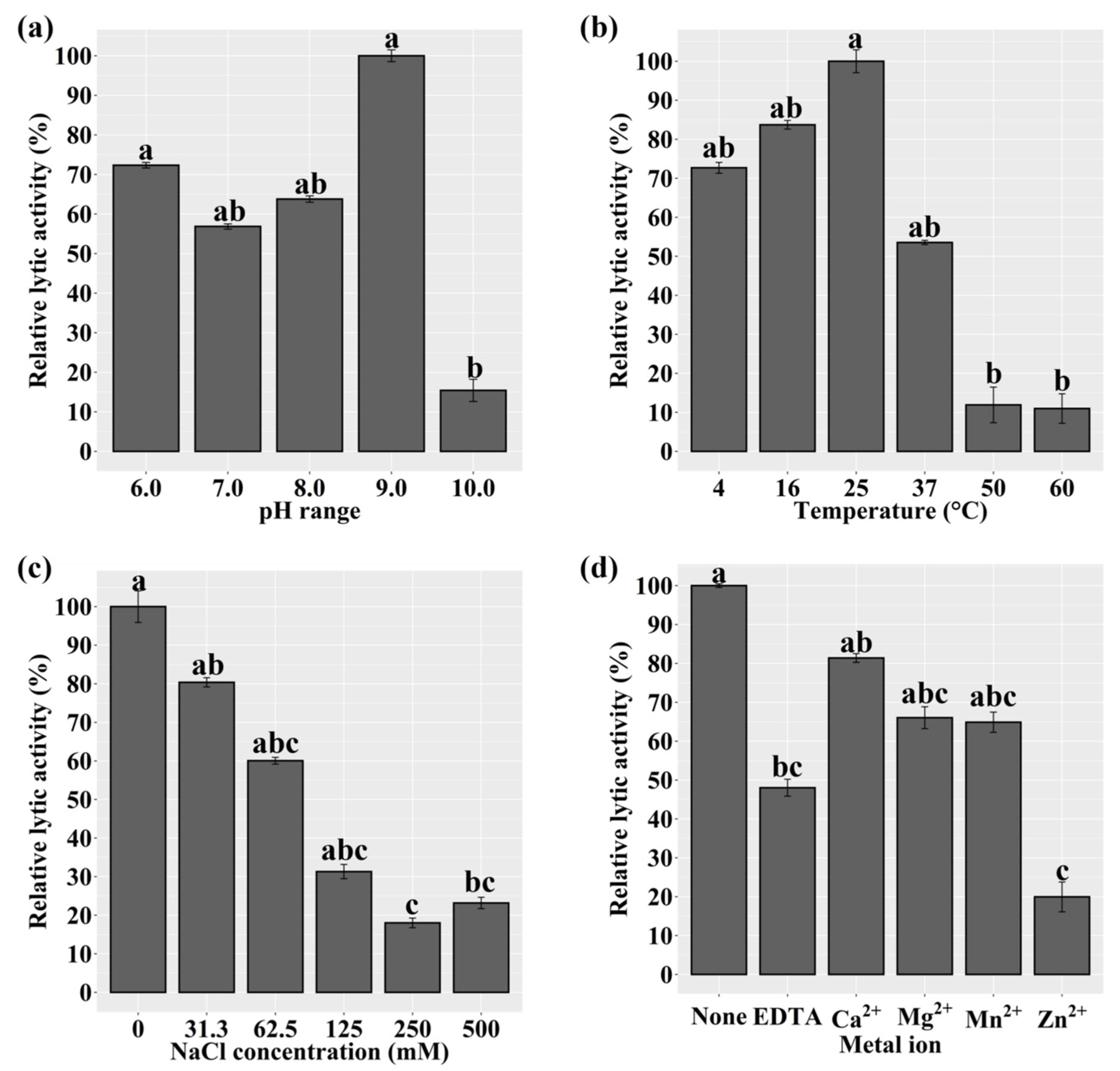
2.7. Optimal Lytic Activity and Lytic Spectrum of LyJH1892
3. Discussion
4. Materials and Methods
4.1. Bacterial Strains and Growth Conditions
4.2. Antibiotic Susceptibility Test
4.3. Collection of Putative Endolysins against MRSA
4.4. Conserved Domain Analysis, Sequence Alignment, and Solubility Prediction of Putative Endolysins
4.5. Identification, Cloning, and Overexpression of Endolysin LyJH1892
4.6. Structure Prediction of LyJH1892
4.7. Characterization and Lytic Spectrum of LyJH1892
4.8. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Haddad Kashani, H.; Schmelcher, M.; Sabzalipoor, H.; Seyed Hosseini, E.; Moniri, R. Recombinant endolysins as potential therapeutics against antibiotic-resistant Staphylococcus aureus: Current status of research and novel delivery strategies. Clin. Microbiol. Rev. 2018, 31, e00071-7. [Google Scholar] [CrossRef] [PubMed]
- Kluytmans, J.; Van Belkum, A.; Verbrugh, H. Nasal carriage of Staphylococcus aureus: Epidemiology, underlying mechanisms, and associated risks. Clin. Microbiol. Rev. 1997, 10, 505–520. [Google Scholar] [CrossRef] [PubMed]
- Gutiérrez, D.; Fernández, L.; Rodríguez, A.; García, P. Are phage lytic proteins the secret weapon to kill Staphylococcus aureus? MBio 2018, 9, e01923-17. [Google Scholar] [CrossRef] [PubMed]
- Wertheim, H.F.L.; Melles, D.C.; Vos, M.C.; van Leeuwen, W.; van Belkum, A.; Verbrugh, H.A.; Nouwen, J.L. The role of nasal carriage in Staphylococcus aureus infections. Lancet Infect. Dis. 2005, 5, 751–762. [Google Scholar] [CrossRef]
- Foster, T.J. Antibiotic resistance in Staphylococcus aureus. Current status and future prospects. FEMS Microbiol. Rev. 2017, 41, 430–449. [Google Scholar] [CrossRef]
- Sakoulas, G.; Moellering, R.C., Jr. Increasing Antibiotic Resistance among Methicillin-Resistant Staphylococcus aureus Strains. Clin. Infect. Dis. 2008, 46, S360–S367. [Google Scholar] [CrossRef]
- Cheng, W.N.; Han, S.G. Bovine mastitis: Risk factors, therapeutic strategies, and alternative treatments—A review. Asian-Australas. J. Anim. Sci. 2020, 33, 1699. [Google Scholar] [CrossRef]
- Guimarães, F.; Manzi, M.; Joaquim, S.; Richini-Pereira, V.; Langoni, H. Outbreak of methicillin-resistant Staphylococcus aureus (MRSA)-associated mastitis in a closed dairy herd. J. Dairy Sci. 2017, 100, 726–730. [Google Scholar] [CrossRef]
- Juhász-Kaszanyitzky, É.; Jánosi, S.; Somogyi, P.; Dán, Á.; vanderGraaf van Bloois, L.; Van Duijkeren, E.; Wagenaar, J.A. MRSA transmission between cows and humans. Emerg. Infect. Dis. 2007, 13, 630. [Google Scholar] [CrossRef]
