8-Oxoguanine DNA Glycosylase 1 Upregulation as a Risk Factor for Obesity and Colorectal Cancer
Abstract
1. Introduction
2. Results
2.1. Baseline and General Characteristics of Participants
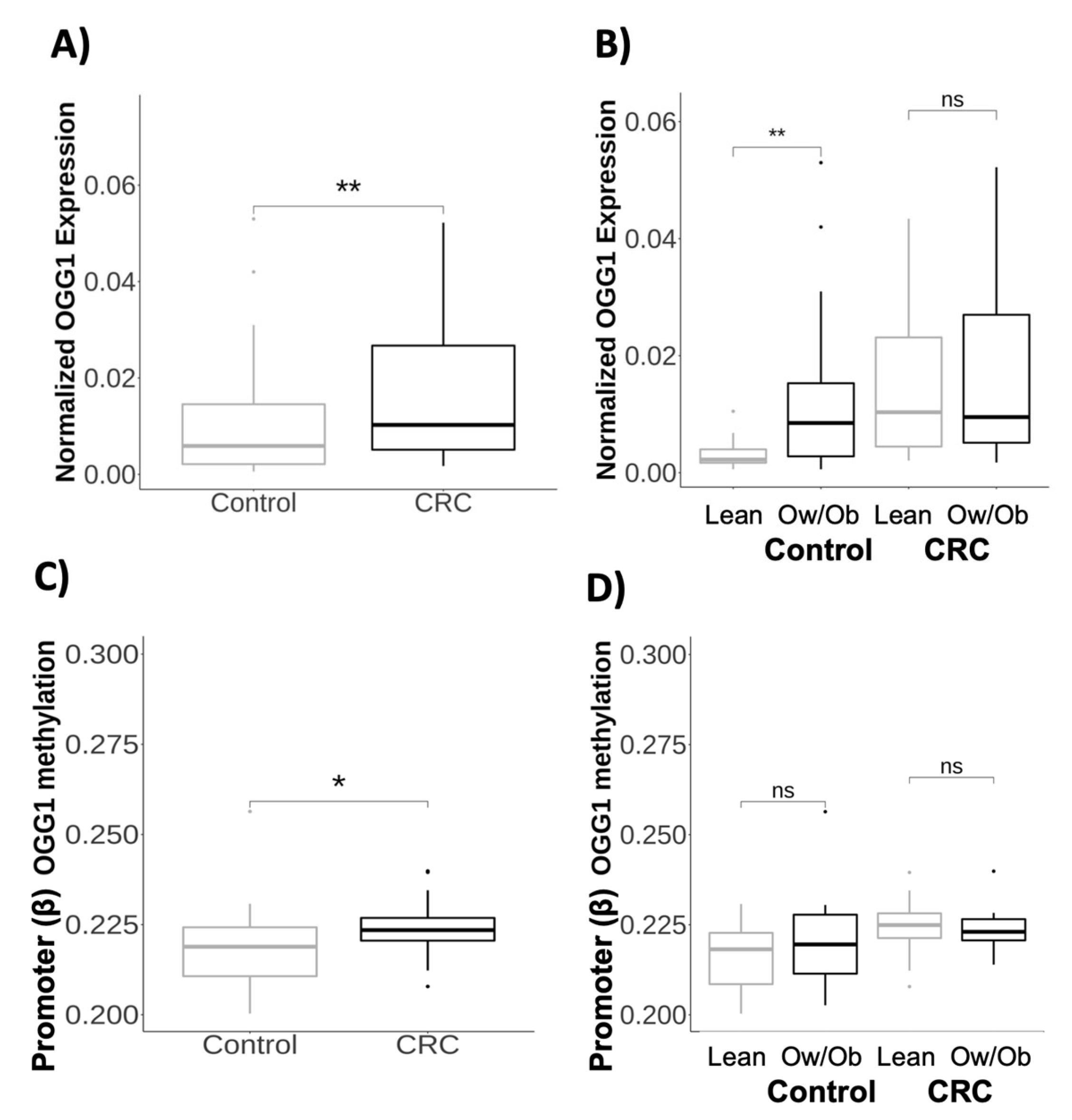
2.2. Transcriptional and Methylation Profiles of the OGG1 Gene in Obesity and Colorectal Cancer
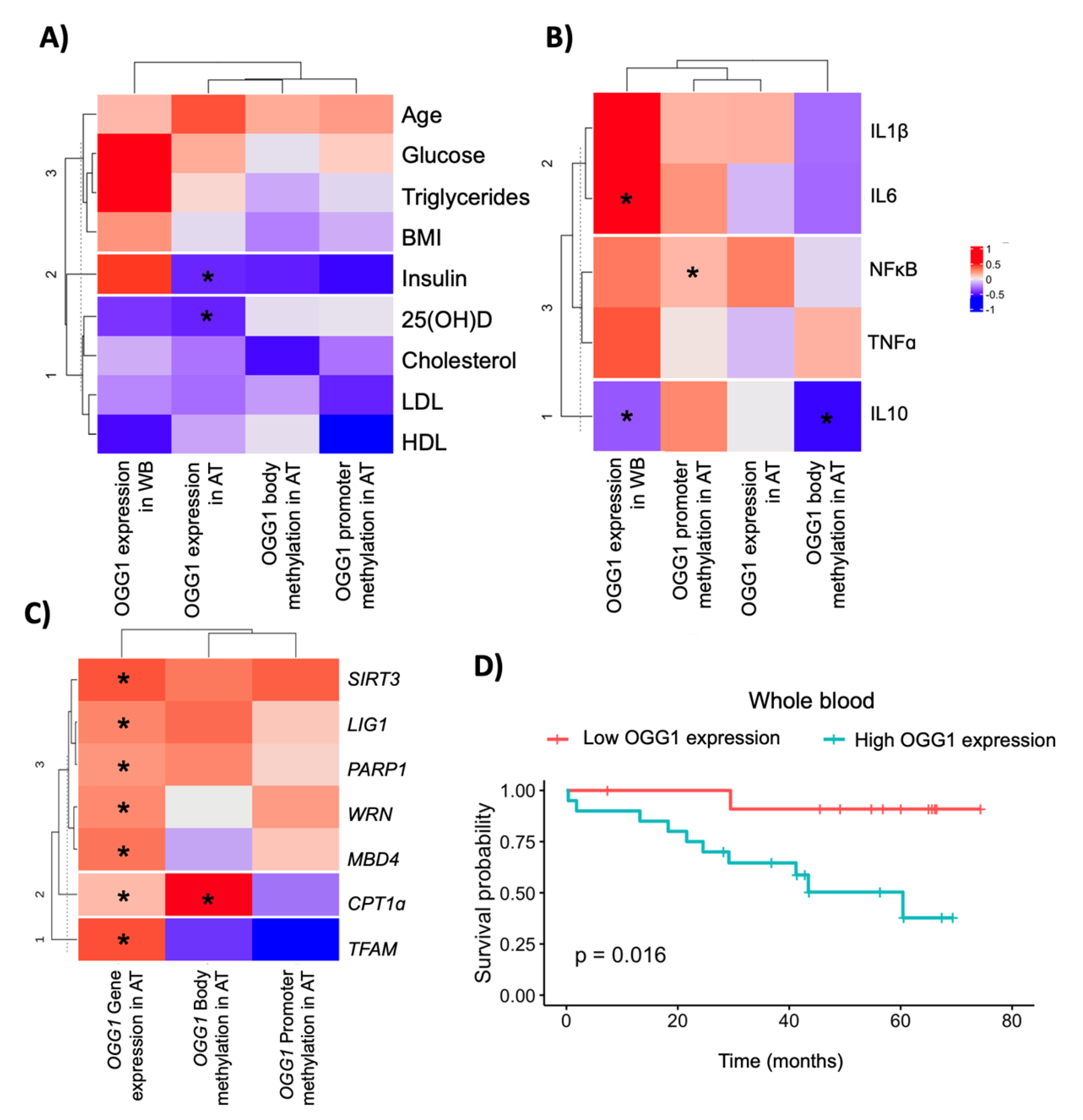
2.3. Association between OGG1 and Metabolic/DNA Repair Genes
2.4. OGG1 as a Potential Candidate Biomarker in Colorectal Cancer Outcomes
2.5. In Vitro Validation of the OGG1 Gene Profile in the Adipose Tissue and Adipocytes
3. Discussion
4. Materials and Methods
4.1. Participants and Study Design
4.2. Biochemical Measurement and Sample Obtaining
4.3. RNA Extraction and qPCR Analysis
4.4. DNA Extraction and Genotyping
4.5. DNA Bisulfite Reaction and DNA Methylation Array
4.6. Cell Culture and Adipose Tissue Explants
4.7. Statistical and Bioinformatic Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Cabrera-Mulero, A.; Crujeiras, A.B.; Izquierdo, A.G.; Torres, E.; Ayers, D.; Casanueva, F.F.; Tinahones, F.J.; Morcillo, S.; Macias-Gonzalez, M. Novel SFRP2 DNA Methylation Profile Following Neoadjuvant Therapy in Colorectal Cancer Patients with Different Grades of BMI. J. Clin. Med. 2019, 8, 1041. [Google Scholar] [CrossRef] [PubMed]
- Sung, H.; Ferlay, J.; Siegel, R.L.; Laversanne, M.; Soerjomataram, I.; Jemal, A.; Bray, F. Global Cancer Statistics 2020: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA Cancer J. Clin. 2021, 71, 209–249. [Google Scholar] [CrossRef] [PubMed]
- Gathirua-Mwangi, W.G.; Monahan, P.; Song, Y.; Zollinger, T.W.; Champion, V.L.; Stump, T.E.; Imperiale, T.F. Changes in Adult BMI and Waist Circumference Are Associated with Increased Risk of Advanced Colorectal Neoplasia. Dig. Dis. Sci. 2017, 62, 3177–3185. [Google Scholar] [CrossRef] [PubMed]
- Silveira, E.A.; Vaseghi, G.; Santos, A.S.D.C.; Kliemann, N.; Masoudkabir, F.; Noll, M.; Mohammadifard, N.; Sarrafzadegan, N.; De Oliveira, C. Visceral Obesity and Its Shared Role in Cancer and Cardiovascular Disease: A Scoping Review of the Pathophysiology and Pharmacological Treatments. Int. J. Mol. Sci. 2020, 21, 9042. [Google Scholar] [CrossRef]
