Turning a Tumor Microenvironment Pitfall into Opportunity: Discovery of Benzamidoxime as PD-L1 Ligand with pH-Dependent Potency
Abstract
1. Introduction
2. Results
2.1. Selection of Virtual Hit Compounds
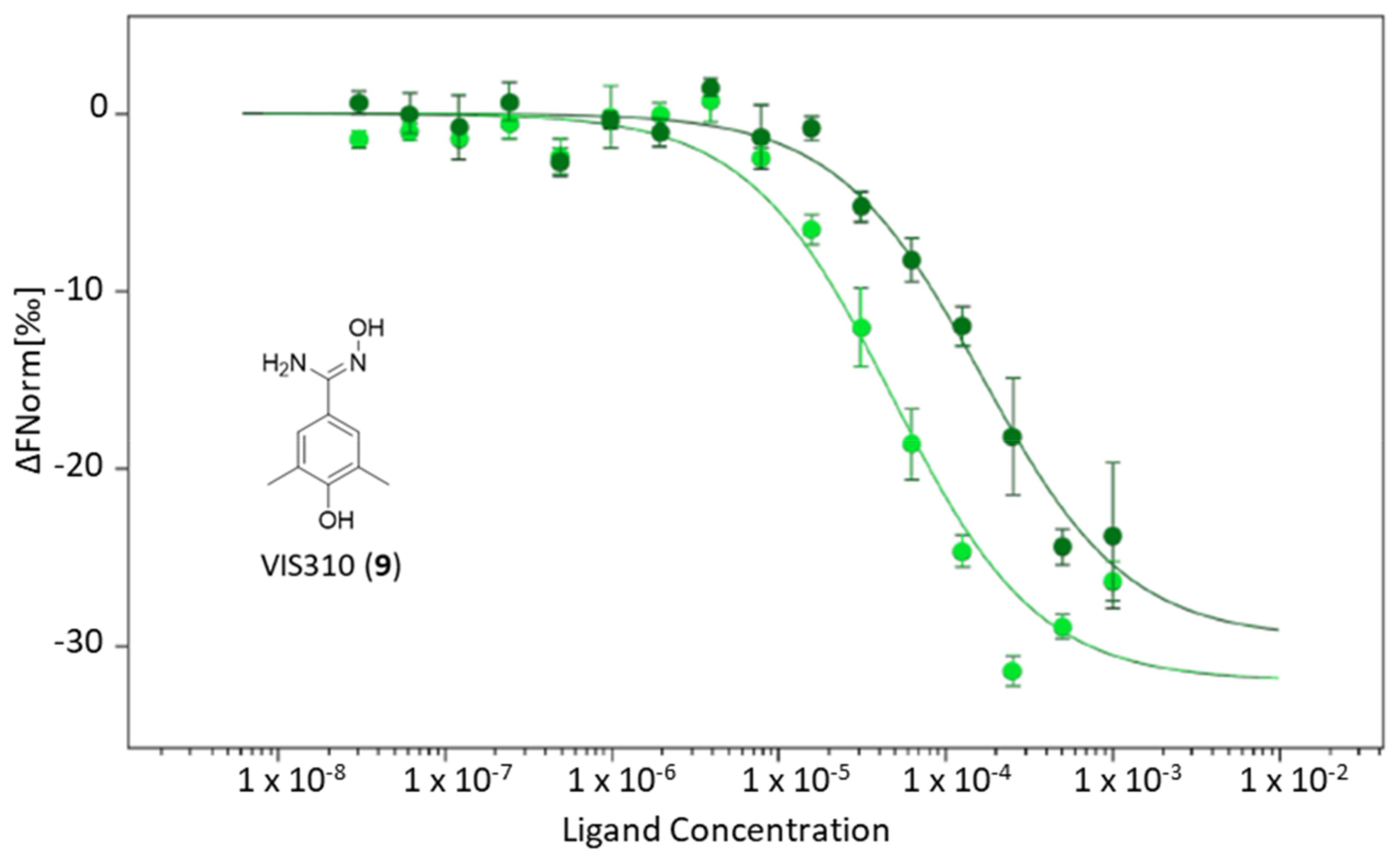
2.2. Identification of VIS310 as Hit Compound
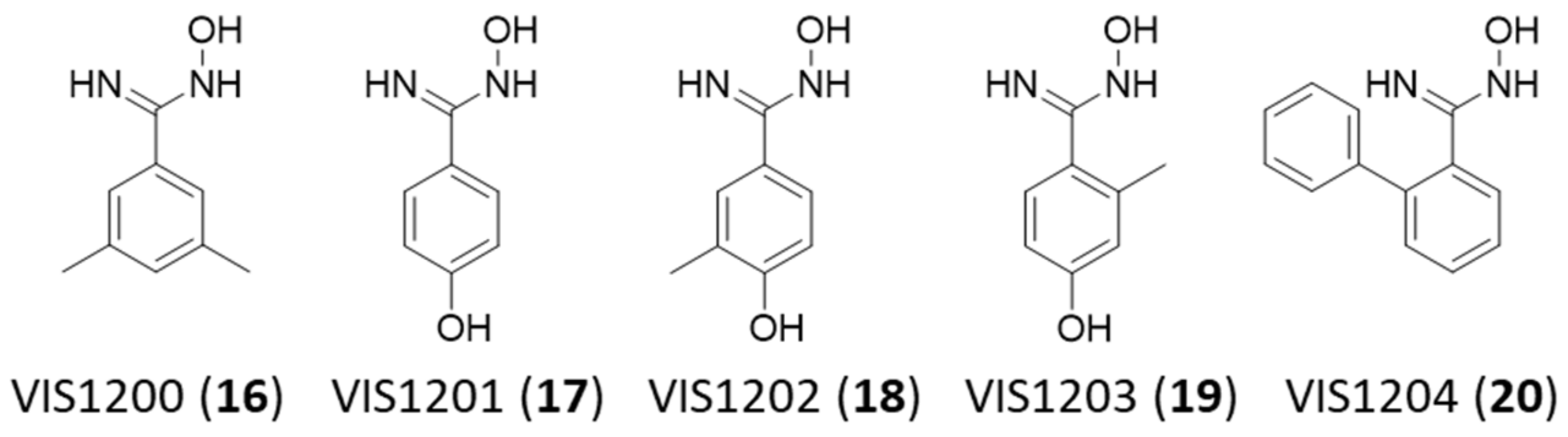
2.3. VIS310 Analogues and Structure–Activity Relationships
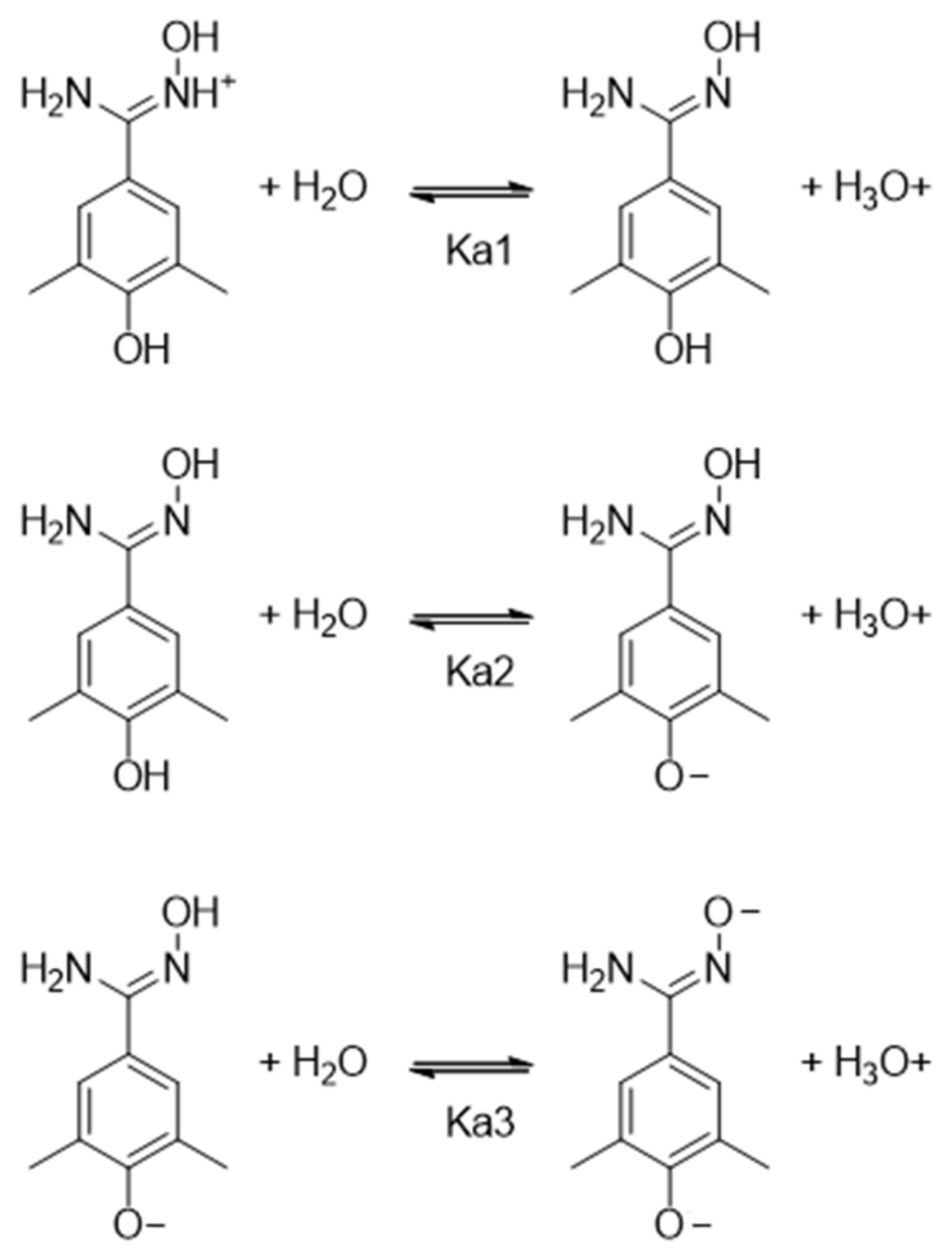
2.4. pKa Measurements
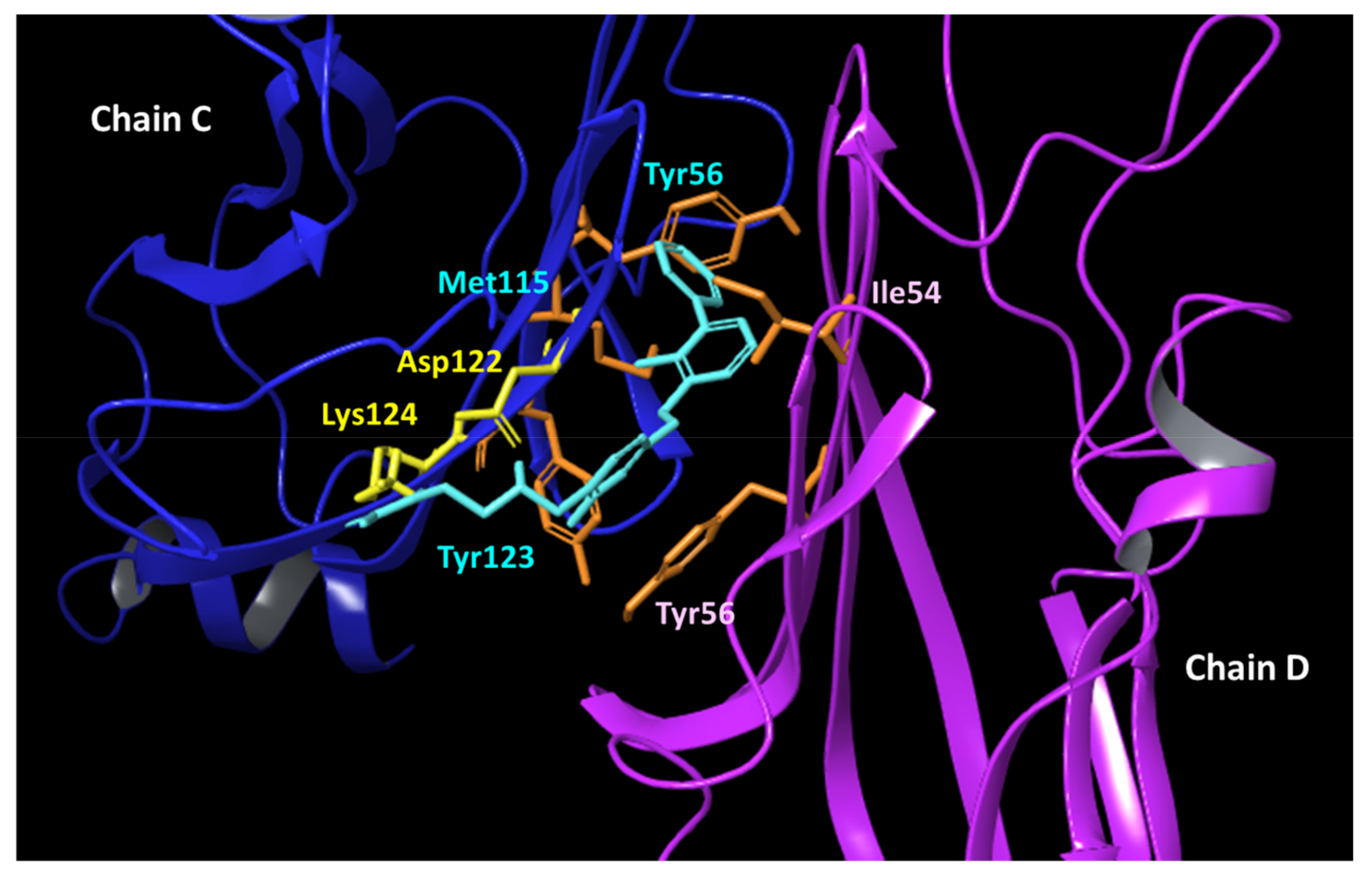
2.5. Binding Modes of VIS310 Analogues to PD-L1
2.6. Ligand Binding Displacement Assay of PD-1/PD-L1 Complex
3. Discussion
4. Materials and Methods
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Chinai, J.M.; Janakiram, M.; Chen, F.; Chen, W.; Kaplan, M.; Zang, X. New immunotherapies targeting the PD-1 pathway. Trends Pharmacol. Sci. 2015, 36, 587–595. [Google Scholar] [CrossRef]
- Boussiotis, V.A. Molecular and Biochemical Aspects of the PD-1 Checkpoint Pathway. N. Engl. J. Med. 2016, 375, 1767–1778. [Google Scholar] [CrossRef] [PubMed]
- Okazaki, T.; Honjo, T. PD-1 and PD-1 ligands: From discovery to clinical application. Int. Immunol. 2007, 19, 813–824. [Google Scholar] [CrossRef] [PubMed]
- Dong, H.; Strome, S.E.; Salomao, D.R.; Tamura, H.; Hirano, F.; Flies, D.B.; Roche, P.C.; Lu, J.; Zhu, G.; Tamada, K.; et al. Tumor-associated B7-H1 promotes T-cell apoptosis: A potential mechanism of immune evasion. Nat. Med. 2002, 8, 793–800. [Google Scholar] [CrossRef] [PubMed]
- Riella, L.V.; Paterson, A.M.; Sharpe, A.H.; Chandraker, A. Role of the PD-1 pathway in the immune response. Am. J. Transplant. 2012, 12, 2575–2587. [Google Scholar] [CrossRef] [PubMed]
