Development and Application of an Optogenetic Manipulation System to Suppress Actomyosin Activity in Ciona Epidermis
Abstract
1. Introduction
2. Results
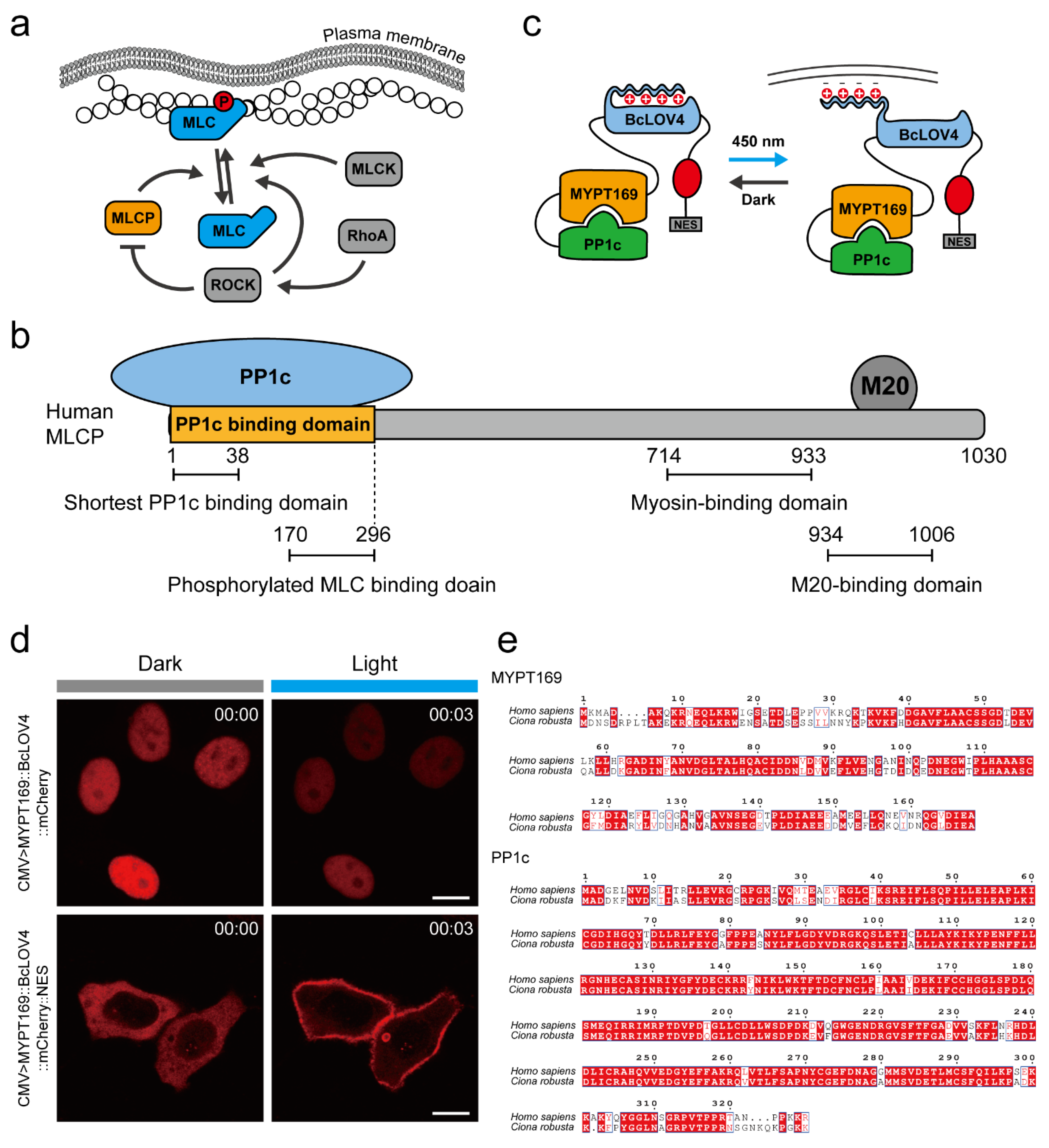
2.1. Design of an MLCP-BcLOV4 System to Suppress the Activity of Myosin II at the Cellular Level
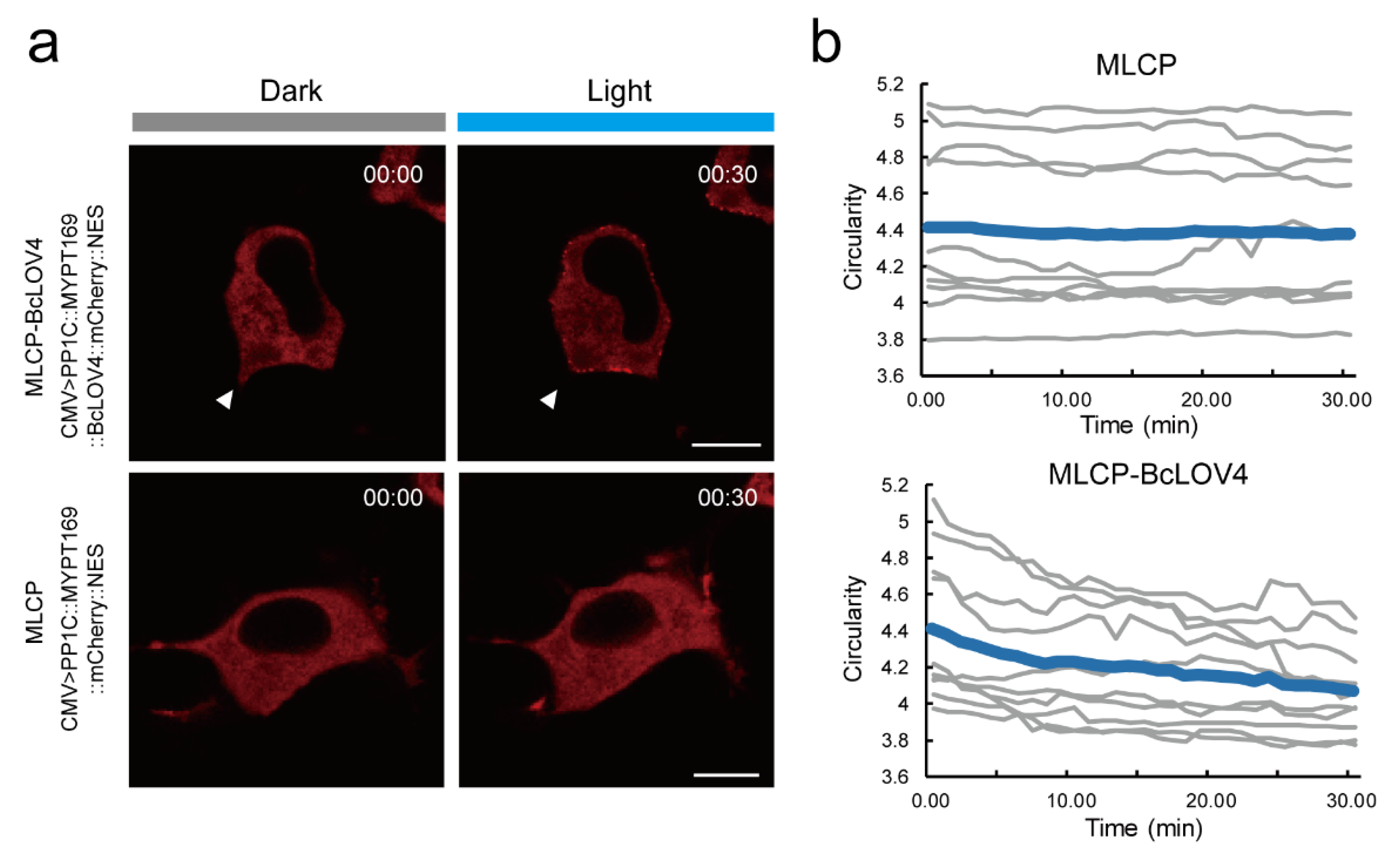
2.2. Validation of the MLCP-BcLOV4 System in HeLa Cells
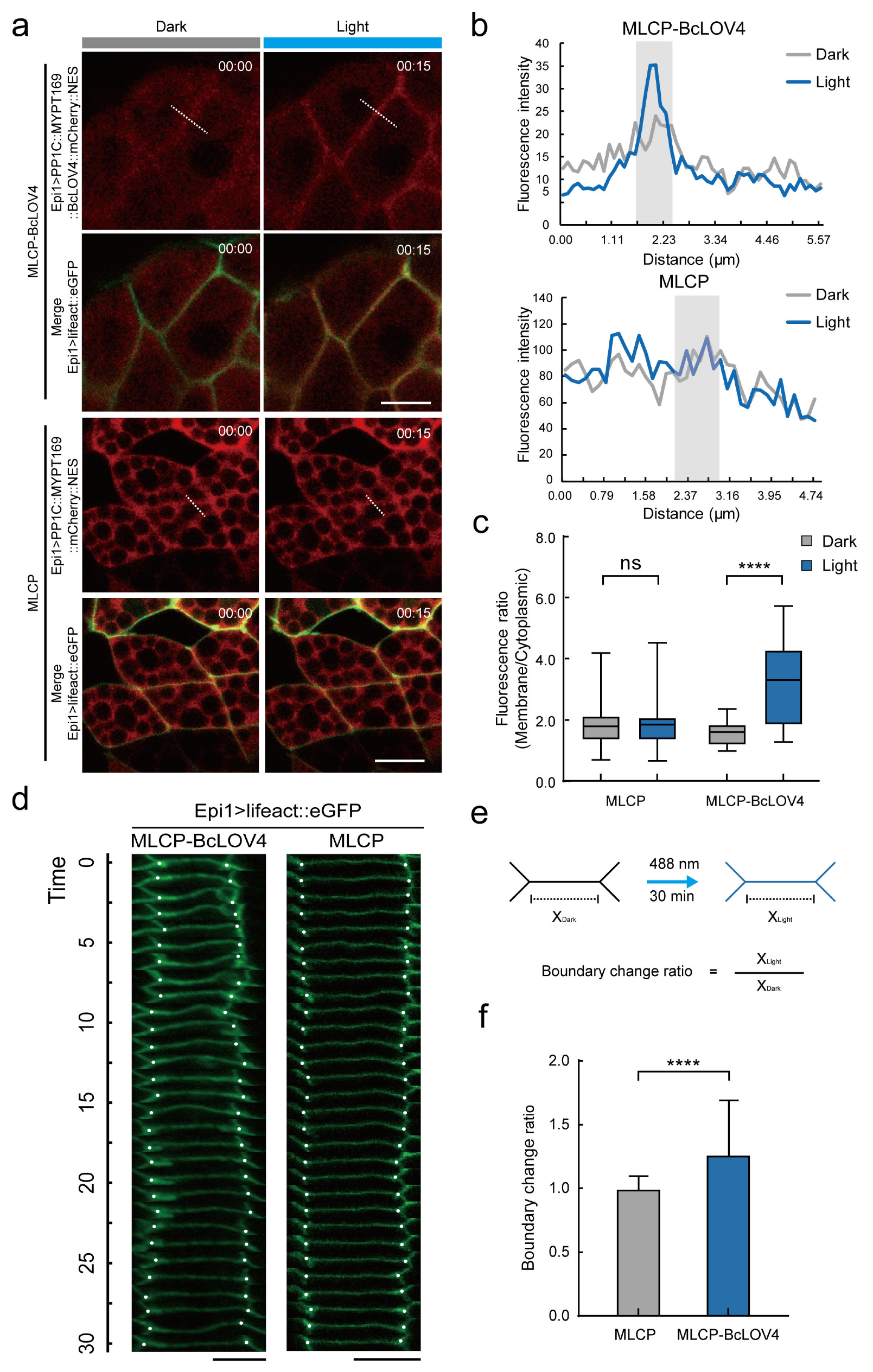
2.3. MLCP-BcLOV4 System Suppressed the Contractility of Epidermal Cells in Ciona Embryos
2.4. MLCP-BcLOV4 System Disrupted Atrial Siphon Invagination via Abolishing the Apical Contraction in Ciona Larvae
3. Discussion
4. Materials and Methods
4.1. Experimental Animal Preparation and Electroporation
4.2. Plasmids Construction
4.3. Cell Transfection
4.4. Immunofluorescence
4.5. Imaging and Optogenetics
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Appendix A

References
- Chan, C.J.; Heisenberg, C.P.; Hiiragi, T. Coordination of morphogenesis and cell-fate specification in development. Curr. Biol. 2017, 27, R1024–R1035. [Google Scholar] [CrossRef]
- Alt, S.; Ganguly, P.; Salbreux, G. Vertex models: From cell mechanics to tissue morphogenesis. Philos. Trans. R. Soc. Lond. B Biol. Sci. 2017, 372, 20150520. [Google Scholar] [CrossRef] [PubMed]
- Collinet, C.; Lecuit, T. Programmed and self-organized flow of information during morphogenesis. Nat. Rev. Mol. Cell Biol. 2021, 22, 245–265. [Google Scholar] [CrossRef] [PubMed]
- Lecuit, T.; Lenne, P.F.; Munro, E. Force generation, transmission, and integration during cell and tissue morphogenesis. Annu. Rev. Cell Dev. Biol. 2011, 27, 157–184. [Google Scholar] [CrossRef] [PubMed]
- Lee, J.Y.; Harland, R.M. Actomyosin contractility and microtubules drive apical constriction in Xenopus bottle cells. Dev. Biol. 2007, 311, 40–52. [Google Scholar] [CrossRef] [PubMed]
- Zartman, J.J.; Shvartsman, S.Y. Unit operations of tissue development: Epithelial folding. Annu. Rev. Chem. Biomol. Eng. 2010, 1, 231–246. [Google Scholar] [CrossRef]
