Anoxia Rapidly Induces Changes in Expression of a Large and Diverse Set of Genes in Endothelial Cells
Abstract
1. Introduction
2. Results and Discussion
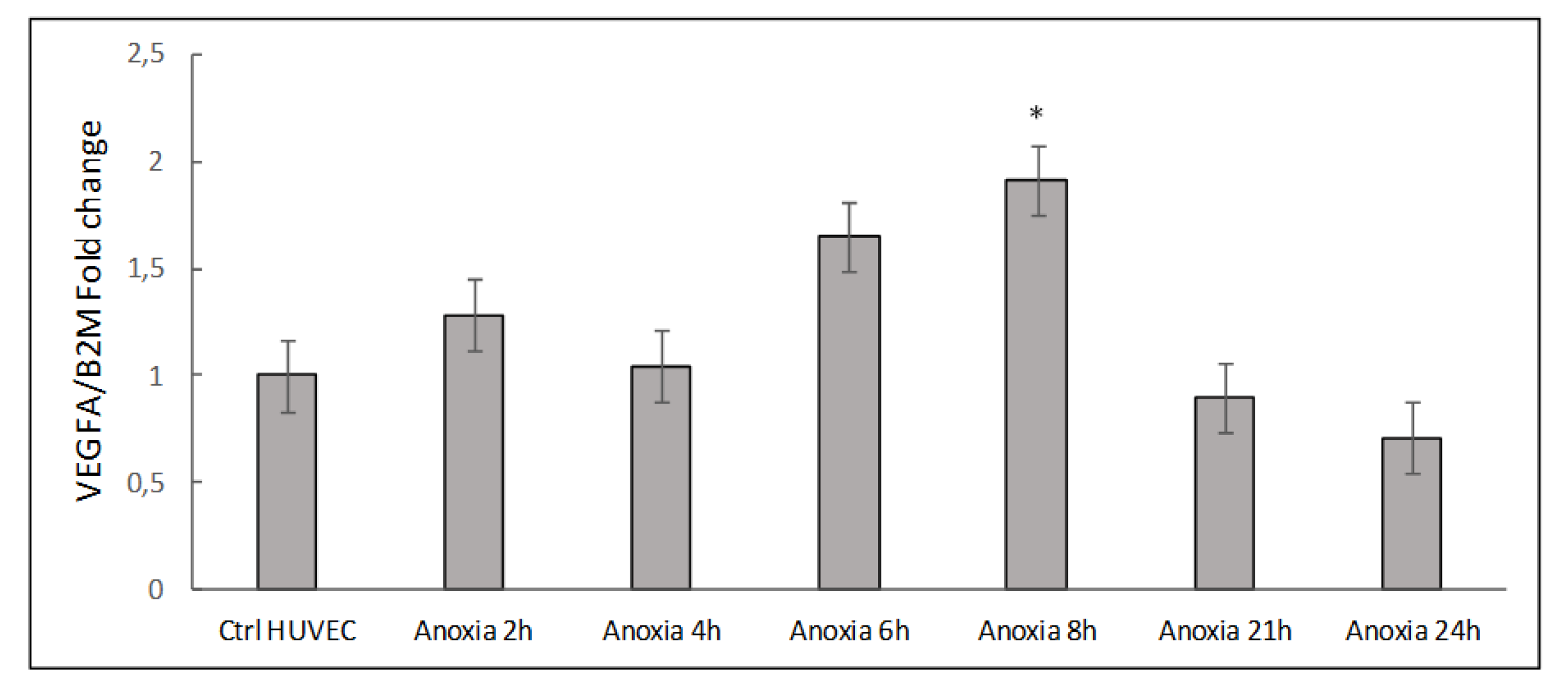
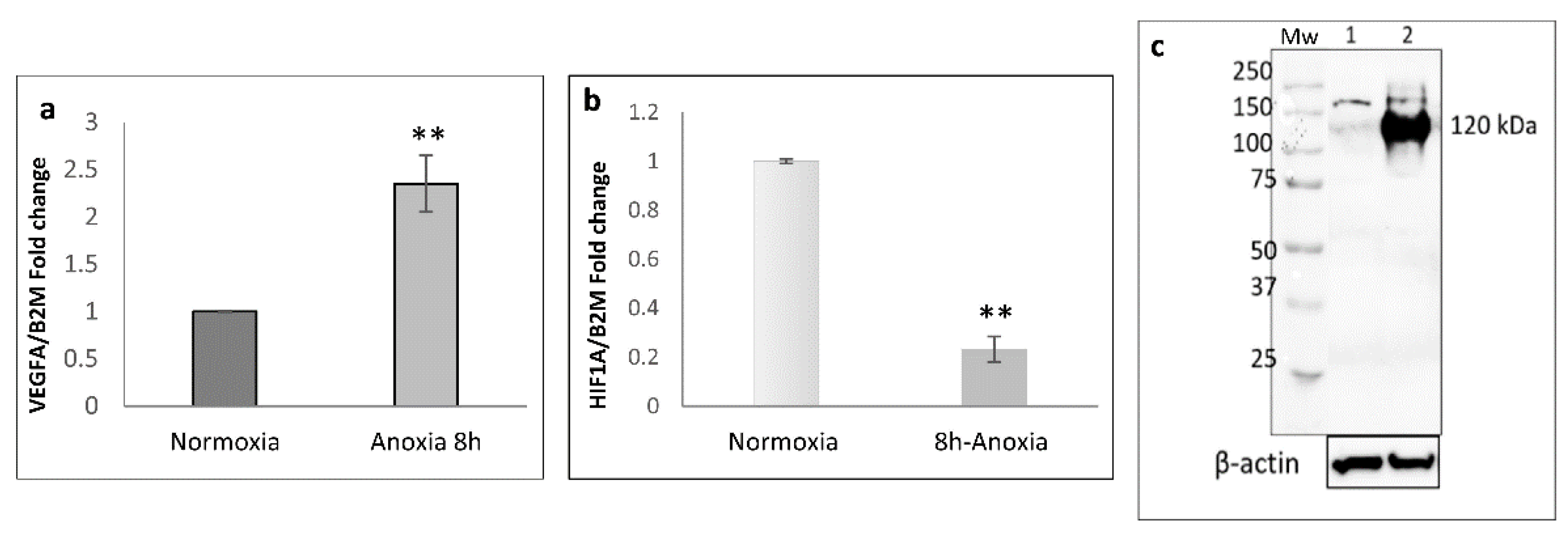
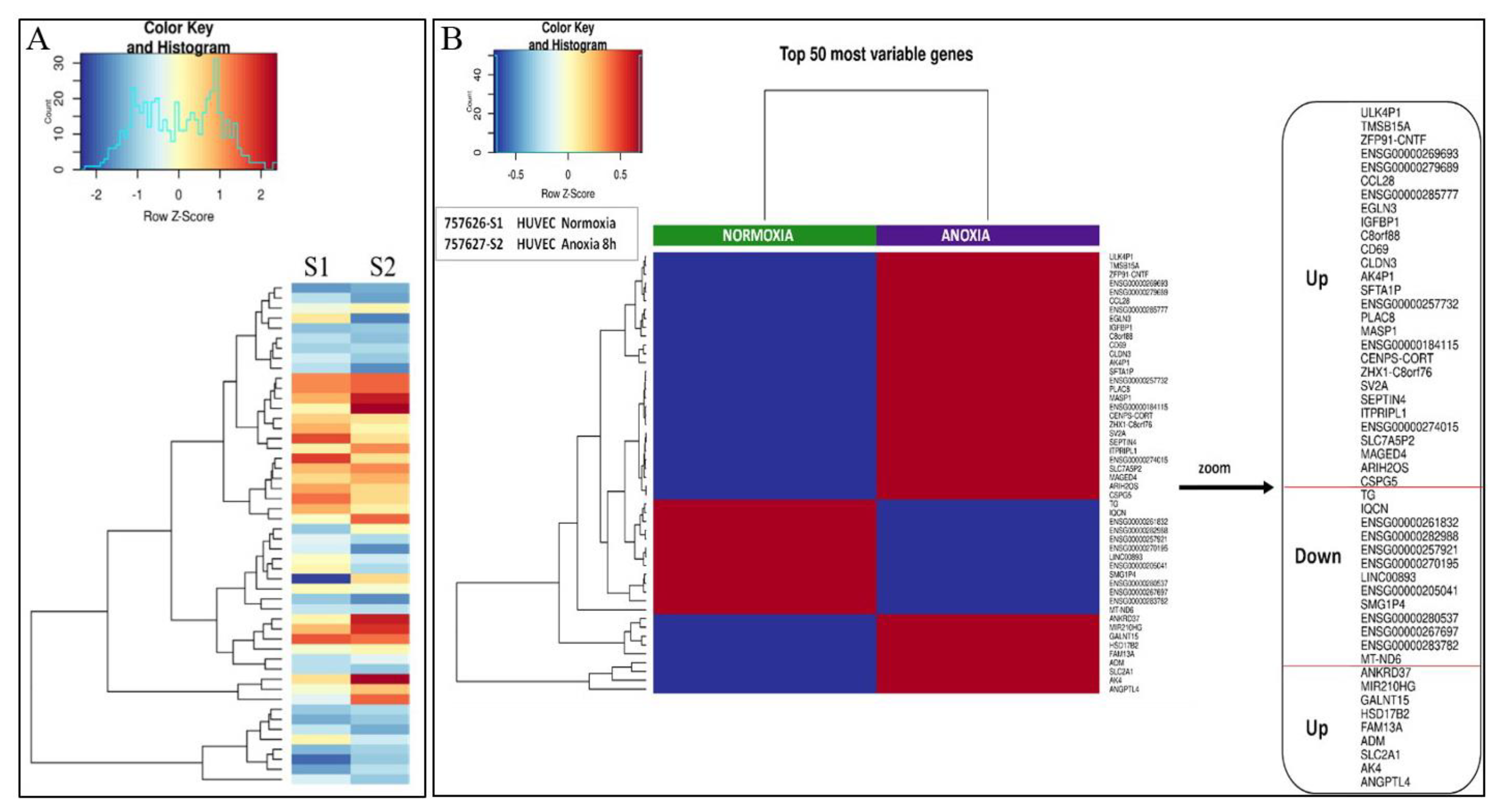
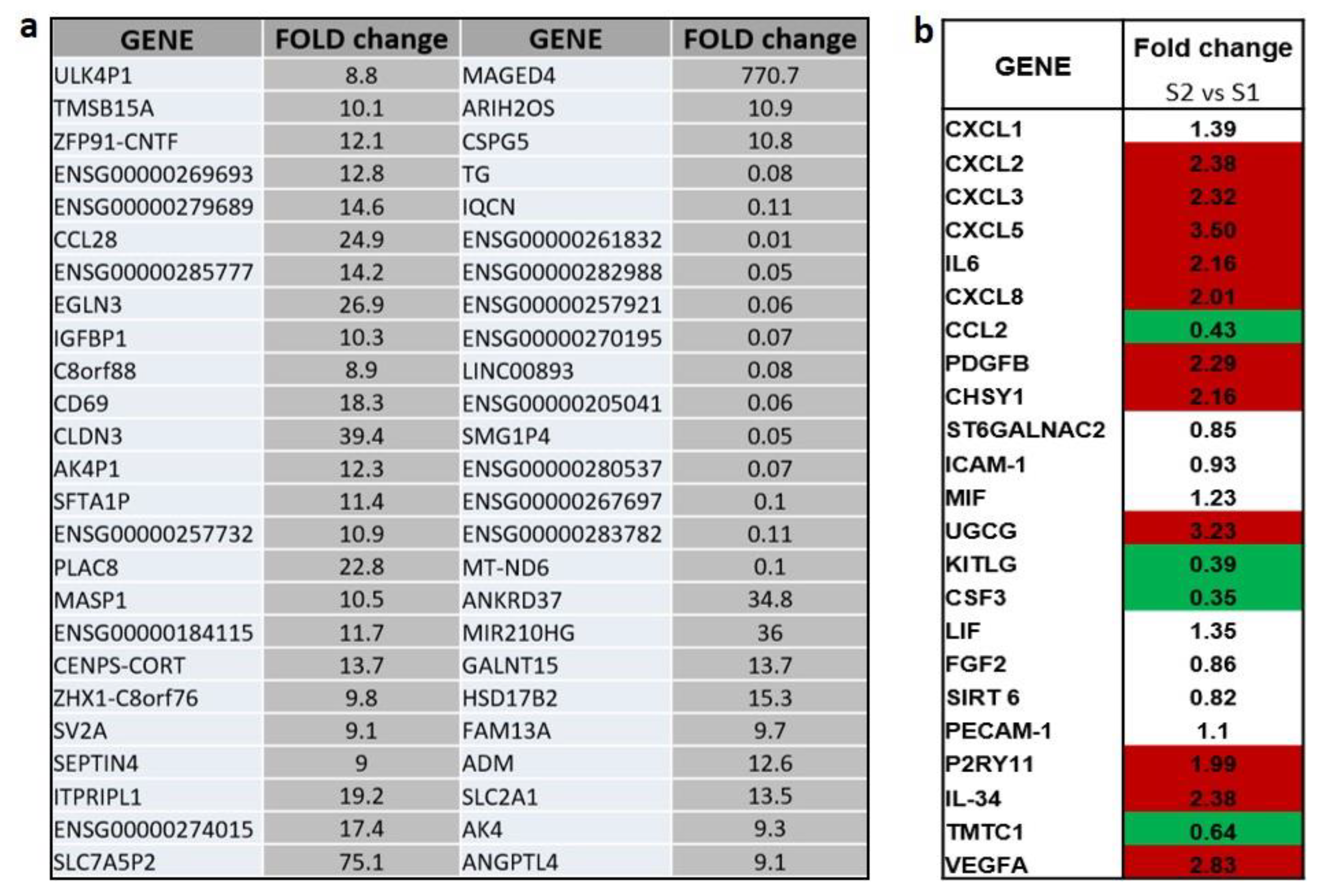
2.1. Transcriptional Analysis and Evaluation of Chemokine Release
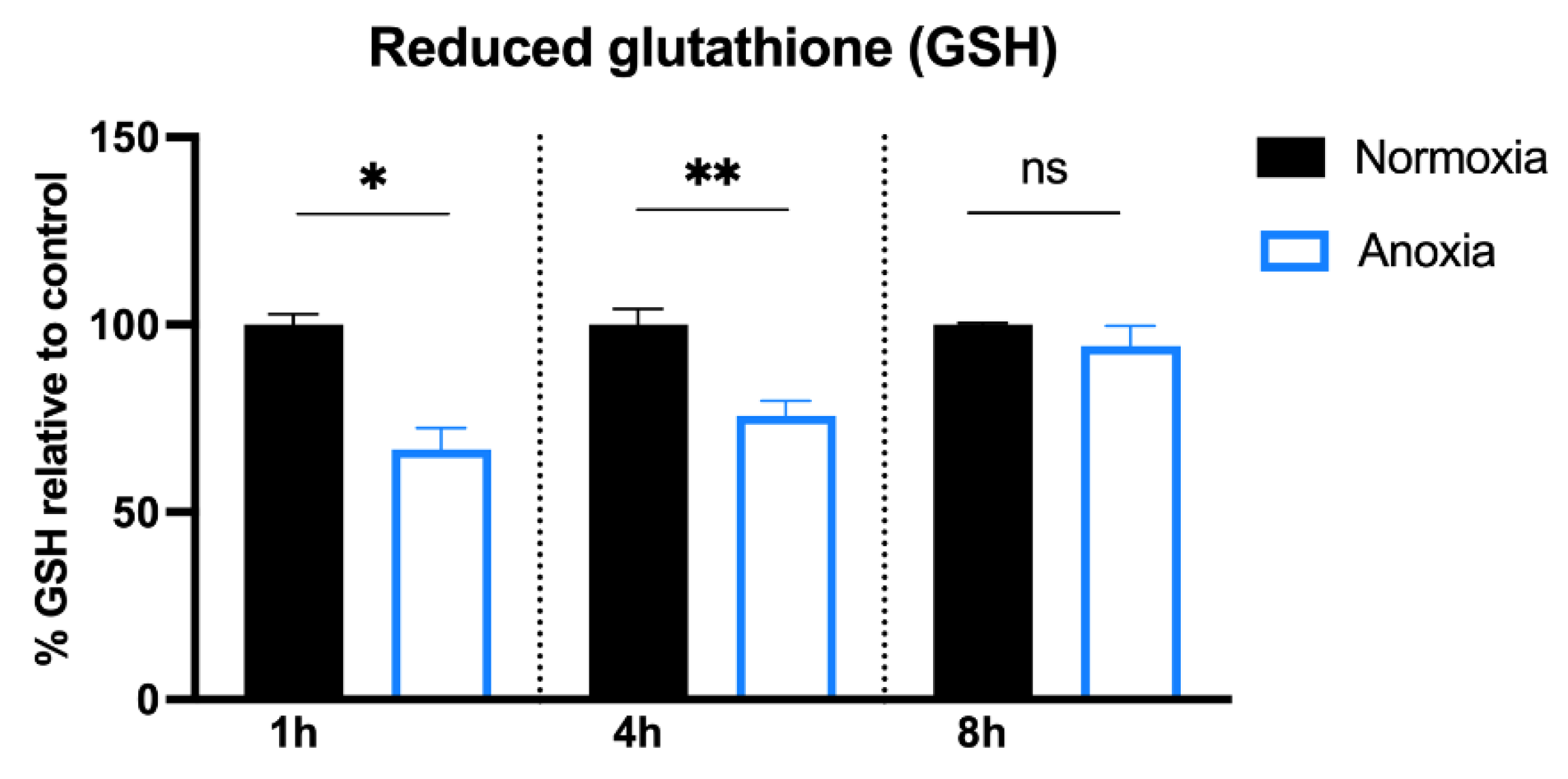
2.2. GSH Content in HUVECs after Anoxia
2.3. Reactome Functional Interaction Network
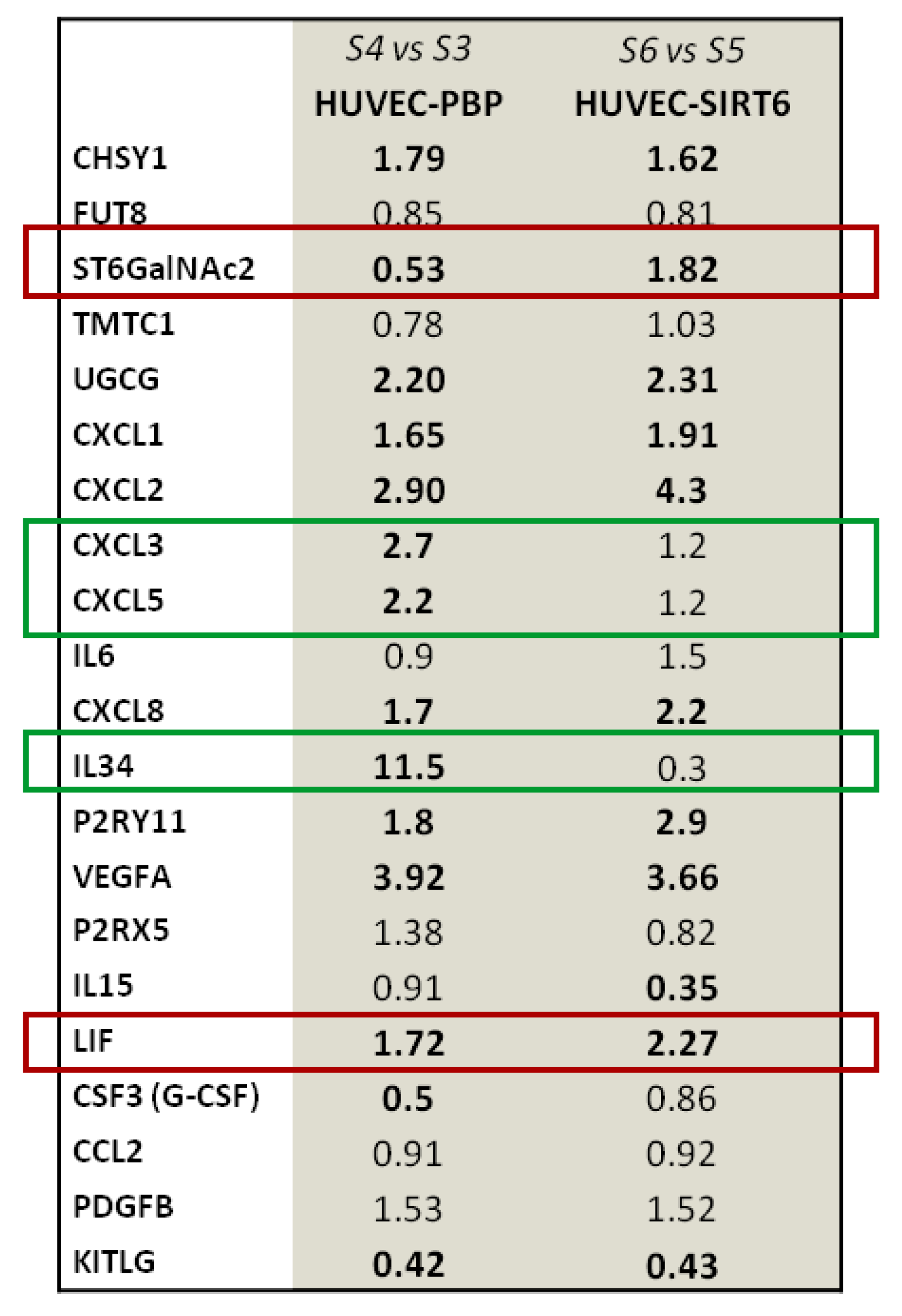
2.4. Results on HUVEC-SIRT6 Transfected Cells after Anoxia Treatment
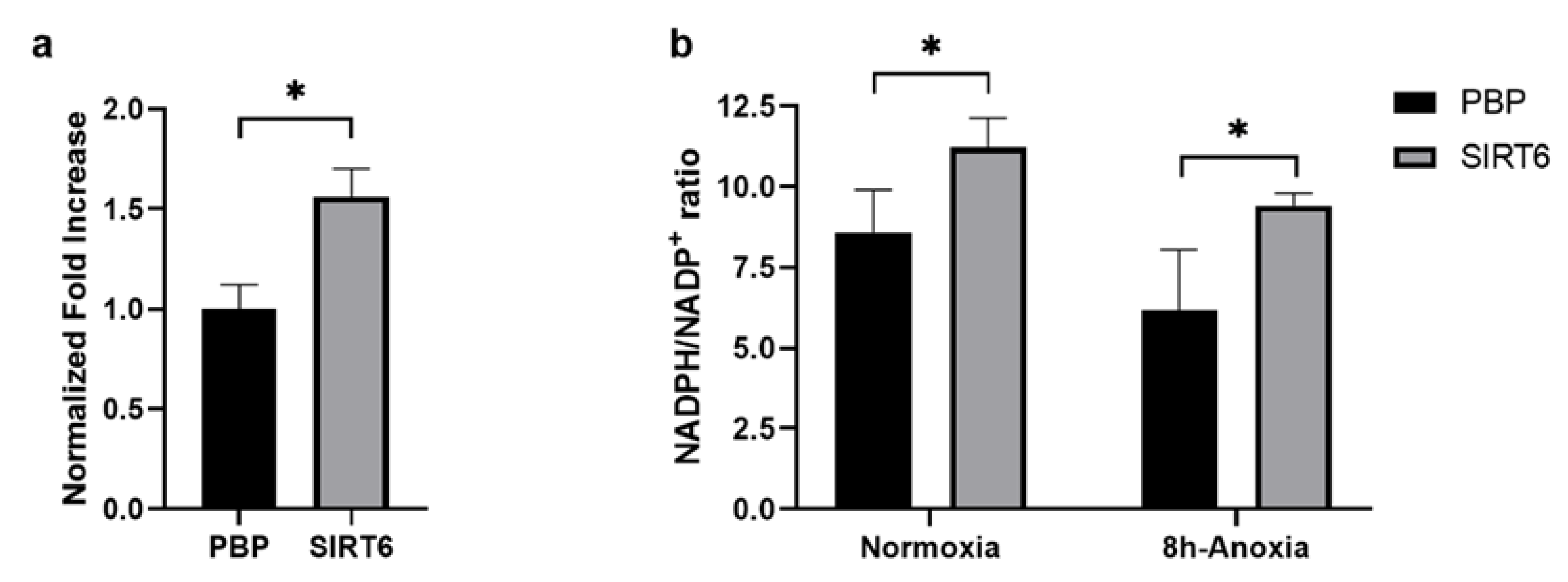
2.5. G6PDH, NADH and ROS Evaluation in HUVEC-SIRT6 Cells Exposed to Anoxic Conditions
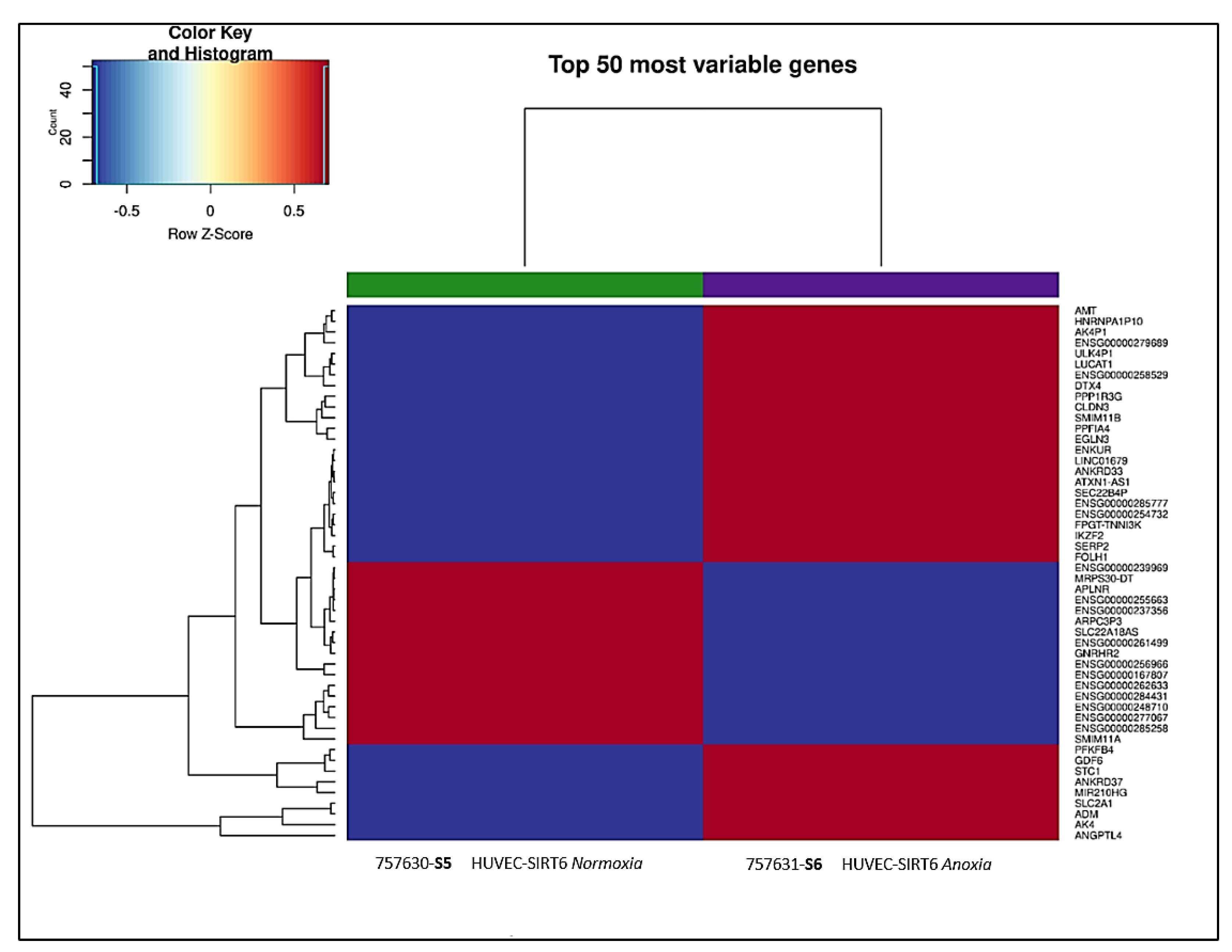
2.6. Transcriptional Analysis of Cell Extracts from 8 h Anoxia-Treated HUVEC-SIRT6 Cells
3. Materials and Methods
3.1. Cell Cultures
3.2. Western Blotting Analysis
3.3. Real-Time Quantitative PCR
3.4. mRNA-Seq Analysis
3.5. Determination of Cytokine and Interleukin Release
3.6. NADP(H) Evaluation
3.7. Assay of Glucose-6-Phosphate Dehydrogenase Activity
3.8. ROS Assay
3.9. Glutathione Detection
3.10. Statistical Analysis
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Richter, R.; Forssmann, W.; Henschler, R. Current Developments in Mobilization of Hematopoietic Stem and Progenitor Cells and Their Interaction with Niches in Bone Marrow. Transfus. Med. Hemother. 2017, 44, 151–164. [Google Scholar] [CrossRef]
- Antonelli, A.; Scarpa, E.S.; Magnani, M. Human Red Blood Cells Modulate Cytokine Expression in Monocytes/Macrophages Under Anoxic Conditions. Front. Physiol. 2021, 12, e632682. [Google Scholar] [CrossRef]
- Chow, D.C.; Wenning, L.A.; Miller, W.M.; Papoutsakis, E.T. Modeling pO2 Distributions in the Bone Marrow Hematopoietic Compartment. I. Krogh’s Model. Biophys. J. 2001, 81, 675–684. [Google Scholar] [CrossRef]
- Namiki, A.; Brogi, E.; Kearney, M.; Kim, E.A.; Wu, T.; Couffinhal, T.; Varticovski, L.; Isner, J.M. Hypoxia Induces Vascular Endothelial Growth Factor in Cultured Human Endothelial Cells. J. Biol. Chem. 1995, 270, 31189–31195. [Google Scholar] [CrossRef]