- Zou, W.; Wolchok, J.D.; Chen, L. PD-L1 (B7-H1) and PD-1 pathway blockade for cancer therapy: Mechanisms, response biomarkers, and combinations. Sci. Transl. Med. 2016, 8, 328rv4. [Google Scholar] [CrossRef] [PubMed]
- Kalantari, N.K.; Ghahremanloo, A.; Hashemy, S.I. Role of tumor microenvironment in the regulation of PD-L1: A novel role in resistance to cancer immunotherapy. J. Cell Physiol. 2020, 235, 6496–6506. [Google Scholar] [CrossRef] [PubMed]
- Herbst, R.S.; Soria, J.C.; Kowanetz, M.; Fine, G.D.; Hamid, O.; Gordon, M.S.; Sosman, J.A.; McDermott, D.F.; Powderly, J.D.; Gettinger, S.N.; et al. Predictive correlates of response to the anti-PD-L1 antibody MPDL3280A in cancer patients. Nature 2014, 515, 563–567. [Google Scholar] [CrossRef]
- Zarganes-Tzitzikas, T.; Konstantinidou, M.; Gao, Y.; Krzemien, D.; Zak, K.; Dubin, G.; Holak, T.A.; Dömling, A. Inhibitors of programmed cell death 1 (PD-1): A patent review (2010–2015). Expert Opin. Ther. Pat. 2016, 26, 973–977. [Google Scholar] [CrossRef]
- Twomey, J.D.; Zhang, B. Cancer Immunotherapy Update: FDA-Approved Checkpoint Inhibitors and Companion Diagnostics. AAPS J. 2021, 23, 39. [Google Scholar] [CrossRef]
- Yi, M.; Zheng, X.; Niu, M.; Zhu, S.; Ge, H.; Wu, K. Combination strategies with PD-1/PD-L1 blockade: Current advances and future directions. Mol. Cancer 2022, 21, 28. [Google Scholar] [CrossRef] [PubMed]
- Maute, R.L.; Gordon, S.R.; Mayer, A.T.; McCracken, M.N.; Natarajan, A.; Ring, N.G.; Kimura, R.; Tsai, J.M.; Manglik, A.; Kruse, A.C.; et al. Engineering high-affinity PD-1 variants for optimized immunotherapy and immuno-PET imaging. Proc. Natl. Acad. Sci. USA 2015, 112, E6506–E6514. [Google Scholar] [CrossRef]
- O’Donnell, J.S.; Long, G.V.; Scolyer, R.A.; Teng, M.W.; Smyth, M.J. Resistance to PD1/PDL1 checkpoint inhibition. Cancer Treat. Rev. 2017, 52, 71–81. [Google Scholar] [CrossRef]