- Fristrom, D. The cellular basis of epithelial morphogenesis. A review. Tissue Cell 1988, 20, 645–690. [Google Scholar] [CrossRef] [PubMed]
- Chung, S.; Kim, S.; Andrew, D.J. Uncoupling apical constriction from tissue invagination. ELife 2017, 6, e22235. [Google Scholar] [CrossRef]
- Vicente-Manzanares, M.; Ma, X.; Adelstein, R.S.; Horwitz, A.R. Non-muscle myosin II takes centre stage in cell adhesion and migration. Nat. Rev. Mol. Cell Biol. 2009, 10, 778–790. [Google Scholar] [CrossRef]
- Heissler, S.M.; Manstein, D.J. Nonmuscle myosin-2: Mix and match. Cell. Mol. Life Sci. 2013, 70, 1–21. [Google Scholar] [CrossRef]
- Even-Ram, S.; Doyle, A.D.; Conti, M.A.; Matsumoto, K.; Adelstein, R.S.; Yamada, K.M. Myosin IIA regulates cell motility and actomyosin-microtubule crosstalk. Nat. Cell Biol. 2007, 9, 299–309. [Google Scholar] [CrossRef] [PubMed]
- Ito, M.; Nakano, T.; Erdodi, F.; Hartshorne, D.J. Myosin phosphatase: Structure, regulation and function. Mol. Cell. Biochem. 2004, 259, 197–209. [Google Scholar] [CrossRef] [PubMed]
- Khasnis, M.; Nakatomi, A.; Gumpper, K.; Eto, M. Reconstituted human myosin light chain phosphatase reveals distinct roles of two inhibitory phosphorylation sites of the regulatory subunit, MYPT1. Biochemistry 2014, 53, 2701–2709. [Google Scholar] [CrossRef]
- Matsumura, F. Regulation of myosin II during cytokinesis in higher eukaryotes. Trends Cell Biol. 2005, 15, 371–377. [Google Scholar] [CrossRef]
- Watanabe, T.; Hosoya, H.; Yonemura, S. Regulation of myosin II dynamics by phosphorylation and dephosphorylation of its light chain in epithelial cells. Mol. Biol. Cell 2007, 18, 605–616. [Google Scholar] [CrossRef] [PubMed]
- Zhao, L.; Gao, F.; Gao, S.; Liang, Y.; Long, H.; Lv, Z.; Su, Y.; Ye, N.; Zhang, L.; Zhao, C.; et al. Biodiversity-based development and evolution: The emerging research systems in model and non-model organisms. Sci. China. Life Sci. 2021, 64, 1236–1280. [Google Scholar] [CrossRef]
- Peng, H.; Qiao, R.; Dong, B. Polarity establishment and maintenance in ascidian notochord. Front. Cell Dev. Biol. 2020, 8, 597446. [Google Scholar] [CrossRef]