- Donovan, D.M. Bacteriophage and peptidoglycan degrading enzymes with antimicrobial applications. Recent Pat. Biotechnol. 2007, 1, 113–122. [Google Scholar] [CrossRef]
- Love, M.J.; Bhandari, D.; Dobson, R.C.; Billington, C. Potential for bacteriophage endolysins to supplement or replace antibiotics in food production and clinical care. Antibiotics 2018, 7, 17. [Google Scholar] [CrossRef]
- Cisani, G.; Varaldo, P.E.; Grazi, G.; Soro, O. High-level potentiation of lysostaphin anti-staphylococcal activity by lysozyme. Antimicrob. Agents Chemother. 1982, 21, 531–535. [Google Scholar] [CrossRef]
- Kumar, J.K. Lysostaphin: An antistaphylococcal agent. Appl. Microbiol. Biotechnol. 2008, 80, 555–561. [Google Scholar] [CrossRef]
- Sugai, M.; Fujiwara, T.; Akiyama, T.; Ohara, M.; Komatsuzawa, H.; Inoue, S.; Suginaka, H. Purification and molecular characterization of glycylglycine endopeptidase produced by Staphylococcus capitis EPK1. J. Bacteriol. 1997, 179, 1193–1202. [Google Scholar] [CrossRef]
- Becker, S.C.; Foster-Frey, J.; Donovan, D.M. The phage K lytic enzyme LysK and lysostaphin act synergistically to kill MRSA. FEMS Microbiol. Lett. 2008, 287, 185–191. [Google Scholar] [CrossRef]
- Sundarrajan, S.; Raghupatil, J.; Vipra, A.; Narasimhaswamy, N.; Saravanan, S.; Appaiah, C.; Poonacha, N.; Desai, S.; Nair, S.; Bhatt, R.N. Bacteriophage-derived CHAP domain protein, P128, kills Staphylococcus cells by cleaving interpeptide cross-bridge of peptidoglycan. Microbiology 2014, 160, 2157–2169. [Google Scholar] [CrossRef]
- Chang, Y.; Ryu, S. Characterization of a novel cell wall binding domain-containing Staphylococcus aureus endolysin LysSA97. Appl. Microbiol. Biotechnol. 2017, 101, 147–158. [Google Scholar] [CrossRef]
- Altermann, E.; Schofield, L.R.; Ronimus, R.S.; Beattie, A.K.; Reilly, K. Inhibition of rumen methanogens by a novel archaeal lytic enzyme displayed on tailored bionanoparticles. Front. Microbiol. 2018, 9, 2378. [Google Scholar] [CrossRef]
- Swift, S.M.; Waters, J.J.; Rowley, D.T.; Oakley, B.B.; Donovan, D.M. Characterization of two glycosyl hydrolases, putative prophage endolysins, that target Clostridium perfringens. FEMS Microbiol. Lett. 2018, 365, fny179. [Google Scholar] [CrossRef]
- Kim, H.; Lee, H.G.; Kwon, I.; Seo, J. Characterization of endolysin LyJH307 with antimicrobial activity against Streptococcus bovis. Animals 2020, 10, 963. [Google Scholar] [CrossRef]
- Vollmer, W.; Blanot, D.; De Pedro, M.A. Peptidoglycan structure and architecture. FEMS Microbiol. Rev. 2008, 32, 149–167. [Google Scholar] [CrossRef] [PubMed]
- Sauvage, E.; Kerff, F.; Terrak, M.; Ayala, J.A.; Charlier, P. The penicillin-binding proteins: Structure and role in peptidoglycan biosynthesis. FEMS Microbiol. Rev. 2008, 32, 234–258. [Google Scholar] [CrossRef] [PubMed]