- Srinivas, U.S.; Tan, B.W.Q.; Vellayappan, B.A.; Jeyasekharan, A.D. ROS and the DNA damage response in cancer. Redox Biol. 2019, 25, 101084. [Google Scholar] [CrossRef]
- Tian, G.; Katchur, S.R.; Jiang, Y.; Briand, J.; Schaber, M.; Kreatsoulas, C.; Schwartz, B.; Thrall, S.; Davis, A.M.; Duvall, S.; et al. Small molecule-mediated allosteric activation of the base excision repair enzyme 8-oxoguanine DNA glycosylase and its impact on mitochondrial function. Sci. Rep. 2022, 12, 1–18. [Google Scholar] [CrossRef]
- Kabziński, J.; Majsterek, I. Association of base excision repair pathway genes OGG1, XRCC1 and mutyh polymorphisms and the level of 8-oxo-guanine with increased risk of colorectal cancer occurrence. Int. J. Occup. Med. Environ. Health 2022, 35, 625–633. [Google Scholar] [CrossRef]
- Obtułowicz, T.; Swoboda, M.; Speina, E.; Gackowski, D.; Rozalski, R.; Siomek, A.; Janik, J.; Janowska, B.; Cieśla, J.M.; Jawien, A.; et al. Oxidative stress and 8-oxoguanine repair are enhanced in colon adenoma and carcinoma patients. Mutagenesis 2010, 25, 463–471. [Google Scholar] [CrossRef]
- D’Augustin, O.; Huet, S.; Campalans, A.; Radicella, J. Lost in the Crowd: How Does Human 8-Oxoguanine DNA Glycosylase 1 (OGG1) Find 8-Oxoguanine in the Genome? Int. J. Mol. Sci. 2020, 21, 8360. [Google Scholar] [CrossRef]
- Ba, X.; Boldogh, I. 8-Oxoguanine DNA glycosylase 1: Beyond repair of the oxidatively modified base lesions. Redox Biol. 2017, 14, 669–678. [Google Scholar] [CrossRef]
- Karahalil, B.; Engin, A.B.; Coskun, E. Could 8-oxoguanine DNA glycosylase 1 Ser326Cys polymorphism be a biomarker of susceptibility in cancer? Toxicol. Ind. Health 2012, 30, 814–825. [Google Scholar] [CrossRef]
- Sliwinski, T.; Krupa, R.; Wisniewska-Jarosinska, M.; Pawlowska, E.; Lech, J.; Chojnacki, J.; Blasiak, J. Common Polymorphisms in the XPD and hOGG1 Genes Are Not Associated with the Risk of Colorectal Cancer in a Polish Population. Tohoku J. Exp. Med. 2009, 218, 185–191. [Google Scholar] [CrossRef][Green Version]