- Lu, Q.; Gao, Y.; Fu, Y.; Peng, H.; Shi, W.; Li, B.; Lv, Z.; Feng, X.Q.; Dong, B. Ciona embryonic tail bending is driven by asymmetrical notochord contractility and coordinated by epithelial proliferation. Development 2020, 147, dev185868. [Google Scholar] [CrossRef]
- Sherrard, K.; Robin, F.; Lemaire, P.; Munro, E. Sequential activation of apical and basolateral contractility drives ascidian endoderm invagination. Curr. Biol. 2010, 20, 1499–1510. [Google Scholar] [CrossRef]
- Fiuza, U.M.; Lemaire, P. Mechanical and genetic control of ascidian endoderm invagination during gastrulation. Semin. Cell Dev. Biol. 2021, 120, 108–118. [Google Scholar] [CrossRef]
- Straight, A.F.; Cheung, A.; Limouze, J.; Chen, I.; Westwood, N.J.; Sellers, J.R.; Mitchison, T.J. Dissecting temporal and spatial control of cytokinesis with a myosin II inhibitor. Science 2003, 299, 1743–1747. [Google Scholar] [CrossRef] [PubMed]
- Wang, T.; Kang, W.; Du, L.; Ge, S. Rho-kinase inhibitor Y-27632 facilitates the proliferation, migration and pluripotency of human periodontal ligament stem cells. J. Cell. Mol. Med. 2017, 21, 3100–3112. [Google Scholar] [CrossRef]
- Huang, C.; Zhang, Z.; Wang, L.; Liu, J.; Gong, X.; Zhang, C. ML-7 attenuates airway inflammation and remodeling via inhibiting the secretion of Th2 cytokines in mice model of asthma. Mol. Med. Rep. 2018, 17, 6293–6300. [Google Scholar] [CrossRef] [PubMed]
- Stephan, L.; Jakoby, M.; Das, A.; Koebke, E.; Hülskamp, M. Unravelling the molecular basis of the dominant negative effect of myosin XI tails on P-bodies. PLoS ONE 2021, 16, e0252327. [Google Scholar] [CrossRef]
- Oh, T.J.; Fan, H.; Skeeters, S.S.; Zhang, K. Steering molecular activity with optogenetics: Recent advances and perspectives. Adv. Biol. 2021, 5, e2000180. [Google Scholar] [CrossRef]
- Yizhar, O.; Fenno, L.E.; Davidson, T.J.; Mogri, M.; Deisseroth, K. Optogenetics in neural systems. Neuron 2011, 71, 9–34. [Google Scholar] [CrossRef] [PubMed]