- Fishovitz, J.; Hermoso, J.A.; Chang, M.; Mobashery, S. Penicillin-binding protein 2a of methicillin-resistant Staphylococcus aureus. IUBMB Life 2014, 66, 572–577. [Google Scholar] [CrossRef] [PubMed]
- Blázquez, B.; Llarrull, L.I.; Luque-Ortega, J.R.; Alfonso, C.; Boggess, B.; Mobashery, S. Regulation of the expression of the β-lactam antibiotic-resistance determinants in methicillin-resistant Staphylococcus aureus (MRSA). Biochemistry 2014, 53, 1548–1550. [Google Scholar] [CrossRef] [PubMed]
- Ryffel, C.; Kayser, F.H.; Berger-Bächi, B. Correlation between regulation of mecA transcription and expression of methicillin resistance in staphylococci. Antimicrob. Agents Chemother. 1992, 36, 25–31. [Google Scholar] [CrossRef]
- Francois, P.; Bento, M.; Renzi, G.; Harbarth, S.; Pittet, D.; Schrenzel, J. Evaluation of three molecular assays for rapid identification of methicillin-resistant Staphylococcus aureus. J. Clin. Microbiol. 2007, 45, 2011–2013. [Google Scholar] [CrossRef]
- Wu, M.; Tong, X.; Liu, S.; Wang, D.; Wang, L.; Fan, H. Prevalence of methicillin-resistant Staphylococcus aureus in healthy Chinese population: A system review and meta-analysis. PLoS ONE 2019, 14, e0223599. [Google Scholar] [CrossRef]
- De Kraker, M.E.A.; Jarlier, V.; Monen, J.C.M.; Heuer, O.E.; van de Sande, N.; Grundmann, H. The changing epidemiology of bacteraemias in Europe: Trends from the European Antimicrobial Resistance Surveillance System. Clin. Microbiol. Infect. 2013, 19, 860–868. [Google Scholar] [CrossRef]
- Garoy, E.Y.; Gebreab, Y.B.; Achila, O.O.; Tekeste, D.G.; Kesete, R.; Ghirmay, R.; Kiflay, R.; Tesfu, T. Methicillin-resistant Staphylococcus aureus (MRSA): Prevalence and antimicrobial sensitivity pattern among patients—A multicenter study in Asmara, Eritrea. Can. J. Infect. Dis. Med. Microbiol. 2019, 2019, 8321834. [Google Scholar] [CrossRef]
- Guzmán-Blanco, M.; Mejía, C.; Isturiz, R.; Alvarez, C.; Bavestrello, L.; Gotuzzo, E.; Labarca, J.; Luna, C.M.; Rodríguez-Noriega, E.; Salles, M.J. Epidemiology of meticillin-resistant Staphylococcus aureus (MRSA) in Latin America. Int. J. Antimicrob. Agents 2009, 34, 304–308. [Google Scholar] [CrossRef]
- Jiménez, J.N.; Ocampo, A.M.; Vanegas, J.M.; Rodriguez, E.A.; Mediavilla, J.R.; Chen, L.; Muskus, C.E.; Vélez, L.A.; Rojas, C.; Restrepo, A.V. CC8 MRSA strains harboring SCC mec type IVc are predominant in Colombian hospitals. PLoS ONE 2012, 7, e38576. [Google Scholar] [CrossRef]
- Falagas, M.E.; Karageorgopoulos, D.E.; Leptidis, J.; Korbila, I.P. MRSA in Africa: Filling the global map of antimicrobial resistance. PLoS ONE 2013, 8, e68024. [Google Scholar] [CrossRef]