- Lim, S.Y.; Rizos, H. Immune cell profiling in the age of immune checkpoint inhibitors: Implications for biomarker discovery and understanding of resistance mechanisms. Mamm. Genome 2018, 29, 866–878. [Google Scholar] [CrossRef] [PubMed]
- Wu, Q.; Jiang, L.; Li, S.C.; He, Q.J.; Yang, B.; Cao, J. Small molecule inhibitors targeting the PD-1/PD-L1 signaling pathway. Acta Pharmacol. Sin. 2021, 42, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Awadasseid, A.; Wu, Y.; Zhang, W. Advance investigation on synthetic small-molecule inhibitors targeting PD-1/PD-L1 signaling pathway. Life Sci. 2021, 282, 119813. [Google Scholar] [CrossRef] [PubMed]
- Yamaguchi, H.; Hsu, J.M.; Yang, W.H.; Hung, M.C. Mechanisms regulating PD-L1 expression in cancers and associated opportunities for novel small-molecule therapeutics. Nat. Rev. Clin. Oncol. 2022, 19, 287–305. [Google Scholar] [CrossRef] [PubMed]
- Guzik, K.; Zak, K.M.; Grudnik, P.; Magiera, K.; Musielak, B.; Törner, R.; Skalniak, L.; Dömling, A.; Dubin, G.; Holak, T.A. Small-Molecule Inhibitors of the Programmed Cell Death-1/Programmed Death-Ligand 1 (PD-1/PD-L1) Interaction via Transiently Induced Protein States and Dimerization of PD-L1. J. Med. Chem. 2017, 60, 5857–5867. [Google Scholar] [CrossRef]
- Skalniak, L.; Zak, K.M.; Guzik, K.; Magiera, K.; Musielak, B.; Pachota, M.; Szelazek, B.; Kocik, J.; Grudnik, P.; Tomala, M.; et al. Small-molecule inhibitors of PD-1/PD-L1 immune checkpoint alleviate the PD-L1-induced exhaustion of T-cells. Oncotarget 2017, 8, 72167–72181. [Google Scholar] [CrossRef]