- Shibuya, M. Vascular endothelial growth factor (VEGF) and its receptor (VEGFR) signaling in angiogenesis: A crucial target for anti- and pro-angiogenic therapies. Genes Cancer 2011, 2, 1097–1105. [Google Scholar] [CrossRef]
- Rehn, M.; Olsson, A.; Reckzeh, K.; Diffner, E.; Carmeliet, P.; Landberg, G.; Cammenga, J. Hypoxic induction of vascular endothelial growth factor regulates murine hematopoietic stem cell function in the low-oxygenic niche. Blood 2011, 118, 1534–1543. [Google Scholar] [CrossRef]
- Geindreau, M.; Bruchard, M.; Vegran, F. Role of Cytokines and Chemokines in Angiogenesis in a Tumor Context. Cancers 2022, 14, 2446. [Google Scholar] [CrossRef]
- Loboda, A.; Jozkowicz, A.; Dulak, J. IF-1 versus HIF-2 is one more important than the other? Vasc. Pharmacol. 2012, 20156, 245–251. [Google Scholar] [CrossRef]
- Zhong, L.; D’Urso, A.; Toiber, D.; Sebastian, C.; Henry, R.E.; Vadysirisack, D.D.; Guimaraes, A.; Marinelli, B.; Wikstrom, J.D.; Nir, T.; et al. The Histone Deacetylase Sirt6 Regulates Glucose Homeostasis via Hif1α. Cell 2010, 140, 280–293. [Google Scholar] [CrossRef]
- Yang, Z.; Huang, Y.; Zhu, L.; Yang, K.; Liang, K.; Tan, J.; Yu, B. SIRT6 promotes angiogenesis and hemorrhage of carotid plaque via regulating HIF-1α and reactive oxygen species. Cell Death Dis. 2021, 12, 1–15. [Google Scholar] [CrossRef]
- Suda, T.; Takubo, K.; Semenza, G.L. Metabolic Regulation of Hematopoietic Stem Cells in the Hypoxic Niche. Cell Stem Cell 2011, 9, 298–310. [Google Scholar] [CrossRef]
- Schofield, C.J.; Ratcliffe, P.J. Oxygen sensing by HIF hydroxylases. Nat. Rev. Mol. Cell Biol. 2004, 5, 343–354. [Google Scholar] [CrossRef]
- Ginouves, A.; Ilc, K.; Macias, N.; Pouyssegur, J.; Berra, E. PHDs overactivation during chronic hypoxia “desensitizes” HIFalpha and protects cells from necrosis. Proc. Natl. Acad. Sci. USA 2008, 105, 4745–4750. [Google Scholar] [CrossRef]
- Demidenko, Z.; Rapisarda, A.; Garayoa, M. Accumulation of hypoxia-inducible factor-1α is limited by transcription-dependent depletion. Oncogene 2005, 24, 4829–4838. [Google Scholar] [CrossRef]