- Bateman, A.; Rawlings, N.D. The CHAP domain: A large family of amidases including GSP amidase and peptidoglycan hydrolases. Trends Biochem. Sci. 2003, 28, 234–237. [Google Scholar] [CrossRef]
- Oliveira, H.; Melo, L.D.; Santos, S.B.; Nóbrega, F.L.; Ferreira, E.C.; Cerca, N.; Azeredo, J.; Kluskens, L.D. Molecular aspects and comparative genomics of bacteriophage endolysins. J. Virol. 2013, 87, 4558–4570. [Google Scholar] [CrossRef]
- Mayer, B.J.; Eck, M.J. SH3 domains: Minding your p’s and q’s. Curr. Biol. 1995, 5, 364–367. [Google Scholar] [CrossRef]
- Becker, S.C.; Foster-Frey, J.; Stodola, A.J.; Anacker, D.; Donovan, D.M. Differentially conserved staphylococcal SH3b_5 cell wall binding domains confer increased staphylolytic and streptolytic activity to a streptococcal prophage endolysin domain. Gene 2009, 443, 32–41. [Google Scholar] [CrossRef]
- Ponting, C.P.; Aravind, L.; Schultz, J.; Bork, P.; Koonin, E.V. Eukaryotic Signalling Domain Homologues in Archaea and Bacteria. Ancient Ancestry and Horizontal Gene Transfer. J. Mol. Biol. 1999, 289, 729–745. [Google Scholar] [CrossRef]
- Baba, T.; Schneewind, O. Target cell specificity of a bacteriocin molecule: A C-terminal signal directs lysostaphin to the cell wall of Staphylococcus aureus. EMBO J. 1996, 15, 4789–4797. [Google Scholar] [CrossRef]
- Low, L.Y.; Yang, C.; Perego, M.; Osterman, A.; Liddington, R.C. Structure and lytic activity of a Bacillus anthracis prophage endolysin. J. Biol. Chem. 2005, 280, 35433–35439. [Google Scholar] [CrossRef]
- Loessner, M.J.; Kramer, K.; Ebel, F.; Scherer, S. C-terminal domains of Listeria monocytogenes bacteriophage murein hydrolases determine specific recognition and high-affinity binding to bacterial cell wall carbohydrates. Mol. Microbiol. 2002, 44, 335–349. [Google Scholar] [CrossRef]
- Schmelcher, M.; Donovan, D.M.; Loessner, M.J. Bacteriophage endolysins as novel antimicrobials. Future Microbiol. 2012, 7, 1147–1171. [Google Scholar] [CrossRef] [PubMed]
- Fischetti, V.A. Bacteriophage lysins as effective antibacterials. Curr. Opin. Microbiol. 2008, 11, 393–400. [Google Scholar] [CrossRef] [PubMed]
- Díez-Martínez, R.; de Paz, H.; Bustamante, N.; García, E.; Menéndez, M.; García, P. Improving the lethal effect of Cpl-7, a pneumococcal phage lysozyme with broad bactericidal activity, by inverting the net charge of its cell wall-binding module. Antimicrob. Agents Chemother. 2013, 57, 5355–5365. [Google Scholar] [CrossRef] [PubMed]
- Fang, Y.; Fang, J. Discrimination of soluble and aggregation-prone proteins based on sequence information. Mol. BioSyst. 2013, 9, 806–811. [Google Scholar] [CrossRef] [PubMed]
