Cardiac Hamp mRNA Is Predominantly Expressed in the Right Atrium and Does Not Respond to Iron
Abstract
1. Introduction
2. Results
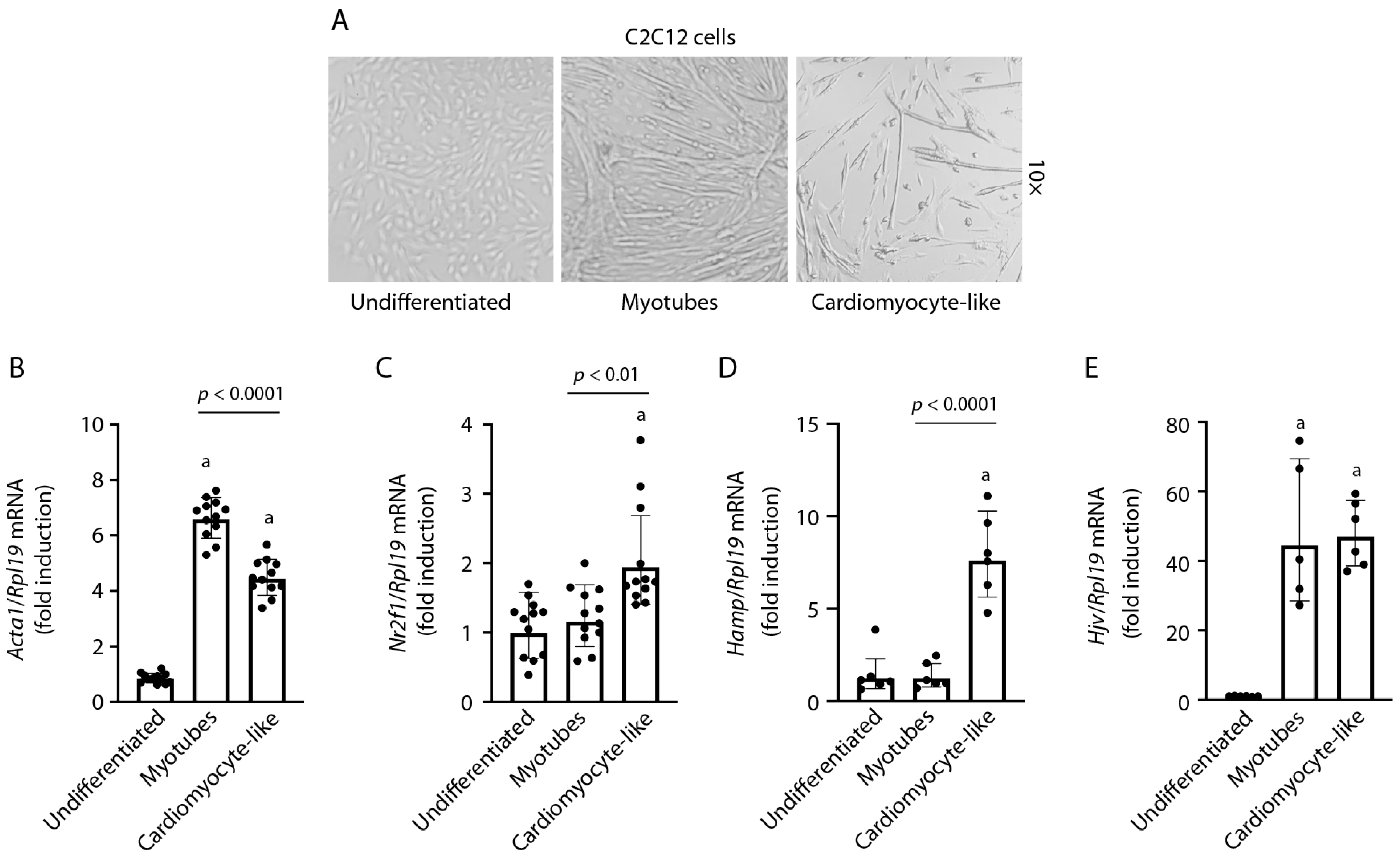
2.1. Differentiation of C2C12 Cells into a Cardiomyocyte-like Phenotype
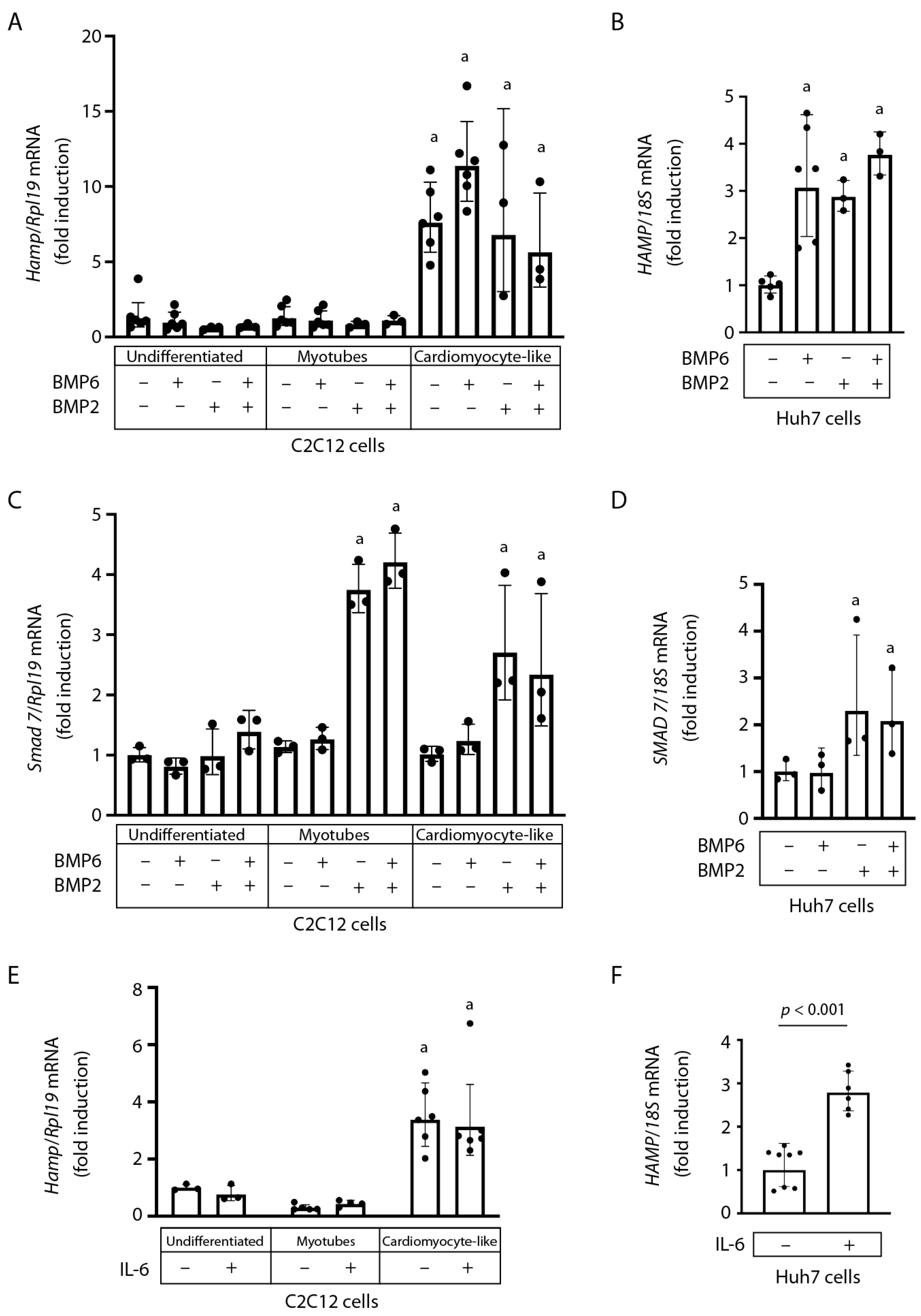
2.2. Hamp mRNA Is Induced during C2C12 Cardiomyocyte Differentiation and Does Not Respond to BMP6, BMP2, or IL-6
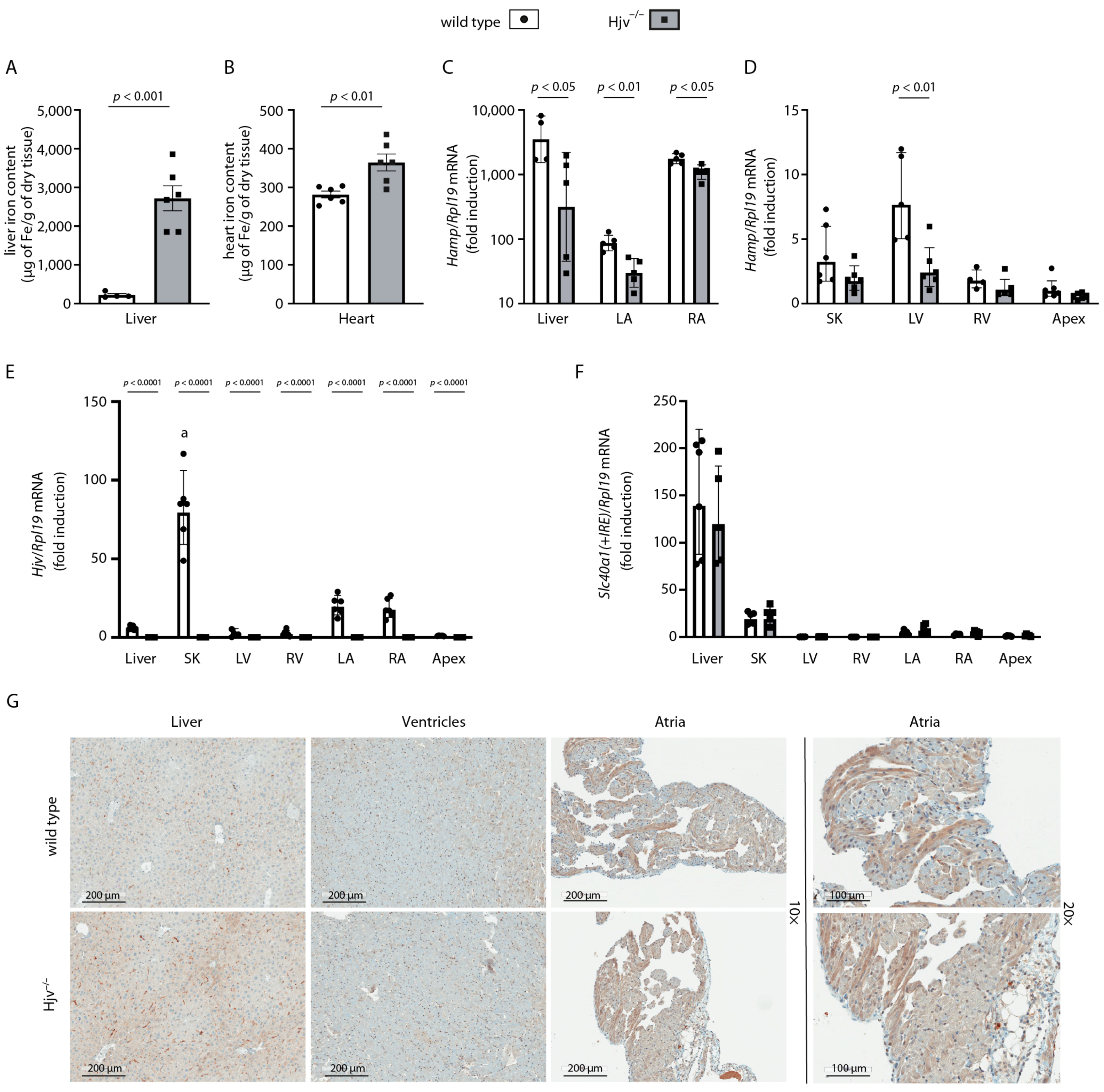
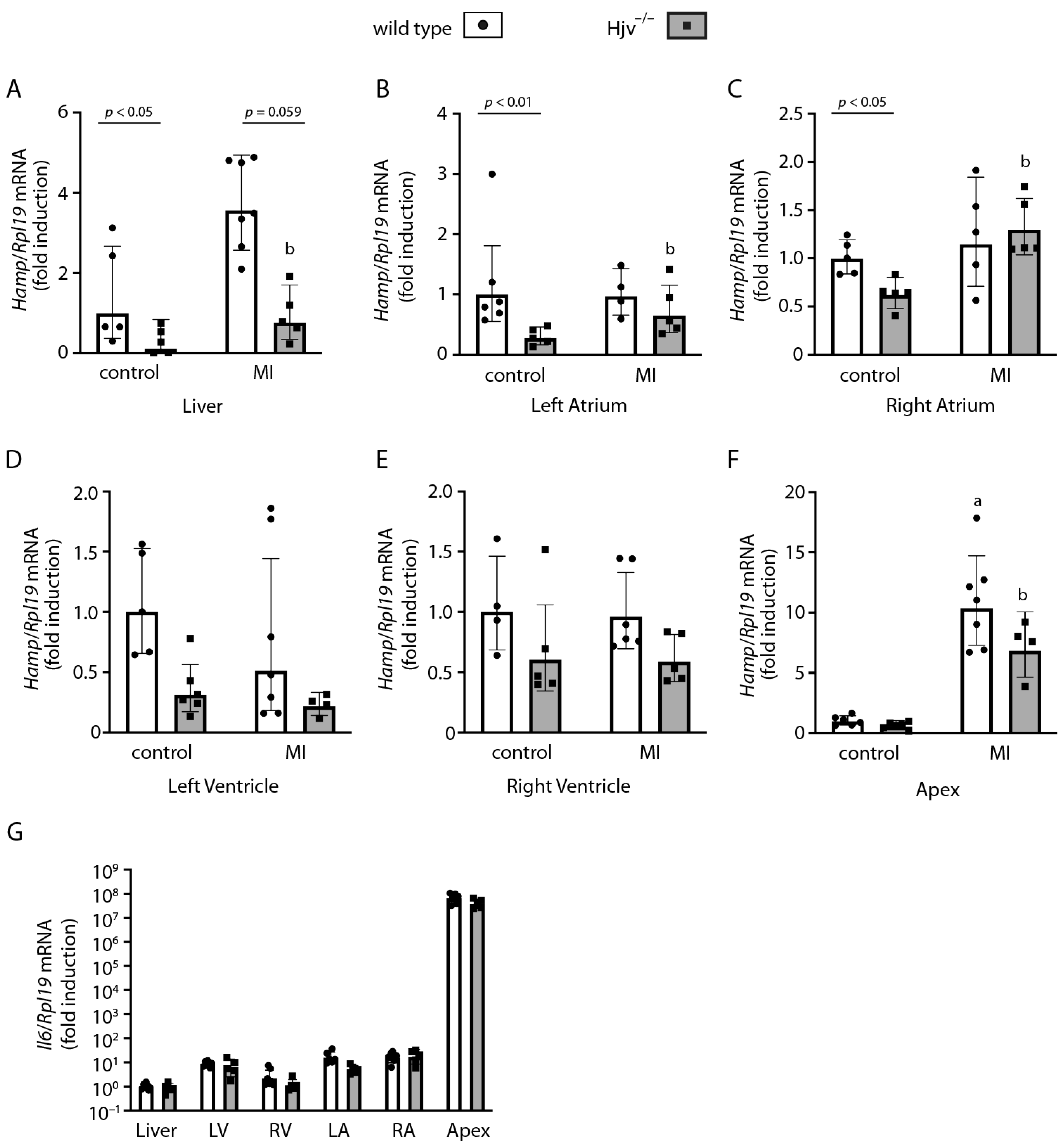
2.3. Cardiac Hamp mRNA Is Predominantly Expressed in the Right Atrium and Only Partially Depends on Hjv
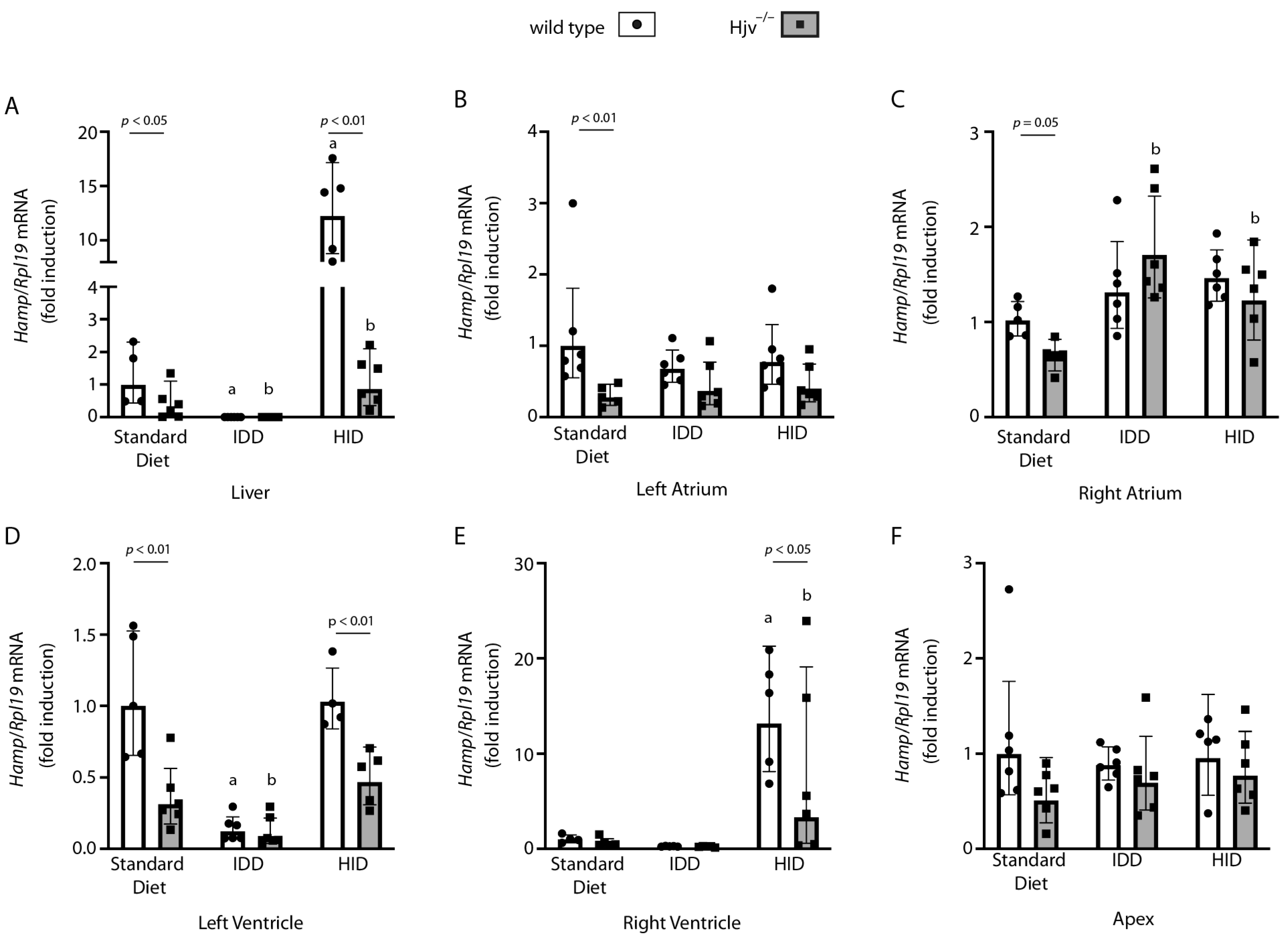
2.4. Atrial Hamp mRNA Does Not Respond to Dietary Iron Manipulations in Wild-Type Mice
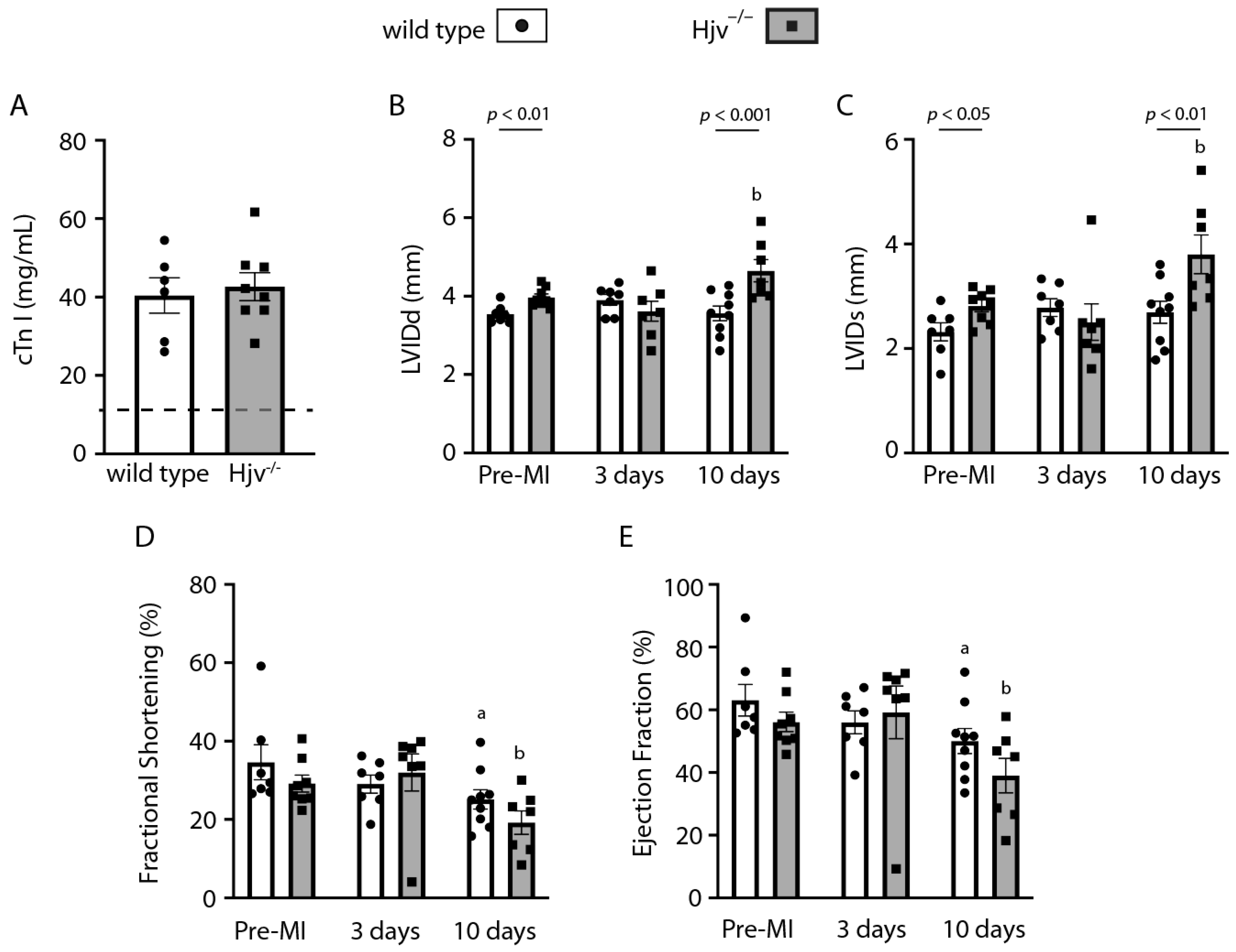
2.5. Myocardial Infarction Studies in Wild-Type and Hjv−/− Mice
3. Discussion
4. Materials and Methods
4.1. Cell Culture
4.2. Animals
4.3. Myocardial Infarction Surgery
4.4. Echocardiography
4.5. qPCR Analysis
4.6. Iron Assays
4.7. Immunohistochemistry
4.8. Statistical Analysis
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Camaschella, C. Iron-deficiency anemia. N. Engl. J. Med. 2015, 372, 1832–1843. [Google Scholar] [CrossRef] [PubMed]
- Anand, I.S.; Gupta, P. Anemia and Iron Deficiency in Heart Failure: Current Concepts and Emerging Therapies. Circulation 2018, 138, 80–98. [Google Scholar] [CrossRef] [PubMed]
- Galaris, D.; Barbouti, A.; Pantopoulos, K. Iron homeostasis and oxidative stress: An intimate relationship. Biochim. Biophys. Acta Mol. Cell Res. 2019, 1866, 118535. [Google Scholar] [CrossRef] [PubMed]