- Kabzinski, J.; Walczak, A.; Dziki, A.; Mik, M.; Majsterek, I. Impact of the Ser326Cys polymorphism of the OGG1 gene on the level of oxidative DNA damage in patients with colorectal cancer. Ann. Surg. 2018, 90, 13–15. [Google Scholar] [CrossRef]
- Su, Y.; Xu, A.; Zhu, J. The effect of oxoguanine glycosylase 1 rs1052133 polymorphism on colorectal cancer risk in Caucasian population. Tumor Biol. 2013, 35, 513–517. [Google Scholar] [CrossRef]
- Kang, S.W.; Kim, S.K.; Park, H.J.; Chung, J.-H.; Ban, J.Y. Human 8-oxoguanine DNA glycosylase gene polymorphism (Ser326Cys) and cancer risk: Updated meta-analysis. Oncotarget 2017, 8, 44761–44775. [Google Scholar] [CrossRef]
- Leu, M.; Riebeling, T.; Dröge, L.; Hubert, L.; Guhlich, M.; Wolff, H.; Brockmöller, J.; Gaedcke, J.; Rieken, S.; Schirmer, M. 8-Oxoguanine DNA Glycosylase (OGG1) Cys326 Variant: Increased Risk for Worse Outcome of Patients with Locally Advanced Rectal Cancer after Multimodal Therapy. Cancers 2021, 13, 2805. [Google Scholar] [CrossRef]
- Sampath, H.; Vartanian, V.; Rollins, M.R.; Sakumi, K.; Nakabeppu, Y.; Lloyd, R.S. 8-Oxoguanine DNA Glycosylase (OGG1) Deficiency Increases Susceptibility to Obesity and Metabolic Dysfunction. PLoS ONE 2012, 7, e51697. [Google Scholar] [CrossRef]
- Ouni, M.; Schürmann, A. Epigenetic contribution to obesity. Mamm. Genome 2020, 31, 134–145. [Google Scholar] [CrossRef]
- Włodarczyk, M.; Nowicka, G. Obesity, DNA Damage, and Development of Obesity-Related Diseases. Int. J. Mol. Sci. 2019, 20, 1146. [Google Scholar] [CrossRef]
- Usman, M.; Volpi, E.V. DNA damage in obesity: Initiator, promoter and predictor of cancer. Mutat. Res. Mol. Mech. Mutagen. 2018, 778, 23–37. [Google Scholar] [CrossRef]
- Kompella, P.; Vasquez, K.M. Obesity and cancer: A mechanistic overview of metabolic changes in obesity that impact genetic instability. Mol. Carcinog. 2019, 58, 1531–1550. [Google Scholar] [CrossRef] [PubMed]