- Madhavan, A.; Sindhu, R.; Binod, P.; Sukumaran, R.K.; Pandey, A. Strategies for design of improved biocatalysts for industrial applications. Bioresour. Technol. 2017, 245, 1304–1313. [Google Scholar] [CrossRef]
- Chan, W.-C.; Liang, P.-H.; Shih, Y.-P.; Yang, U.-C.; Lin, W.-C.; Hsu, C.-N. Learning to predict expression efficacy of vectors in recombinant protein production. BMC Bioinform. 2010, 11, S21. [Google Scholar] [CrossRef]
- Musil, M.; Konegger, H.; Hon, J.; Bednar, D.; Damborsky, J. Computational design of stable and soluble biocatalysts. ACS Catal. 2018, 9, 1033–1054. [Google Scholar] [CrossRef]
- Hebditch, M.; Carballo-Amador, M.A.; Charonis, S.; Curtis, R.; Warwicker, J. Protein–Sol: A web tool for predicting protein solubility from sequence. Bioinformatics 2017, 33, 3098–3100. [Google Scholar] [CrossRef]
- Hon, J.; Marusiak, M.; Martinek, T.; Kunka, A.; Zendulka, J.; Bednar, D.; Damborsky, J. SoluProt: Prediction of soluble protein expression in Escherichia coli. Bioinformatics 2021, 37, 23–28. [Google Scholar] [CrossRef]
- Raimondi, D.; Orlando, G.; Fariselli, P.; Moreau, Y. Insight into the protein solubility driving forces with neural attention. PLoS Comput. Biol. 2020, 16, e1007722. [Google Scholar] [CrossRef]
- Wang, X.; Li, L.; Li, S.; Huang, J.; Fan, Y.; Yao, Z.; Ye, X.; Chen, S. Phenotypic and molecular characteristics of Staphylococcus aureus and methicillin-resistant Staphylococcus aureus in slaughterhouse pig-related workers and control workers in Guangdong Province, China. Epidemiol. Infect. 2017, 145, 1843–1851. [Google Scholar] [CrossRef]
- Ye, X.; Liu, W.; Fan, Y.; Wang, X.; Zhou, J.; Yao, Z.; Chen, S. Frequency-risk and duration-risk relations between occupational livestock contact and methicillin-resistant Staphylococcus aureus carriage among workers in Guangdong, China. Am. J. Infect. Control 2015, 43, 676–681. [Google Scholar] [CrossRef]
- Clinical Laboratory Standards Institute. Performance Standards for Antimicrobial Susceptibility Testing, M100, 31st ed.; Clinical Laboratory Standards Institute: Malvern, PA, USA, 2021. [Google Scholar]
- Udo, E.E.; Boswihi, S.S.; Mathew, B.; Noronha, B.; Verghese, T. Resurgence of Chloramphenicol Resistance in Methicillin-Resistant Staphylococcus aureus Due to the Acquisition of a Variant Florfenicol Exporter (fexAv)-Mediated Chloramphenicol Resistance in Kuwait Hospitals. Antibiotics 2021, 10, 1250. [Google Scholar] [CrossRef]
- Kehrenberg, C.; Schwarz, S. Distribution of florfenicol resistance genes fexA and cfr among chloramphenicol-resistant Staphylococcus isolates. Antimicrob. Agents Chemother. 2006, 50, 1156–1163. [Google Scholar] [CrossRef]