- Diez-Lopez, C.; Comin-Colet, J.; Gonzalez-Costello, J. Iron overload cardiomyopathy: From diagnosis to management. Curr. Opin. Cardiol. 2018, 33, 334–340. [Google Scholar] [CrossRef]
- Mehta, K.J.; Farnaud, S.J.; Sharp, P.A. Iron and liver fibrosis: Mechanistic and clinical aspects. World J. Gastroenterol. 2019, 25, 521–538. [Google Scholar] [CrossRef]
- Fargion, S.; Valenti, L.; Fracanzani, A.L. Role of iron in hepatocellular carcinoma. Clin. Liver Dis. 2014, 3, 108–110. [Google Scholar] [CrossRef] [PubMed]
- Nemeth, E.; Ganz, T. Hepcidin-Ferroportin Interaction Controls Systemic Iron Homeostasis. Int. J. Mol. Sci. 2021, 22, 6493. [Google Scholar] [CrossRef]
- Fisher, A.L.; Babitt, J.L. Coordination of iron homeostasis by bone morphogenetic proteins: Current understanding and unanswered questions. Dev. Dyn. 2022, 251, 26–46. [Google Scholar] [CrossRef]
- Camaschella, C.; Nai, A.; Silvestri, L. Iron metabolism and iron disorders revisited in the hepcidin era. Haematologica 2020, 105, 260–272. [Google Scholar] [CrossRef]
- Zumerle, S.; Mathieu, J.R.; Delga, S.; Heinis, M.; Viatte, L.; Vaulont, S.; Peyssonnaux, C. Targeted disruption of hepcidin in the liver recapitulates the hemochromatotic phenotype. Blood 2014, 123, 3646–3650. [Google Scholar] [CrossRef]
- Pigeon, C.; Ilyin, G.; Courselaud, B.; Leroyer, P.; Turlin, B.; Brissot, P.; Loreal, O. A new mouse liver-specific gene, encoding a protein homologous to human antimicrobial peptide hepcidin, is overexpressed during iron overload. J. Biol. Chem. 2001, 276, 7811–7819. [Google Scholar] [CrossRef] [PubMed]