- Bryder, D.; Jacobsen, S. Interleukin-3 supports expansion of long-term multilineage repopulating activity after multiple stem cell divisions in vitro. Blood 2000, 96, 1748–1755. [Google Scholar] [CrossRef]
- Gazzola, M.V.; Collins, N.H.; Tafuri, A.; Keever, C. Recombinant interleukin 3 induces interleukin 2 receptor expression on early myeloid cells in normal human bone marrow. Exp. Hematol. 1992, 20, 201–208. [Google Scholar] [PubMed]
- Giampaolo, S.; Wójcik, G.; Serfling, E.; Patra, A.K. Interleukin-2-regulatory T cell axis critically regulates maintenance of hematopoietic stem cells. Oncotarget 2017, 8, 29625–29642. [Google Scholar] [CrossRef]
- Dela Paz, N.G.; Walshe, T.E.; Leach, L.L.; Saint-Geniez, M.; D′Amore, P.A. Role of shear-stress-induced VEGF expression in endothelial cell survival. J. Cell Sci. 2012, 125, 831–843. [Google Scholar] [CrossRef]
- Martin, D.; Galisteo, R.; Silvio, J. Gutkind. CXCL8/IL8 Stimulates Vascular Endothelial Growth Factor (VEGF) Expression and the Autocrine Activation of VEGFR2 in Endothelial Cells by Activating NF B through the CBM (Carma3/Bcl10/Malt1) Complex. J. Biol. Chem. 2009, 284, 6038–6042. [Google Scholar] [CrossRef]
- Ségaliny, A.I.; Mohamadi, A.; Dizier, B.; Lokajczyk, A.; Brion, R.; Lanel, R.; Amiaud, J.; Charrier, C.; Boisson-Vidal, C.; Heymann, D. Interleukin-34 promotes tumor progression and metastatic process in osteosarcoma through induction of angiogenesis and macrophage recruitment. Int. J. Cancer 2015, 137, 73–85. [Google Scholar] [CrossRef]
- Baghdadi, M.; Umeyama, Y.; Hama, N.; Kobayashi, T.; Han, N.; Wada, H.; Seino, K.I. Interleukin-34, a comprehensive review. J. Leukoc. Biol. 2018, 104, 931–951. [Google Scholar] [CrossRef] [PubMed]
- Eda, H.; Zhang, J.; Keith, R.H.; Michener, M.; Beidler, D.R.; Monahan, J.B. Macrophage-colony stimulating factor and interleukin-34 induce chemokines in human whole blood. Cytokine 2010, 52, 215–220. [Google Scholar] [CrossRef] [PubMed]
- Lee, E.; Seo, M.; Juhnn, Y.-S.; Kim, J.Y.; Hong, Y.J.; Lee, Y.J.; Song, Y.W. Potential role and mechanism of IFN-gamma inducible protein-10 on receptor activator of nuclear factor kappa-B ligand (RANKL) expression in rheumatoid arthritis. Thromb. Haemost. 2011, 13, R104–R108. [Google Scholar] [CrossRef] [PubMed]