- Huck, B.R.; Kötzner, L.; Urbahns, K. Small Molecules Drive Big Improvements in Immuno-Oncology Therapies. Angew. Chem. Int. Ed. Engl. 2018, 57, 4412–4428. [Google Scholar] [CrossRef]
- Ganesan, A.; Ahmed, M.; Okoye, I.; Arutyunova, E.; Babu, D.; Turnbull, W.L.; Kundu, J.K.; Shields, J.; Agopsowicz, K.C.; Xu, L.; et al. Comprehensive in vitro characterization of PD-L1 small molecule inhibitors. Sci. Rep. 2019, 9, 12392. [Google Scholar] [CrossRef]
- Zhai, W.; Zhou, X.; Du, J.; Gao, Y. In vitro assay for the development of small molecule inhibitors targeting PD-1/PD-L1. Methods Enzymol. 2019, 629, 361–381. [Google Scholar]
- Guzik, K.; Tomala, M.; Muszak, D.; Konieczny, M.; Hec, A.; Błaszkiewicz, U.; Pustuła, M.; Butera, R.; Dömling, A.; Holak, T.A. Development of the Inhibitors that Target the PD-1/PD-L1 Interaction-A Brief Look at Progress on Small Molecules, Peptides and Macrocycles. Molecules 2019, 24, 2071. [Google Scholar] [CrossRef] [PubMed]
- Zak, K.M.; Grudnik, P.; Guzik, K.; Zieba, B.J.; Musielak, B.; Dömling, A.; Dubin, G.; Holak, T.A. Structural basis for small molecule targeting of the programmed death ligand 1 (PD-L1). Oncotarget 2016, 7, 30323–30335. [Google Scholar] [CrossRef] [PubMed]
- Boedtkjer, E.; Pedersen, S.F. The Acidic Tumor Microenvironment as a Driver of Cancer. Annu. Rev. Physiol. 2020, 82, 103–126. [Google Scholar] [CrossRef]
- Wang, J.X.; Choi, S.Y.C.; Niu, X.; Kang, N.; Xue, H.; Killam, J.; Wang, Y. Lactic Acid and an Acidic Tumor Microenvironment suppress Anticancer Immunity. Int. J. Mol. Sci. 2020, 21, 8363. [Google Scholar] [CrossRef] [PubMed]
- Riccio, A.; Coletti, A.; Dolciami, D.; Mammoli, A.; Cerra, B.; Moretti, S.; Gioiello, A.; Ferlin, S.; Puxeddu, E.; Macchiarulo, A. The Stone Guest: How Does pH Affect Binding Properties of PD-1/PD-L1 Inhibitors? ChemMedChem 2021, 16, 568–577. [Google Scholar] [CrossRef]