- Deyno, S.; Fekadu, S.; Astatkie, A. Resistance of Staphylococcus aureus to antimicrobial agents in Ethiopia: A meta-analysis. Antimicrob. Resist. Infect. Control 2017, 6, 85. [Google Scholar] [CrossRef]
- Shlezinger, M.; Khalifa, L.; Houri-Haddad, Y.; Coppenhagen-Glazer, S.; Resch, G.; Que, Y.-A.; Beyth, S.; Dorfman, E.; Hazan, R.; Beyth, N. Phage therapy: A new horizon in the antibacterial treatment of oral pathogens. Curr. Top. Med. Chem. 2017, 17, 1199–1211. [Google Scholar] [CrossRef]
- Azam, A.H.; Tanji, Y. Bacteriophage-host arm race: An update on the mechanism of phage resistance in bacteria and revenge of the phage with the perspective for phage therapy. Appl. Microbiol. Biotechnol. 2019, 103, 2121–2131. [Google Scholar] [CrossRef]
- Nilsson, A.S. Phage therapy—Constraints and possibilities. Upsala J. Med. Sci. 2014, 119, 192–198. [Google Scholar] [CrossRef]
- Vanderhaeghen, W.; Piepers, S.; Leroy, F.; Van Coillie, E.; Haesebrouck, F.; De Vliegher, S. Invited review: Effect, persistence, and virulence of coagulase-negative Staphylococcus species associated with ruminant udder health. J. Dairy Sci. 2014, 97, 5275–5293. [Google Scholar] [CrossRef]
- Thorberg, B.-M.; Danielsson-Tham, M.-L.; Emanuelson, U.; Waller, K.P. Bovine subclinical mastitis caused by different types of coagulase-negative staphylococci. J. Dairy Sci. 2009, 92, 4962–4970. [Google Scholar] [CrossRef]
- Supré, K.; Haesebrouck, F.; Zadoks, R.; Vaneechoutte, M.; Piepers, S.; De Vliegher, S. Some coagulase-negative Staphylococcus species affect udder health more than others. J. Dairy Sci. 2011, 94, 2329–2340. [Google Scholar] [CrossRef] [PubMed]
- De Visscher, A.; Piepers, S.; Haesebrouck, F.; De Vliegher, S. Intramammary infection with coagulase-negative staphylococci at parturition: Species-specific prevalence, risk factors, and effect on udder health. J. Dairy Sci. 2016, 99, 6457–6469. [Google Scholar] [CrossRef] [PubMed]
- Schleifer, K.H.; Kandler, O. Peptidoglycan types of bacterial cell walls and their taxonomic implications. Bacteriol. Rev. 1972, 36, 407–477. [Google Scholar] [CrossRef] [PubMed]
- Lu, J.Z.; Fujiwara, T.; Komatsuzawa, H.; Sugai, M.; Sakon, J. Cell wall-targeting domain of glycylglycine endopeptidase distinguishes among peptidoglycan cross-bridges. J. Biol. Chem. 2006, 281, 549–558. [Google Scholar] [CrossRef]
- Gründling, A.; Schneewind, O. Cross-linked peptidoglycan mediates lysostaphin binding to the cell wall envelope of Staphylococcus aureus. J. Bacteriol. 2006, 188, 2463–2472. [Google Scholar] [CrossRef]
- Donovan, D.M.; Lardeo, M.; Foster-Frey, J. Lysis of staphylococcal mastitis pathogens by bacteriophage phi11 endolysin. FEMS Microbiol. Lett. 2006, 265, 133–139. [Google Scholar] [CrossRef]