- Pike, J.W.; Meyer, M.B. Regulation of mouse Cyp24a1 expression via promoter-proximal and downstream-distal enhancers highlights new concepts of 1,25-dihydroxyvitamin D3 action. Arch. Biochem. Biophys. 2012, 523, 2–8. [Google Scholar] [CrossRef] [PubMed]
- Sæbø, M.; Skjelbred, C.F.; Nexø, B.A.; Wallin, H.; Hansteen, I.-L.; Vogel, U.; Kure, E.H. Increased mRNA expression levels of ERCC1, OGG1 and RAI in colorectal adenomas and carcinomas. BMC Cancer 2006, 6, 208. [Google Scholar] [CrossRef] [PubMed]
- Leguisamo, N.M.; Gloria, H.C.; Kalil, A.N.; Martins, T.V.; Azambuja, D.B.; Meira, L.B.; Saffi, J. Base excision repair imbalance in colorectal cancer has prognostic value and modulates response to chemotherapy. Oncotarget 2017, 8, 54199–54214. [Google Scholar] [CrossRef] [PubMed]
- Slyskova, J.; Naccarati, A.; Pardini, B.; Polakova, V.; Vodickova, L.; Smerhovsky, Z.; Levy, M.; Lipska, L.; Liska, V.; Vodicka, P. Differences in nucleotide excision repair capacity between newly diagnosed colorectal cancer patients and healthy controls. Mutagenesis 2012, 27, 225–232. [Google Scholar] [CrossRef] [PubMed]
- Santos, J.C.; Funck, A.; Silva-Fernandes, I.J.L.; Rabenhorst, S.H.B.; Martinez, C.A.R.; Ribeiro, M.L. Effect of APE1 T2197G (Asp148Glu) Polymorphism on APE1, XRCC1, PARP1 and OGG1 Expression in Patients with Colorectal Cancer. Int. J. Mol. Sci. 2014, 15, 17333–17343. [Google Scholar] [CrossRef]
- Slyskova, J.; Korenkova, V.; Collins, A.R.; Prochazka, P.; Vodickova, L.; Svec, J.; Lipska, L.; Levy, M.; Schneiderova, M.; Liska, V.; et al. Functional, Genetic, and Epigenetic Aspects of Base and Nucleotide Excision Repair in Colorectal Carcinomas. Clin. Cancer Res. 2012, 18, 5878–5887. [Google Scholar] [CrossRef]
- Dhar, G.A.; Saha, S.; Mitra, P.; Chaudhuri, R.N. DNA methylation and regulation of gene expression: Guardian of our health. Nucl. 2021, 64, 259–270. [Google Scholar] [CrossRef]
- Komakula, S.S.B.; Tumova, J.; Kumaraswamy, D.; Burchat, N.; Vartanian, V.; Ye, H.; Dobrzyn, A.; Lloyd, R.S.; Sampath, H. The DNA Repair Protein OGG1 Protects Against Obesity by Altering Mitochondrial Energetics in White Adipose Tissue. Sci. Rep. 2018, 8, 1–15. [Google Scholar] [CrossRef]