- Merle, U.; Fein, E.; Gehrke, S.G.; Stremmel, W.; Kulaksiz, H. The iron regulatory peptide hepcidin is expressed in the heart and regulated by hypoxia and inflammation. Endocrinology 2007, 148, 2663–2668. [Google Scholar] [CrossRef] [PubMed]
- Lakhal-Littleton, S.; Wolna, M.; Chung, Y.J.; Christian, H.C.; Heather, L.C.; Brescia, M.; Ball, V.; Diaz, R.; Santos, A.; Biggs, D.; et al. An essential cell-autonomous role for hepcidin in cardiac iron homeostasis. eLife 2016, 5, e19804. [Google Scholar] [CrossRef] [PubMed]
- Berezovsky, B.; Frydlova, J.; Gurieva, I.; Rogalsky, D.W.; Vokurka, M.; Krijt, J. Heart Ferroportin Protein Content Is Regulated by Heart Iron Concentration and Systemic Hepcidin Expression. Int. J. Mol. Sci. 2022, 23, 5899. [Google Scholar] [CrossRef] [PubMed]
- Haddad, S.; Wang, Y.; Galy, B.; Korf-Klingebiel, M.; Hirsch, V.; Baru, A.M.; Rostami, F.; Reboll, M.R.; Heineke, J.; Flogel, U.; et al. Iron-regulatory proteins secure iron availability in cardiomyocytes to prevent heart failure. Eur. Heart J. 2017, 38, 362–372. [Google Scholar] [CrossRef] [PubMed]
- Blau, H.M.; Chiu, C.P.; Webster, C. Cytoplasmic activation of human nuclear genes in stable heterocaryons. Cell 1983, 32, 1171–1180. [Google Scholar] [CrossRef]
- Zebedin, E.; Mille, M.; Speiser, M.; Zarrabi, T.; Sandtner, W.; Latzenhofer, B.; Todt, H.; Hilber, K. C2C12 skeletal muscle cells adopt cardiac-like sodium current properties in a cardiac cell environment. Am. J. Physiol. Heart Circ. Physiol. 2007, 292, H439–H450. [Google Scholar] [CrossRef]