- Donovan, D.M.; Dong, S.; Garrett, W.; Rousseau, G.M.; Moineau, S.; Pritchard, D.G. Peptidoglycan Hydrolase Fusions Maintain Their Parental Specificities. Appl. Environ. Microbiol. 2006, 72, 2988–2996. [Google Scholar] [CrossRef]
- De Jonge, B.; Chang, Y.-S.; Gage, D.; Tomasz, A. Peptidoglycan composition of a highly methicillin-resistant Staphylococcus aureus strain. The role of penicillin binding protein 2A. J. Biol. Chem. 1992, 267, 11248–11254. [Google Scholar] [CrossRef]
- Aziz, R.K.; Bartels, D.; Best, A.A.; DeJongh, M.; Disz, T.; Edwards, R.A.; Formsma, K.; Gerdes, S.; Glass, E.M.; Kubal, M. The RAST Server: Rapid annotations using subsystems technology. BMC Genom. 2008, 9, 75. [Google Scholar] [CrossRef]
- Marchler-Bauer, A.; Derbyshire, M.K.; Gonzales, N.R.; Lu, S.; Chitsaz, F.; Geer, L.Y.; Geer, R.C.; He, J.; Gwadz, M.; Hurwitz, D.I. CDD: NCBI’s conserved domain database. Nucleic Acids Res. 2015, 43, D222–D226. [Google Scholar] [CrossRef]
- Wickham, H. ggplot2: Elegant Graphics for Data Analysis; Springer: Berlin/Heidelberg, Germany, 2016. [Google Scholar]
- The R Development Core Team. R: A Language and Environment for Statistical Computing; R Foundation for Statistical Computing: Vienna, Austria, 2013. [Google Scholar]
- Katoh, K.; Standley, D.M. MAFFT multiple sequence alignment software version 7: Improvements in performance and usability. Mol. Biol. Evol. 2013, 30, 772–780. [Google Scholar] [CrossRef]
- Price, M.N.; Dehal, P.S.; Arkin, A.P. FastTree 2–approximately maximum-likelihood trees for large alignments. PLoS ONE 2010, 5, e9490. [Google Scholar] [CrossRef]
- Yu, G.; Smith, D.K.; Zhu, H.; Guan, Y.; Lam, T.T.Y. ggtree: An R package for visualization and annotation of phylogenetic trees with their covariates and other associated data. Methods Ecol. Evol. 2017, 8, 28–36. [Google Scholar] [CrossRef]
- Mirdita, M.; Schütze, K.; Moriwaki, Y.; Heo, L.; Ovchinnikov, S.; Steinegger, M. ColabFold-Making protein folding accessible to all. Nat. Methods 2021, 19, 679–682. [Google Scholar] [CrossRef]
- Pettersen, E.F.; Goddard, T.D.; Huang, C.C.; Meng, E.C.; Couch, G.S.; Croll, T.I.; Morris, J.H.; Ferrin, T.E. UCSF ChimeraX: Structure visualization for researchers, educators, and developers. Protein Sci. 2021, 30, 70–82. [Google Scholar] [CrossRef]
- Mónico, A.; Martínez-Senra, E.; Zorrilla, S.; Pérez-Sala, D. Drawbacks of dialysis procedures for removal of EDTA. PLoS ONE 2017, 12, e0169843. [Google Scholar] [CrossRef]
- Ogle, D.H.; Doll, J.C.; Wheeler, A.P.; Dinno, A. FSA: Simple Fisheries Stock Assessment Methods. R Package Version 0.9.4. Available online: https:/fishr-core-team.github.io/FSA/ (accessed on 14 March 2023).