- Komakula, S.; Blaze, B.; Ye, H.; Dobrzyn, A.; Sampath, H. A Novel Role for the DNA Repair Enzyme 8-Oxoguanine DNA Glycosylase in Adipogenesis. Int. J. Mol. Sci. 2021, 22, 1152. [Google Scholar] [CrossRef]
- Li, G.; Yuan, K.; Yan, C.; Fox, J.; Gaid, M.; Breitwieser, W.; Bansal, A.K.; Zeng, H.; Gao, H.; Wu, M. 8-Oxoguanine-DNA glycosylase 1 deficiency modifies allergic airway inflammation by regulating STAT6 and IL-4 in cells and in mice. Free. Radic. Biol. Med. 2012, 52, 392–401. [Google Scholar] [CrossRef]
- Visnes, T.; Grube, M.; Hanna, B.M.F.; Benitez-Buelga, C.; Cázares-Körner, A.; Helleday, T. Targeting BER enzymes in cancer therapy. DNA Repair 2018, 71, 118–126. [Google Scholar] [CrossRef]
- Wele, P.; Wu, X.; Shi, H. Sex-Dependent Differences in Colorectal Cancer: With a Focus on Obesity. Cells 2022, 11, 3688. [Google Scholar] [CrossRef]
- Kim, I.-J.; Ku, J.-L.; Kang, H.C.; Park, J.-H.; Yoon, K.-A.; Shin, Y.; Park, H.-W.; Jang, S.G.; Lim, S.-K.; Han, S.Y.; et al. Mutational analysis of OGG1, MYH, MTH1 in FAP, HNPCC and sporadic colorectal cancer patients: R154H OGG1 polymorphism is associated with sporadic colorectal cancer patients. Hum. Genet. 2004, 115, 498–503. [Google Scholar] [CrossRef]
- Garre, P.; Briceño, V.; Xicola, R.M.; Doyle, B.J.; de la Hoya, M.; Sanz, J.; Llovet, P.; Pescador, P.; Puente, J.; Díaz-Rubio, E.; et al. Analysis of the Oxidative Damage Repair Genes NUDT1, OGG1, and MUTYH in Patients from Mismatch Repair Proficient HNPCC Families (MSS-HNPCC). Clin. Cancer Res. 2011, 17, 1701–1712. [Google Scholar] [CrossRef]
- Hansen, R.; Sæbø, M.; Skjelbred, C.F.; Nexø, B.A.; Hagen, P.C.; Bock, G.; Lothe, I.M.B.; Johnson, E.; Aase, S.; Hansteen, I.-L.; et al. GPX Pro198Leu and OGG1 Ser326Cys polymorphisms and risk of development of colorectal adenomas and colorectal cancer. Cancer Lett. 2005, 229, 85–91. [Google Scholar] [CrossRef]
- Przybylowska, K.; Kabzinski, J.; Sygut, A.; Dziki, L.; Dziki, A.; Majsterek, I. An association selected polymorphisms of XRCC1, OGG1 and MUTYH gene and the level of efficiency oxidative DNA damage repair with a risk of colorectal cancer. Mutat. Res. Mol. Mech. Mutagen. 2013, 745, 6–15. [Google Scholar] [CrossRef]