- Oesterhelt, D.; Stoeckenius, W. Rhodopsin-like protein from the purple membrane of Halobacterium halobium. Nat. New Biol. 1971, 233, 149–152. [Google Scholar] [CrossRef]
- Oesterhelt, D.; Stoeckenius, W. Functions of a new photoreceptor membrane. Proc. Natl. Acad. Sci. USA 1973, 70, 2853–2857. [Google Scholar] [CrossRef]
- Matsuno-Yagi, A.; Mukohata, Y.J.B. Two possible roles of bacteriorhodopsin; a comparative study of strains of Halobacterium halobium differing in pigmentation. Biochem. Biophys. Res. Commun. 1977, 78, 237–243. [Google Scholar] [CrossRef]
- Lanyi, J.; Oesterhelt, D. Identification of the retinal-binding protein in halorhodopsin. J. Biol. Chem. 1982, 257, 2674–2677. [Google Scholar] [CrossRef]
- Boyden, E.S.; Zhang, F.; Bamberg, E.; Nagel, G.; Deisseroth, K. Millisecond-timescale, genetically targeted optical control of neural activity. Nat. Neurosci. 2005, 8, 1263–1268. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Gutierrez, D.V.; Hanson, M.G.; Han, J.; Mark, M.D.; Chiel, H.; Hegemann, P.; Landmesser, L.T.; Herlitze, S. Fast noninvasive activation and inhibition of neural and network activity by vertebrate rhodopsin and green algae channelrhodopsin. Proc. Natl. Acad. Sci. USA 2005, 102, 17816–17821. [Google Scholar] [CrossRef] [PubMed]
- Vilinsky, I.; Hibbard, K.L.; Johnson, B.R.; Deitcher, D.L. Probing synaptic transmission and behavior in drosophila with optogenetics: A laboratory exercise. J. Undergrad. Neurosci. Educ. 2018, 16, A289–A295. [Google Scholar] [PubMed]
- Krueger, D.; Izquierdo, E.; Viswanathan, R.; Hartmann, J.; Pallares Cartes, C.; De Renzis, S. Principles and applications of optogenetics in developmental biology. Development 2019, 146, dev175067. [Google Scholar] [CrossRef]
- Dwijayanti, A.; Zhang, C.; Poh, C.L.; Lautier, T. Toward multiplexed optogenetic circuits. Front. Bioeng. Biotechnol. 2021, 9, 804563. [Google Scholar] [CrossRef]
- Izquierdo, E.; Quinkler, T.; De Renzis, S. Guided morphogenesis through optogenetic activation of Rho signalling during early Drosophila embryogenesis. Nat. Commun. 2018, 9, 2366. [Google Scholar] [CrossRef]