| Group 2 | Name | Amino Acids | Soluprot | Protein-Sol | SKADE | Sum 7 | |||
|---|---|---|---|---|---|---|---|---|---|
| Solubility 3 | Z-Score 4 | Scaled-sol 5 | Z-Score 4 | Solubility 6 | Z-Score 4 | ||||
| Group 3 | Putative endolysin 177 | 249 | 0.569 | −0.269 | 0.680 | 5.559 | 0.186 | −1.638 | 3.652 |
| Group 1 | Putative endolysin 117 | 481 | 0.735 | 1.114 | 0.354 | −0.276 | 0.327 | 0.903 | 1.741 |
| Group 1 | Putative endolysin 129 | 481 | 0.776 | 1.465 | 0.343 | −0.473 | 0.311 | 0.610 | 1.601 |
| Group 1 | Putative endolysin 121 | 481 | 0.749 | 1.239 | 0.332 | −0.670 | 0.330 | 0.951 | 1.520 |
| Group 1 | Putative endolysin 159 | 481 | 0.718 | 0.973 | 0.332 | −0.670 | 0.343 | 1.182 | 1.485 |
| Group 1 | Putative endolysin 132 | 481 | 0.750 | 1.241 | 0.343 | −0.473 | 0.312 | 0.631 | 1.398 |
| Group 1 | Putative endolysin 147 | 481 | 0.727 | 1.052 | 0.332 | −0.670 | 0.332 | 0.981 | 1.364 |
| Group 1 | Putative endolysin 143 | 481 | 0.741 | 1.167 | 0.322 | −0.849 | 0.334 | 1.015 | 1.333 |
| Group 1 | Putative endolysin 174 | 481 | 0.722 | 1.006 | 0.332 | −0.670 | 0.332 | 0.986 | 1.322 |
| Group 1 | Putative endolysin 150 | 481 | 0.732 | 1.092 | 0.322 | −0.849 | 0.336 | 1.063 | 1.306 |
| Antibiotic Disc | Antibiotic Susceptibility 1 | |||
|---|---|---|---|---|
| NCCP 16830 | NCCP 1457 | NCCP 14754 | KVCC-BA0500624 | |
| Cefoxitin (30 μg) | Susceptible | Resistant | Resistant | Resistant |
| Oxacillin (1 μg) | Susceptible | Resistant | Resistant | Resistant |
| Penicillin (10 μg) | Resistant | Resistant | Resistant | Resistant |
| Erythromycin (15 μg) | Susceptible | Resistant | Resistant | Resistant |
| Clindamycin (2 μg) | Susceptible | Susceptible | Intermediate | Susceptible |
| Chloramphenicol (30 μg) | Susceptible | Susceptible | Susceptible | Susceptible |
| Bacterial Strain | Purpose | Growth Condition 1 |
|---|---|---|
| Escherichia coli DH5α | Cloning host | LB broth |
| E. coli BL21(DE3) | Expression host | LB broth |
| Staphylococcus aureus (NCCP 16830) | Indicator strain | BHI broth |
| MRSA (KVCC-BA0500624) | Lytic spectrum | BHI broth |
| MRSA (NCCP 14567) | Lytic spectrum | BHI broth |
| MRSA (NCCP 14754) | Lytic spectrum | BHI broth |
| Staphylococcus hyicus (KVCC-BA0001860) | Lytic spectrum | BHI broth |
| Staphylococcus epidermidis (KVCC-BA0001452) | Lytic spectrum | BHI broth |
| Staphylococcus haemolyticus (NCCP 14693) | Lytic spectrum | BHI broth |
| Staphylococcus simulans (NCCP 16236) | Lytic spectrum | BHI broth |
| Staphylococcus warneri (NCCP 16234) | Lytic spectrum | BHI broth |
| Staphylococcus chromogenes (KCTC 3579) | Lytic spectrum | BHI broth |
| Staphylococcus xylosus (KCCM 40887) | Lytic spectrum | BHI broth |
| Streptococcus agalactiae (NCCP 14729) | Lytic spectrum | BHI broth |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kim, H.; Seo, J. A Novel Strategy to Identify Endolysins with Lytic Activity against Methicillin-Resistant Staphylococcus aureus. Int. J. Mol. Sci. 2023, 24, 5772. https://doi.org/10.3390/ijms24065772
Kim H, Seo J. A Novel Strategy to Identify Endolysins with Lytic Activity against Methicillin-Resistant Staphylococcus aureus. International Journal of Molecular Sciences. 2023; 24(6):5772. https://doi.org/10.3390/ijms24065772
Chicago/Turabian StyleKim, Hanbeen, and Jakyeom Seo. 2023. "A Novel Strategy to Identify Endolysins with Lytic Activity against Methicillin-Resistant Staphylococcus aureus" International Journal of Molecular Sciences 24, no. 6: 5772. https://doi.org/10.3390/ijms24065772
APA StyleKim, H., & Seo, J. (2023). A Novel Strategy to Identify Endolysins with Lytic Activity against Methicillin-Resistant Staphylococcus aureus. International Journal of Molecular Sciences, 24(6), 5772. https://doi.org/10.3390/ijms24065772