- Wiesinger, A.; Boink, G.J.J.; Christoffels, V.M.; Devalla, H.D. Retinoic acid signaling in heart development: Application in the differentiation of cardiovascular lineages from human pluripotent stem cells. Stem Cell Rep. 2021, 16, 2589–2606. [Google Scholar] [CrossRef]
- Devalla, H.D.; Schwach, V.; Ford, J.W.; Milnes, J.T.; El-Haou, S.; Jackson, C.; Gkatzis, K.; Elliott, D.A.; Chuva de Sousa Lopes, S.M.; Mummery, C.L.; et al. Atrial-like cardiomyocytes from human pluripotent stem cells are a robust preclinical model for assessing atrial-selective pharmacology. EMBO Mol. Med. 2015, 7, 394–410. [Google Scholar] [CrossRef]
- Papanikolaou, G.; Samuels, M.E.; Ludwig, E.H.; MacDonald, M.L.; Franchini, P.L.; Dube, M.P.; Andres, L.; MacFarlane, J.; Sakellaropoulos, N.; Politou, M.; et al. Mutations in HFE2 cause iron overload in chromosome 1q-linked juvenile hemochromatosis. Nat. Genet. 2004, 36, 77–82. [Google Scholar] [CrossRef]
- Huang, F.W.; Pinkus, J.L.; Pinkus, G.S.; Fleming, M.D.; Andrews, N.C. A mouse model of juvenile hemochromatosis. J. Clin. Investig. 2005, 115, 2187–2191. [Google Scholar] [CrossRef] [PubMed]
- Gkouvatsos, K.; Fillebeen, C.; Daba, A.; Wagner, J.; Sebastiani, G.; Pantopoulos, K. Iron-dependent regulation of hepcidin in Hjv−/− mice: Evidence that hemojuvelin is dispensable for sensing body iron levels. PLoS ONE 2014, 9, e85530. [Google Scholar] [CrossRef] [PubMed]
- Lindsey, M.L.; Kassiri, Z.; Virag, J.A.I.; de Castro Bras, L.E.; Scherrer-Crosbie, M. Guidelines for measuring cardiac physiology in mice. Am. J. Physiol. Heart Circ. Physiol. 2018, 314, H733–H752. [Google Scholar] [CrossRef]