- Fang, F.; Lin, L.; Zhang, Q.; Lu, M.; Skvortsova, M.Y.; Podolec, R.; Zhang, Q.; Pi, J.; Zhang, C.; Ulm, R.; et al. Mechanisms of UV-B light-induced photoreceptor UVR8 nuclear localization dynamics. New Phytol. 2022, 236, 1824–1837. [Google Scholar] [CrossRef]
- Glantz, S.T.; Berlew, E.E.; Jaber, Z.; Schuster, B.S.; Gardner, K.H.; Chow, B.Y. Directly light-regulated binding of RGS-LOV photoreceptors to anionic membrane phospholipids. Pro. Natl. Acad. Sci. USA 2018, 115, E7720–E7727. [Google Scholar] [CrossRef]
- Losi, A.; Gärtner, W. Solving blue light riddles: New lessons from flavin-binding LOV photoreceptors. Photochem. Photobiol. 2017, 93, 141–158. [Google Scholar] [CrossRef]
- Harper, S.M.; Neil, L.C.; Gardner, K.H. Structural basis of a phototropin light switch. Science 2003, 301, 1541–1544. [Google Scholar] [CrossRef]
- Herrou, J.; Crosson, S. Function, structure and mechanism of bacterial photosensory LOV proteins. Nat. Rev. Microbiol. 2011, 9, 713–723. [Google Scholar] [CrossRef] [PubMed]
- Glantz, S.T.; Carpenter, E.J.; Melkonian, M.; Gardner, K.H.; Boyden, E.S.; Wong, G.K.; Chow, B.Y. Functional and topological diversity of LOV domain photoreceptors. Proc. Natl. Acad. Sci. USA 2016, 113, E1442–E1451. [Google Scholar] [CrossRef] [PubMed]
- Zayner, J.P.; Antoniou, C.; Sosnick, T.R. The amino-terminal helix modulates light-activated conformational changes in AsLOV2. J. Mol. Biol. 2012, 419, 61–74. [Google Scholar] [CrossRef] [PubMed]
- Berlew, E.E.; Kuznetsov, I.A.; Yamada, K.; Bugaj, L.J.; Chow, B.Y. Optogenetic Rac1 engineered from membrane lipid-binding RGS-LOV for inducible lamellipodia formation. Photochem. Photobiol. Sci. 2020, 19, 353–361. [Google Scholar] [CrossRef] [PubMed]
- Yamamoto, K.; Miura, H.; Ishida, M.; Sawai, S.; Kondo, Y.; Aoki, K. Optogenetic relaxation of actomyosin contractility uncovers mechanistic roles of cortical tension during cytokinesis. Nat. Commun. 2021, 12, 7145. [Google Scholar] [CrossRef]