- Corbet, C.; Feron, O. Tumour acidosis: From the passenger to the driver’s seat. Nat. Rev. Cancer 2017, 17, 577–593. [Google Scholar] [CrossRef]
- Webb, B.A.; Chimenti, M.; Jacobson, M.P.; Barber, D.L. Dysregulated pH: A perfect storm for cancer progression. Nat. Rev. Cancer 2011, 11, 671–677. [Google Scholar] [CrossRef]
- Javadrashid, D.; Baghbanzadeh, A.; Derakhshani, A.; Leone, P.; Silvestris, N.; Racanelli, V.; Solimando, A.G.; Baradaran, B. Pancreatic Cancer Signaling Pathways, Genetic Alterations, and Tumor Microenvironment: The Barriers Affecting the Method of Treatment. Biomedicines 2021, 9, 373. [Google Scholar] [CrossRef]
- Magiera-Mularz, K.; Skalniak, L.; Zak, K.M.; Musielak, B.; Rudzinska-Szostak, E.; Berlicki, Ł.; Kocik, J.; Grudnik, P.; Sala, D.; Zarganes-Tzitzikas, T.; et al. Bioactive Macrocyclic Inhibitors of the PD-1/PD-L1 Immune Checkpoint. Angew. Chem. Int. Ed. Engl. 2017, 56, 13732–13735. [Google Scholar] [CrossRef] [PubMed]
- Backman, T.W.; Cao, Y.; Girke, T. ChemMine tools: An online service for analyzing and clustering small molecules. Nucleic Acids Res. 2011, 39, W486–W491. [Google Scholar] [CrossRef] [PubMed]
- Mehio, N.; Lashely, M.A.; Nugent, J.W.; Tucker, L.; Correia, B.; Do-Thanh, C.L.; Dai, S.; Hancock, R.D.; Bryantsev, V.S. Acidity of the amidoxime functional group in aqueous solution: A combined experimental and computational study. J. Phys. Chem. B 2015, 119, 3567–3576. [Google Scholar] [CrossRef] [PubMed]
- Dardonville, C. Automated techniques in pKa determination: Low, medium and high-throughput screening methods. Drug Discov. Today Technol. 2018, 27, 49–58. [Google Scholar] [CrossRef] [PubMed]