- Dufour, J.H.; Dziejman, M.; Liu, M.T.; Leung, J.H.; Lane, T.E.; Luster, A.D. IFN-γ-Inducible Protein 10 (IP-10; CXCL10)-Deficient Mice Reveal a Role for IP-10 in Effector T Cell Generation and Trafficking. J. Immunol. 2002, 168, 3195–3204. [Google Scholar] [CrossRef] [PubMed]
- Xia, J.B.; Liu, G.H.; Chen, Z.Y.; Cheng-Zhou Mao, C.Z.; Deng-Cheng Zhou, D.C.; Wu, H.Y.; Park, K.S.; Zhao, H.; Kim, S.K.; Cai, D.Q.; et al. Hypoxia/ischemia promotes CXCL10 expression in cardiac microvascular endothelial cells by NFkB activation. Cytokine 2016, 81, 63–70. [Google Scholar] [CrossRef]
- Deshmane, S.L.; Kremlev, S.; Amini, S.; Sawaya, B.E. Monocyte Chemoattractant Protein-1 (MCP-1): An Overview. J. Interf. Cytokine Res. 2009, 29, 313–326. [Google Scholar] [CrossRef]
- Ou, X.; Wen, T.; Ying, J.; He, Q.; Xuan, A.; Ruan, D. MCP 1/CCR2 axis inhibits the chondrogenic differentiation of human nucleus pulposus mesenchymal stem cells. Mol. Med. Rep. 2022, 26, 277. [Google Scholar] [CrossRef]
- Park, C.-Y.; Choi, S.C.; Kim, J.H.; Choi, J.H.; Joo, H.J.; Hong, S.J.; Lim, D.S. Cardiac Stem Cell Secretome Protects Cardiomyocytes from Hypoxic Injury Partly via Monocyte Chemotactic Protein-1-Dependent Mechanism. Int. J. Mol. Sci. 2016, 17, 800. [Google Scholar] [CrossRef]
- Didion, S.P. Cellular and Oxidative Mechanisms Associated with Interleukin-6 Signaling in the Vasculature. Int. J. Mol. Sci. 2017, 18, 2563. [Google Scholar] [CrossRef]
- Noetzli, L.J.; French, S.L.; Machlus, K. New Insights into the Differentiation of Megakaryocytes from Hematopoietic Progenitors. Arter. Thromb. Vasc. Biol. 2019, 39, 1288–1300. [Google Scholar] [CrossRef]
- Chen, Y.F.; Wu, Z.M.; Xie, C.; Bai, S.; Zhao, L.D. Expression Level of IL-6 Secreted by Bone Marrow Stromal Cells in Mice with Aplastic Anemia. ISRN Hematol. 2013, 2013, 986219. [Google Scholar] [CrossRef] [PubMed]
- Blaser, B.W.; Moore, J.; Li, B.; Tamplin, O.J.; Binder, V.; Zon, L.I. IL-8 and CXCR1 Remodel the Vascular Niche to Promote Hematopoietic Stem and Progenitor Cell Engraftment. Blood 2015, 126, 783. [Google Scholar] [CrossRef]