- Gao, X.M.; Dart, A.M.; Dewar, E.; Jennings, G.; Du, X.J. Serial echocardiographic assessment of left ventricular dimensions and function after myocardial infarction in mice. Cardiovasc. Res. 2000, 45, 330–338. [Google Scholar] [CrossRef] [PubMed]
- Kuninger, D.; Kuns-Hashimoto, R.; Kuzmickas, R.; Rotwein, P. Complex biosynthesis of the muscle-enriched iron regulator RGMc. J. Cell Sci. 2006, 119, 3273–3283. [Google Scholar] [CrossRef] [PubMed]
- Zhang, A.S.; Anderson, S.A.; Meyers, K.R.; Hernandez, C.; Eisenstein, R.S.; Enns, C.A. Evidence that inhibition of hemojuvelin shedding in response to iron is mediated through neogenin. J. Biol. Chem. 2007, 282, 12547–12556. [Google Scholar] [CrossRef]
- Katagiri, T.; Yamaguchi, A.; Komaki, M.; Abe, E.; Takahashi, N.; Ikeda, T.; Rosen, V.; Wozney, J.M.; Fujisawa-Sehara, A.; Suda, T. Bone morphogenetic protein-2 converts the differentiation pathway of C2C12 myoblasts into the osteoblast lineage. J. Cell. Biol. 1994, 127, 1755–1766. [Google Scholar] [CrossRef]
- Charlebois, E.; Pantopoulos, K. Iron overload inhibits BMP/SMAD and IL-6/STAT3 signaling to hepcidin in cultured hepatocytes. PLoS ONE 2021, 16, e0253475. [Google Scholar] [CrossRef]
- Brandenburg, S.; Arakel, E.C.; Schwappach, B.; Lehnart, S.E. The molecular and functional identities of atrial cardiomyocytes in health and disease. Biochim. Biophys. Acta 2016, 1863, 1882–1893. [Google Scholar] [CrossRef]