- Cheng, T.; Li, X.; Li, Y.; Liu, Z.; Wang, R. Comparative assessment of scoring functions on a diverse test set. J. Chem. Inf. Model. 2009, 49, 1079–1093. [Google Scholar] [CrossRef]
- Sándor, M.; Kiss, R.; Keseru, G.M. Virtual fragment docking by Glide: A validation study on 190 protein-fragment complexes. J. Chem. Inf. Model. 2010, 50, 1165–1172. [Google Scholar] [CrossRef]
- Mammoli, A.; Bianconi, E.; Ruta, L.; Riccio, A.; Bigiotti, C.; Souma, M.; Carotti, A.; Rossini, S.; Suvieri, C.; Pallotta, M.T.; et al. Critical Assessment of a Structure-Based Screening Campaign for IDO1 Inhibitors: Tips and Pitfalls. Int. J. Mol. Sci. 2022, 23, 3981. [Google Scholar] [CrossRef]
- Berman, H.M.; Westbrook, J.; Feng, Z.; Gilliland, G.; Bhat, T.N.; Weissig, H.; Shindyalov, I.N.; Bourne, P.E. The Protein Data Bank. Nucleic Acids Res. 2000, 28, 235–242. [Google Scholar] [CrossRef] [PubMed]
- Friesner, R.A.; Banks, J.L.; Murphy, R.B.; Halgren, T.A.; Klicic, J.J.; Mainz, D.T.; Repasky, M.P.; Knoll, E.H.; Shelley, M.; Perry, J.K.; et al. Glide: A new approach for rapid, accurate docking and scoring. 1. Method and as-sessment of docking accuracy. J. Med. Chem. 2004, 47, 1739–1749. [Google Scholar] [CrossRef]
- Polak, E.; Ribiere, G. Note sur la Convergence de Methodes de Directions Conjuguees. Revue Fr. Inf. Rech. Oper. Ser. Rouge 1969, 3, 35–43. [Google Scholar] [CrossRef]
- Scopes, R.K. Measurement of protein by spectrophotometry at 205 nm. Anal. Biochem. 1974, 59, 277–282. [Google Scholar] [CrossRef] [PubMed]
- Duhr, S.; Braun, D. Why molecules move along a temperature gradient. Proc. Natl. Acad. Sci. USA 2006, 103, 19678–19682. [Google Scholar] [CrossRef] [PubMed]