- Karakurum, M.; Shreeniwas, R.; Chen, J.; Pinsky, D.; Yan, S.D.; Anderson, M.; Sunouchi, K.; Major, J.; Hamilton, T.; Kuwabara, K. Hypoxic induction of interleukin-8 gene expression in human endothelial cells. J. Clin. Investig. 1994, 93, 1564–1570. [Google Scholar] [CrossRef] [PubMed]
- Florczyk, U.; Czauderna, S.; Stachurska, A.; Tertil, M.; Nowak, W.; Kozakowska, M.; Poellinger, L.; Jozkowicz, A.; Loboda, A.; Dulak, J. Opposite effects of HIF-1α and HIF-2α on the regulation of IL-8 expression in endothelial cells. Free. Radic. Biol. Med. 2011, 51, 1882–1892. [Google Scholar] [CrossRef] [PubMed]
- Zhang, P.; Zhang, C.; Li, J.; Han, J.; Liu, X.; Yang, H. The physical microenvironment of hematopoietic stem cells and its emerging roles in engineering applications. Stem Cell Res. Ther. 2019, 10, 1–13. [Google Scholar] [CrossRef]
- D′Atri, L.P.; Pozner, R.G.; Nahmod, K.A.; Landoni, V.I.; Isturiz, M.; Negrotto, S. Paracrine regulation of megakaryo/thrombopoiesis by macrophages. Exp. Hematol. 2011, 39, 763–772. [Google Scholar] [CrossRef]
- Dănilă, M.-D.; Piollet, M.; Aburel, O.-M.; Angoulvant, D.; Lefort, C.; Chadet, S.; Roger, S.; Muntean, M.-D.; Ivanes, F. Modulation of P2Y11-related purinergic signaling in inflammation and cardio-metabolic diseases. Eur. J. Pharmacol. 2020, 876, 173060. [Google Scholar] [CrossRef]
- Dreisig, K.; Kornum, B.R. A critical look at the function of the P2Y11 receptor. Purinergic Signal. 2016, 12, 427–437. [Google Scholar] [CrossRef]
- Piollet, M.; Sturza, A.; Chadet, S.; Gabillard-Lefort, C.; Benoist, L.; Muntean, D.-M.; Aburel, O.-M.; Angoulvant, D.; Ivanes, F. P2Y11 Agonism Prevents Hypoxia/Reoxygenation- and Angiotensin II-Induced Vascular Dysfunction and Intimal Hyperplasia Development. Int. J. Mol. Sci. 2021, 22, 855. [Google Scholar] [CrossRef]
- Lefort, C.; Benoist, L.; Chadet, S.; Piollet, M.; Heraud, A.; Babuty, D.; Baron, C.; Ivanes, F.; Angoulvant, D. Stimulation of P2Y11 receptor modulates cardiac fibroblasts secretome toward immunomodulatory and protective roles after Hypoxia/Reoxygenation injury. J. Mol. Cell. Cardiol. 2018, 121, 212–222. [Google Scholar] [CrossRef]