- Berlew, E.E.; Kuznetsov, I.A.; Yamada, K.; Bugaj, L.J.; Boerckel, J.D.; Chow, B.Y. Single-component optogenetic tools for inducible RhoA GTPase signaling. Adv. boil. 2021, 5, e2100810. [Google Scholar] [CrossRef]
- Berlew, E.E.; Yamada, K.; Kuznetsov, I.A.; Rand, E.A.; Ochs, C.C.; Jaber, Z.; Gardner, K.H.; Chow, B.Y. Designing single-component optogenetic membrane recruitment systems: The Rho-family GTPase signaling toolbox. ACS Synth. Boil. 2022, 11, 515–521. [Google Scholar] [CrossRef]
- Glantz, S.T.; Berlew, E.E.; Chow, B.Y. Synthetic cell-like membrane interfaces for probing dynamic protein-lipid interactions. Methods Enzymol. 2019, 622, 249–270. [Google Scholar]
- Chang, A.N.; Gao, N.; Liu, Z.; Huang, J.; Nairn, A.C.; Kamm, K.E.; Stull, J.T. The dominant protein phosphatase PP1c isoform in smooth muscle cells, PP1cβ, is essential for smooth muscle contraction. J. Biol. Chem. 2018, 293, 16677–16686. [Google Scholar] [CrossRef]
- Wu, Y.; Murányi, A.; Erdodi, F.; Hartshorne, D.J. Localization of myosin phosphatase target subunit and its mutants. J. Muscle Res. Cell Motil. 2005, 26, 123–134. [Google Scholar] [CrossRef]
- Riedl, J.; Crevenna, A.H.; Kessenbrock, K.; Yu, J.H.; Neukirchen, D.; Bista, M.; Bradke, F.; Jenne, D.; Holak, T.A.; Werb, Z.; et al. Lifeact: A versatile marker to visualize F-actin. Nat. Methods 2008, 5, 605–607. [Google Scholar] [CrossRef]
- Flores, L.R.; Keeling, M.C.; Zhang, X.; Sliogeryte, K.; Gavara, N. Lifeact-GFP alters F-actin organization, cellular morphology and biophysical behaviour. Sci. Rep. 2019, 9, 3241. [Google Scholar] [CrossRef]
- Du, J.; Fan, Y.L.; Chen, T.L.; Feng, X.Q. Lifeact and Utr230 induce distinct actin assemblies in cell nuclei. Cytoskeleton 2015, 72, 570–575. [Google Scholar] [CrossRef] [PubMed]
- Sliogeryte, K.; Thorpe, S.D.; Wang, Z.; Thompson, C.L.; Gavara, N.; Knight, M.M. Differential effects of LifeAct-GFP and actin-GFP on cell mechanics assessed using micropipette aspiration. J. Biomech. 2016, 49, 310–317. [Google Scholar] [CrossRef] [PubMed]
- Belyy, A.; Merino, F.; Sitsel, O.; Raunser, S. Structure of the Lifeact-F-actin complex. PLoS Biol. 2020, 18, e3000925. [Google Scholar] [CrossRef] [PubMed]