- Oexle, H.; Gnaiger, E.; Weiss, G. Iron-dependent changes in cellular energy metabolism: Influence on citric acid cycle and oxidative phosphorylation. Biochim. Biophys. Acta 1999, 1413, 99–107. [Google Scholar] [CrossRef]
- Hoes, M.F.; Grote Beverborg, N.; Kijlstra, J.D.; Kuipers, J.; Swinkels, D.W.; Giepmans, B.N.G.; Rodenburg, R.J.; van Veldhuisen, D.J.; de Boer, R.A.; van der Meer, P. Iron deficiency impairs contractility of human cardiomyocytes through decreased mitochondrial function. Eur. J. Heart Fail 2018, 20, 910–919. [Google Scholar] [CrossRef] [PubMed]
- Xu, W.; Barrientos, T.; Mao, L.; Rockman, H.A.; Sauve, A.A.; Andrews, N.C. Lethal Cardiomyopathy in Mice Lacking Transferrin Receptor in the Heart. Cell Rep. 2015, 13, 533–545. [Google Scholar] [CrossRef] [PubMed]
- Lakhal-Littleton, S.; Robbins, P.A. The interplay between iron and oxygen homeostasis with a particular focus on the heart. J. Appl. Physiol. 2017, 123, 967–973. [Google Scholar] [CrossRef] [PubMed]
- McMahon, S.; Grondin, F.; McDonald, P.P.; Richard, D.E.; Dubois, C.M. Hypoxia-enhanced expression of the proprotein convertase furin is mediated by hypoxia-inducible factor-1: Impact on the bioactivation of proproteins. J. Biol. Chem. 2005, 280, 6561–6569. [Google Scholar] [CrossRef]
- Simonis, G.; Mueller, K.; Schwarz, P.; Wiedemann, S.; Adler, G.; Strasser, R.H.; Kulaksiz, H. The iron-regulatory peptide hepcidin is upregulated in the ischemic and in the remote myocardium after myocardial infarction. Peptides 2010, 31, 1786–1790. [Google Scholar] [CrossRef]