- Vultaggio-Poma, V.; Di Virgilio, F. P2 Receptors: Novel Disease Markers and Metabolic Checkpoints in Immune Cells. Biomolecules 2022, 12, 983. [Google Scholar] [CrossRef] [PubMed]
- Schömel, N.; Hancock, S.E.; Gruber, L.; Olzomer, E.M.; Byrne, F.L.; Shah, D.; Hoehn, K.L.; Turner, N.; Grösch, S.; Geisslinger, G.; et al. UGCG influences glutamine metabolism of breast cancer cells. Sci. Rep. 2019, 9, 15665. [Google Scholar] [CrossRef] [PubMed]
- Wegner, M.S.; Gruber, L.; Mattjus, P. The UDP-glucose ceramide glycosyltransferase (UGCG) and the link to multidrug resistance protein 1 (MDR1). BMC Cancer 2018, 18, 153. [Google Scholar] [CrossRef]
- Korbecki, J.; Kojder, K.; Kapczuk, P.; Kupnicka, P.; Gawrońska-Szklarz, B.; Gutowska, I.; Chlubek, D.; Baranowska-Bosiacka, I. The Effect of Hypoxia on the Expression of CXC Chemokines and CXC Chemokine Receptors—A Review of Literature. Int. J. Mol. Sci. 2021, 22, 843. [Google Scholar] [CrossRef]
- Chen, Y.-I.; Wei, P.-C.; Hsu, J.-L.; Su, F.-Y.; Lee, W.-H. NPGPx (GPx7): A novel oxidative stress sensor/transmitter with multiple roles in redox homeostasis. Am. J. Transl. Res. 2016, 8, 1626–1640. [Google Scholar]
- Peppelenbosch, M.P.; Spaander, M.C.W.; Bruno, M.J. Glutathione peroxidase 7 prevents cancer in the oesophagus. Gut 2013, 63, 537–538. [Google Scholar] [CrossRef] [PubMed]
- Chen, Z.; Hu, T.; Zhu, S.; Mukaisho, K.; El-Rifai, W.; Peng, D.-F. Glutathione peroxidase 7 suppresses cancer cell growth and is hypermethylated in gastric cancer. Oncotarget 2017, 8, 54345–54356. [Google Scholar] [CrossRef]
- Guerriero, E.; Capone, F.; Accardo, M.; Sorice, A.; Costantini, M.; Colonna, G.; Castello, G. GPX4 and GPX7 over-expression in human hepatocellular carcinoma tissues. Eur. J. Histochem. 2015, 59, 2540. [Google Scholar] [CrossRef] [PubMed]
- Wei, P.-C.; Hsieh, Y.-H.; Su, M.-I.; Jiang, X.; Hsu, P.-H.; Lo, W.-T.; Weng, J.-Y.; Jeng, Y.-M.; Wang, J.-M.; Chen, P.-L.; et al. Loss of the Oxidative Stress Sensor NPGPx Compromises GRP78 Chaperone Activity and Induces Systemic Disease. Mol. Cell 2012, 48, 747–759. [Google Scholar] [CrossRef]