- Abad-Zapatero, C. Ligand efficiency indices for effective drug discovery. Expert Opin. Drug Discov. 2007, 2, 469–488. [Google Scholar] [CrossRef] [PubMed]




| Virtual Hits | GScore (kcal/mol) | Binder [a] | Kd ± c.v. [b] (μM) | MW (Dalton) | BEI |
|---|---|---|---|---|---|
| BMS-202 (5) | −10.22 | x | 8.13 ± 1.38 | 419 | 12.1 |
| VIS284 (6) | −7.94 | x | >250 | 192 | n.d. [c] |
| VIS285 (7) | −7.05 | x/o | >250 | 175 | n.d. |
| VIS303 (8) | −9.13 | x | >250 | 299 | n.d. |
| VIS310 (9) | −7.22 | x | 163.75 ± 33.61 | 180 | 21.0 |
| VIS545 (10) | −9.09 | x/o | >250 | 269 | n.d. |
| VIS826 (11) | −6.05 | x | >250 | 316 | n.d. |
| VIS877 (12) | −4.76 | x | >250 | 226 | n.d. |
| VIS879 (13) | −4.56 | x | >250 | 254 | n.d. |
| VIS1079 (14) | −7.61 | x/o | >250 | 209 | n.d. |
| VIS32926 (15) | −7.71 | x/o | >250 | 195 | n.d. |
| Virtual Hits | pH 7.2 Kd ± c.v. [a] (μM) | pH 6.2 Kd ± c.v. [a] (μM) | MW (Dalton) | BEI (pH 6.2) |
|---|---|---|---|---|
| BMS-202 (5) | 8.13 ± 1.38 | 4.50 ± 0.56 | 419 | 12.8 |
| VIS310 (9) | 164 ± 33.6 | 47.8 ± 11.5 | 180 | 24.0 |
| VIS1200 (16) | >1000 | 95.6 ± 20.6 | 164 | 24.5 |
| VIS1201 (17) | >1000 | 45.2 ± 10.6 | 152 | 28.6 |
| VIS1202 (18) | >1000 | 55.4 ± 9.86 | 166 | 25.6 |
| VIS1203 (19) | >1000 | 54.9 ± 8.20 | 166 | 25.7 |
| VIS1204 (20) | 148 ± 71.3 | 70.8 ± 12.7 | 212 | 19.6 |
| Compounds | pKa1 | pKa2 | pKa3 |
|---|---|---|---|
| BMS-202 (5) | n.d. [a] | 8.39 ± 0.05 [c] | - |
| VIS310 (9) | 5.65 ± 0.01 [b] | 9.32 ± 0.02 [d] | n.d. [e] |
| VIS1200 (16) | 5.26 ± 0.01 [b] | - | n.d. [e] |
| VIS1201 (17) | 5.35 ± 0.12 [b] | 8.64 ± 0.01 [d] | n.d. [e] |
| VIS1202 (18) | 5.38 ± 0.01 [b] | 8.97 ± 0.03 [d] | n.d. [e] |
| VIS1203 (19) | 5.38 ± 0.01 [b] | 9.17 ± 0.02 [d] | n.d. [e] |
| VIS1204 (20) | 5.30 ± 0.01 [b] | - | n.d. [e] |