- Van Breda, G.F.; Bongartz, L.G.; Zhuang, W.; van Swelm, R.P.; Pertijs, J.; Braam, B.; Cramer, M.J.; Swinkels, D.W.; Doevendans, P.A.; Verhaar, M.C.; et al. Cardiac Hepcidin Expression Associates with Injury Independent of Iron. Am. J. Nephrol. 2016, 44, 368–378. [Google Scholar] [CrossRef]
- Lakhal-Littleton, S.; Wolna, M.; Carr, C.A.; Miller, J.J.; Christian, H.C.; Ball, V.; Santos, A.; Diaz, R.; Biggs, D.; Stillion, R.; et al. Cardiac ferroportin regulates cellular iron homeostasis and is important for cardiac function. Proc. Natl. Acad. Sci. USA 2015, 112, 3164–3169. [Google Scholar] [CrossRef]
- Yang, X.P.; Liu, Y.H.; Rhaleb, N.E.; Kurihara, N.; Kim, H.E.; Carretero, O.A. Echocardiographic assessment of cardiac function in conscious and anesthetized mice. Am. J. Physiol. 1999, 277, H1967–H1974. [Google Scholar] [CrossRef]
- Gao, S.; Ho, D.; Vatner, D.E.; Vatner, S.F. Echocardiography in Mice. Curr. Protoc. Mouse Biol. 2011, 1, 71–83. [Google Scholar]
- Livak, K.J.; Schmittgen, T.D. Analysis of Relative Gene Expression Data Using Real-Time Quantitative PCR and the 2−ΔΔCT Method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef]
- Daba, A.; Gkouvatsos, K.; Sebastiani, G.; Pantopoulos, K. Differences in activation of mouse hepcidin by dietary iron and parenterally administered iron dextran: Compartmentalization is critical for iron sensing. J. Mol. Med. 2013, 91, 95–102. [Google Scholar] [CrossRef] [PubMed]
- Jenkitkasemwong, S.; Wang, C.Y.; Coffey, R.; Zhang, W.; Chan, A.; Biel, T.; Kim, J.S.; Hojyo, S.; Fukada, T.; Knutson, M.D. SLC39A14 Is Required for the Development of Hepatocellular Iron Overload in Murine Models of Hereditary Hemochromatosis. Cell Metab. 2015, 22, 138–150. [Google Scholar] [CrossRef] [PubMed]