- Petrosyan, A.; Casey, C.A.; Cheng, P.W. The role of Rab6a and phosphorylation of non-muscle myosin IIA tailpiece in alcohol-induced Golgi disorganization. Sci. Rep. 2016, 6, 31962. [Google Scholar] [CrossRef]
- Rossman, K.L.; Der, C.J.; Sondek, J. GEF means go: Turning on RHO GTPases with guanine nucleotide-exchange factors. Nat. Rev. Mol. Cell Biol. 2005, 6, 167–180. [Google Scholar] [CrossRef]
- Lv, Z.; Lu, Q.; Dong, B. Morphogenesis: A focus on marine invertebrates. Mar. Life Sci. Technol. 2019, 1, 28–40. [Google Scholar] [CrossRef]
- Sawyer, J.M.; Harrell, J.R.; Shemer, G.; Sullivan-Brown, J.; Roh-Johnson, M.; Goldstein, B. Apical constriction: A cell shape change that can drive morphogenesis. Dev. Biol. 2010, 341, 5–19. [Google Scholar] [CrossRef]
- Christiaen, L.; Wagner, E.; Shi, W.; Levine, M. Electroporation of transgenic DNAs in the sea squirt Ciona. Cold Spring Harb. Protoc. 2009, 2009, pdb.prot5345. [Google Scholar] [CrossRef]


Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Qiao, J.; Peng, H.; Dong, B. Development and Application of an Optogenetic Manipulation System to Suppress Actomyosin Activity in Ciona Epidermis. Int. J. Mol. Sci. 2023, 24, 5707. https://doi.org/10.3390/ijms24065707
Qiao J, Peng H, Dong B. Development and Application of an Optogenetic Manipulation System to Suppress Actomyosin Activity in Ciona Epidermis. International Journal of Molecular Sciences. 2023; 24(6):5707. https://doi.org/10.3390/ijms24065707
Chicago/Turabian StyleQiao, Jinghan, Hongzhe Peng, and Bo Dong. 2023. "Development and Application of an Optogenetic Manipulation System to Suppress Actomyosin Activity in Ciona Epidermis" International Journal of Molecular Sciences 24, no. 6: 5707. https://doi.org/10.3390/ijms24065707
APA StyleQiao, J., Peng, H., & Dong, B. (2023). Development and Application of an Optogenetic Manipulation System to Suppress Actomyosin Activity in Ciona Epidermis. International Journal of Molecular Sciences, 24(6), 5707. https://doi.org/10.3390/ijms24065707