- Moreno, V.; Gemignani, F.; Landi, S.; Gioia-Patricola, L.; Chabrier, A.; Blanco, I.; González, S.; Guino, E.; Capellà, G.; Canzian, F.; et al. Polymorphisms in Genes of Nucleotide and Base Excision Repair: Risk and Prognosis of Colorectal Cancer. Clin. Cancer Res. 2006, 12, 2101–2108. [Google Scholar] [CrossRef]
- Zhang, Y.; He, B.-S.; Pan, Y.-Q.; Xu, Y.-Q.; Wang, S.-K. Association of OGG1 Ser326Cys polymorphism with colorectal cancer risk: A meta-analysis. Int. J. Color. Dis. 2011, 26, 1525–1530. [Google Scholar] [CrossRef]
- Aggarwal, N.; Donald, N.D.; Malik, S.; Selvendran, S.S.; McPhail, M.; Monahan, K.J. The Association of Low-Penetrance Variants in DNA Repair Genes with Colorectal Cancer: A Systematic Review and Meta-Analysis. Clin. Transl. Gastroenterol. 2017, 8, e109. [Google Scholar] [CrossRef]
- Gaiani, F.; Marchesi, F.; Negri, F.; Greco, L.; Malesci, A.; De’Angelis, G.; Laghi, L. Heterogeneity of Colorectal Cancer Progression: Molecular Gas and Brakes. Int. J. Mol. Sci. 2021, 22, 5246. [Google Scholar] [CrossRef] [PubMed]
- Lan, N.; Luo, G.; Yang, X.; Cheng, Y.; Zhang, Y.; Wang, X.; Wang, X.; Xie, T.; Li, G.; Liu, Z.; et al. 25-Hydroxyvitamin D3-Deficiency Enhances Oxidative Stress and Corticosteroid Resistance in Severe Asthma Exacerbation. PLoS ONE 2014, 9, e111599. [Google Scholar] [CrossRef] [PubMed]
- Amirinejad, R.; Shirvani-Farsani, Z.; Gargari, B.N.; Sahraian, M.A.; Soltani, B.M.; Behmanesh, M. Vitamin D changes expression of DNA repair genes in the patients with multiple sclerosis. Gene 2021, 781, 145488. [Google Scholar] [CrossRef] [PubMed]
- Friedewald, W.T.; Levy, R.I.; Fredrickson, D.S. Estimation of the Concentration of Low-Density Lipoprotein Cholesterol in Plasma, Without Use of the Preparative Ultracentrifuge. Clin. Chem. 1972, 18, 499–502. [Google Scholar] [CrossRef]
- Levy, J.C.; Matthews, D.R.; Hermans, M.P. Correct Homeostasis Model Assessment (HOMA) Evaluation Uses the Computer Program. Diabetes Care 1998, 21, 2191–2192. [Google Scholar] [CrossRef]