- Fillebeen, C.; Wilkinson, N.; Charlebois, E.; Katsarou, A.; Wagner, J.; Pantopoulos, K. Hepcidin-mediated hypoferremic response to acute inflammation requiRes. a threshold of Bmp6/Hjv/Smad signaling. Blood 2018, 132, 1829–1841. [Google Scholar] [CrossRef] [PubMed]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Bigorra Mir, M.; Charlebois, E.; Tsyplenkova, S.; Fillebeen, C.; Pantopoulos, K. Cardiac Hamp mRNA Is Predominantly Expressed in the Right Atrium and Does Not Respond to Iron. Int. J. Mol. Sci. 2023, 24, 5163. https://doi.org/10.3390/ijms24065163
Bigorra Mir M, Charlebois E, Tsyplenkova S, Fillebeen C, Pantopoulos K. Cardiac Hamp mRNA Is Predominantly Expressed in the Right Atrium and Does Not Respond to Iron. International Journal of Molecular Sciences. 2023; 24(6):5163. https://doi.org/10.3390/ijms24065163
Chicago/Turabian StyleBigorra Mir, Maria, Edouard Charlebois, Sofiya Tsyplenkova, Carine Fillebeen, and Kostas Pantopoulos. 2023. "Cardiac Hamp mRNA Is Predominantly Expressed in the Right Atrium and Does Not Respond to Iron" International Journal of Molecular Sciences 24, no. 6: 5163. https://doi.org/10.3390/ijms24065163
APA StyleBigorra Mir, M., Charlebois, E., Tsyplenkova, S., Fillebeen, C., & Pantopoulos, K. (2023). Cardiac Hamp mRNA Is Predominantly Expressed in the Right Atrium and Does Not Respond to Iron. International Journal of Molecular Sciences, 24(6), 5163. https://doi.org/10.3390/ijms24065163