| Vehicle Kd ± c.v. [a] (pH 6.2) | VIS1201 (50 μM) Kd ± c.v. [a] (pH 6.2) | VIS1201 (100 μM) Kd ± c.v. [a] (pH 6.2) | |
|---|---|---|---|
| PD-1/PD-L1 | 2.50 ± 0.50 | 9.68 ± 2.57 | >30 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Bianconi, E.; Riccio, A.; Ruta, L.; Bigiotti, C.; Carotti, A.; Moretti, S.; Cerra, B.; Gioiello, A.; Ferlin, S.; Puxeddu, E.; et al. Turning a Tumor Microenvironment Pitfall into Opportunity: Discovery of Benzamidoxime as PD-L1 Ligand with pH-Dependent Potency. Int. J. Mol. Sci. 2023, 24, 5535. https://doi.org/10.3390/ijms24065535
Bianconi E, Riccio A, Ruta L, Bigiotti C, Carotti A, Moretti S, Cerra B, Gioiello A, Ferlin S, Puxeddu E, et al. Turning a Tumor Microenvironment Pitfall into Opportunity: Discovery of Benzamidoxime as PD-L1 Ligand with pH-Dependent Potency. International Journal of Molecular Sciences. 2023; 24(6):5535. https://doi.org/10.3390/ijms24065535
Chicago/Turabian StyleBianconi, Elisa, Alessandra Riccio, Luana Ruta, Carlo Bigiotti, Andrea Carotti, Sonia Moretti, Bruno Cerra, Antimo Gioiello, Simone Ferlin, Efisio Puxeddu, and et al. 2023. "Turning a Tumor Microenvironment Pitfall into Opportunity: Discovery of Benzamidoxime as PD-L1 Ligand with pH-Dependent Potency" International Journal of Molecular Sciences 24, no. 6: 5535. https://doi.org/10.3390/ijms24065535
APA StyleBianconi, E., Riccio, A., Ruta, L., Bigiotti, C., Carotti, A., Moretti, S., Cerra, B., Gioiello, A., Ferlin, S., Puxeddu, E., & Macchiarulo, A. (2023). Turning a Tumor Microenvironment Pitfall into Opportunity: Discovery of Benzamidoxime as PD-L1 Ligand with pH-Dependent Potency. International Journal of Molecular Sciences, 24(6), 5535. https://doi.org/10.3390/ijms24065535