- Zerbino, D.R.; Achuthan, P.; Akanni, W.; Amode, M.R.; Barrell, D.; Bhai, J.; Billis, K.; Cummins, C.; Gall, A.; Girón, C.G.; et al. Ensembl. Nucleic Acids Res. 2018, 46, D754–D761. [Google Scholar] [CrossRef]
- Aryee, M.J.; Jaffe, A.E.; Corrada-Bravo, H.; Ladd-Acosta, C.; Feinberg, A.P.; Hansen, K.D.; Irizarry, R.A. Minfi: A flexible and comprehensive Bioconductor package for the analysis of Infinium DNA methylation microarrays. Bioinformatics 2014, 30, 1363–1369. [Google Scholar] [CrossRef]
- Clemente-Postigo, M.; Muñoz-Garach, A.; Serrano, M.; Garrido-Sánchez, L.; Bernal-López, M.R.; Fernández-García, D.; Moreno-Santos, I.; Garriga, N.; Castellano-Castillo, D.; Camargo, A.; et al. Serum 25-Hydroxyvitamin D and Adipose Tissue Vitamin D Receptor Gene Expression: Relationship With Obesity and Type 2 Diabetes. J. Clin. Endocrinol. Metab. 2015, 100, E591–E595. [Google Scholar] [CrossRef]

| Variable | All | Healthy Participants | Patients with CRC | p |
|---|---|---|---|---|
| N = 310 | N = 230 | N = 80 | ||
| Age (years) | 54.5 (15.0) | 50.1 (13.8) | 67.2 (10.3) | <0.001 * |
| Sex (males/females) | 159/151 | 103/127 | 56/24 | <0.001 * |
| BMI (Kg/m2) | 28.9 (7.33) | 29.5 (8.09) | 27.2 (4.06) | 0.001 * |
| Glucose (mg/dL) | 105 (35.2) | 98.3 (20.3) | 125 (55.9) | <0.001 * |
| HOMAIR | 2.38 (1.82) | 2.50 (1.76) | 2.01 (1.95) | 0.054 |
| Total cholesterol (mg/dL) | 196 (44.7) | 205 (42.7) | 173 (41.3) | <0.001 * |
| Triglycerides (mg/dL) | 134 (67.4) | 123 (58.5) | 163 (79.6) | <0.001 * |
| LDL (mg/dL) | 120 (34.7) | 127 (32.9) | 104 (33.8) | <0.001 * |
| HDL (mg/dL) | 50.0 (15.3) | 53.4 (14.4) | 41.2 (14.1) | <0.001 * |
| 25-Hydroxyvitamin D (ng/mL) | 39.1 (18.9) | 43.5 (20.2) | 30.8 (12.8) | <0.001 * |
| OGG1 Ser302Cys | 0.174 | |||
| C/C | 146 (65.8%) | 100 (66.2%) | 46 (64.8%) | |
| C/G | 61 (27.5%) | 44 (29.1%) | 17 (23.9%) | |
| G/G | 15 (6.76%) | 7 (4.64%) | 8 (11.3%) |
| Variables | β (SE) R2 = 0.41, p < 0.001 | β (SE) R2 = 0.30, p < 0.001 | β (SE) R2 = 0.44, p < 0.001 | β (SE) R2 = 0.40, p < 0.001 |