- Shao, J.; Liu, X.; Zhu, L.; Yen, Y. Targeting ribonucleotide reductase for cancer therapy. Expert Opin. Ther. Targets 2013, 17, 1423–1437. [Google Scholar] [CrossRef]
- Foskolou, I.P.; Jorgensen, C.; Leszczynska, K.B.; Olcina, M.M.; Tarhonskaya, H.; Haisma, B.; D’Angiolella, V.; Myers, W.K.; Domene, C.; Flashman, E.; et al. Ribonucleotide Reductase Requires Subunit Switching in Hypoxia to Maintain DNA Replication. Mol. Cell 2017, 66, 206–220.E9. [Google Scholar] [CrossRef] [PubMed]
- Haffo, L.; Lu, J.; Bykov, V.J.N.; Martin, S.S.; Ren, X.; Coppo, L.; Wiman, K.G.; Holmgren, A. Inhibition of the glutaredoxin and thioredoxin systems and ribonucleotide reductase by mutant p53-targeting compound APR-246. Sci. Rep. 2018, 8, 12671. [Google Scholar] [CrossRef] [PubMed]
- Sengupta, R.; Coppo, L.; Mishra, P.; Holmgren, A. Glutathione-glutaredoxin is an efficient electron donor system for mammalian p53R2–R1-dependent ribonucleotide reductase. J. Biol. Chem. 2019, 294, 12708–12716. [Google Scholar] [CrossRef] [PubMed]
- Bruschi, M.; Vanzolini, T.; Sahu, N.; Balduini, A.; Magnani, M.; Fraternale, A. Functionalized 3D scaffolds for engineering the hematopoietic niche. Front. Bioeng. Biotechnol. 2022, 10, 968086. [Google Scholar] [CrossRef] [PubMed]
- Motterlini, R.; Foresti, R.; Bassi, R.; Calabrese, V.; Clark, J.E.; Green, C.J. Endothelial Heme Oxygenase-1 Induction by Hypoxia. J. Biol. Chem. 2000, 275, 13613–13620. [Google Scholar] [CrossRef] [PubMed]
- Fraternale, A.; De Angelis, M.; De Santis, R.; Amatore, D.; Masini, S.; Monittola, F.; Menotta, M.; Biancucci, F.; Bartoccini, F.; Retini, M.; et al. Targeting SARS-CoV-2 by synthetic dual-acting thiol compounds that inhibit Spike/ACE2 interaction and viral protein production. FASEB J. 2023, 37, e22741. [Google Scholar] [CrossRef] [PubMed]
- Hopman, R.K.; DiPersio, J.F. Advances in stem cell mobilization. Blood Rev. 2014, 28, 31–40. [Google Scholar] [CrossRef]
- Mendelson, A.; Frenette, P.S. Hematopoietic stem cell niche maintenance during homeostasis and regeneration. Nat. Med. 2014, 20, 833–846. [Google Scholar] [CrossRef]