|---|---|---|---|---|
| Age (years) | 0.16 (0.00) *** | 0.01 (0.00) *** | 0.02 (0.00) *** | 0.02 (0.00) *** |
| Sex (males/females) | −0.23 (0.11) | −0.17 (0.08) * | −0.13 (0.12) | −0.15 (0.12) |
| BMI (kg/m2) | 0.00 (0.01) | −0.02 (0.01) * | −0.01 (0.011) | −0.01 (0.01) |
| OGG1 expression in VAT | NA | 7.21 (3.09) * | NA | NA |
| OGG1 expression in whole blood | 0.91 (0.55) | NA | NA | NA |
| Promoter OGG1 methylation | NA | NA | 10.22 (5.53) | NA |
| Body OGG1 methylation | NA | NA | NA | −3.16 (6.91) |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Pilo, J.; García-Flores, L.A.; Clemente-Postigo, M.; Arranz-Salas, I.; Alcaide, J.; Ramos-Fernandez, M.; Lozano, J.; Boughanem, H.; Kompella, P.; Macías-González, M. 8-Oxoguanine DNA Glycosylase 1 Upregulation as a Risk Factor for Obesity and Colorectal Cancer. Int. J. Mol. Sci. 2023, 24, 5488. https://doi.org/10.3390/ijms24065488
Pilo J, García-Flores LA, Clemente-Postigo M, Arranz-Salas I, Alcaide J, Ramos-Fernandez M, Lozano J, Boughanem H, Kompella P, Macías-González M. 8-Oxoguanine DNA Glycosylase 1 Upregulation as a Risk Factor for Obesity and Colorectal Cancer. International Journal of Molecular Sciences. 2023; 24(6):5488. https://doi.org/10.3390/ijms24065488
Chicago/Turabian StylePilo, Jesús, Libia Alejandra García-Flores, Mercedes Clemente-Postigo, Isabel Arranz-Salas, Julia Alcaide, Maria Ramos-Fernandez, José Lozano, Hatim Boughanem, Pallavi Kompella, and Manuel Macías-González. 2023. "8-Oxoguanine DNA Glycosylase 1 Upregulation as a Risk Factor for Obesity and Colorectal Cancer" International Journal of Molecular Sciences 24, no. 6: 5488. https://doi.org/10.3390/ijms24065488
APA StylePilo, J., García-Flores, L. A., Clemente-Postigo, M., Arranz-Salas, I., Alcaide, J., Ramos-Fernandez, M., Lozano, J., Boughanem, H., Kompella, P., & Macías-González, M. (2023). 8-Oxoguanine DNA Glycosylase 1 Upregulation as a Risk Factor for Obesity and Colorectal Cancer. International Journal of Molecular Sciences, 24(6), 5488. https://doi.org/10.3390/ijms24065488