- Nowicki, M.; Szemraj, J.; Wierzbowska, A.; Misiewicz, M.; Małachowski, R.; Pluta, A.; Grzybowska-Izydorczyk, O.; Robak, T.; Szmigielska-Kapłon, A. miRNA-15a, miRNA-16, miRNA-126, miRNA-146a, and miRNA-223 expressions in autologous hematopoietic stem cell transplantation and their impact on engraftment. Eur. J. Haematol. 2018, 100, 426–435. [Google Scholar] [CrossRef]
- Nowicki, M.; Szemraj, J.; Wierzbowska, A.; Pluta, A.; Grzybowska-Izydorczyk, O.; Nowicka, A.; Stelmach, P.; Czemerska, M.; Szmigielska-Kapłon, A. Alterations in microRNA Expression during Hematopoietic Stem Cell Mobilization. Biology 2021, 10, 668. [Google Scholar] [CrossRef]
- Gattazzo, F.; Urciuolo, A.; Bonaldo, P. Extracellular matrix: A dynamic microenvironment for stem cell niche. Biochim. Et Biophys. Acta (BBA)–Gen. Subj. 2014, 1840, 2506–2519. [Google Scholar] [CrossRef] [PubMed]
- Skokowa, J.; Lan, D.; Thakur, B.; Gupta, K.; Cario, G.; Brechlin, A.; Stanulla, M.; Welte, K. NAMPT is essential for the G-CSF-induced myeloid differentiation via a NAD+-sirtuin-1-dependent pathway. Nat. Med. 2009, 221, A64. [Google Scholar] [CrossRef]
- O′Callaghan, C.; Vassilopoulos, A. Sirtuins at the crossroads of stemness, aging, and cancer. Aging Cell 2017, 16, 1208–1218. [Google Scholar] [CrossRef] [PubMed]
- Liszt, G.; Ford, E.; Kurtev, M.; Guarente, L. Mouse Sir2 Homolog SIRT6 Is a Nuclear ADP-ribosyltransferase. J. Biol. Chem. 2005, 280, 21313–21320. [Google Scholar] [CrossRef] [PubMed]
- Bruzzone, S.; Fruscione, F.; Morando, S.; Ferrando, T.; Poggi, A.; Garuti, A.; D’Urso, A.; Selmo, M.; Benvenuto, F.; Cea, M.; et al. Catastrophic NAD+ depletion in activated T lymphocytes through Nampt inhibition reduces demyelination and disability in EAE. PLoS ONE 2009, 4, e7897. [Google Scholar] [CrossRef]
- Kugel, S.; Mostoslavsky, R. Chromatin and beyond: The multitasking roles for SIRT6. Trends Biochem. Sci. 2014, 39, 72–81. [Google Scholar] [CrossRef]
- Mao, Z.; Hine, C.; Tian, X.; Van Meter, M.; Au, M.; Vaidya, A.; Seluanov, A.; Gorbunova, V. SIRT6 Promotes DNA Repair Under Stress by Activating PARP1. Science 2011, 332, 1443–1446. [Google Scholar] [CrossRef]
- Nowicki, M.; Wierzbowska, A.; Stec-Martyna, E.; Kulczycka-Wojdala, D.; Nowicki, G.; Szmigielska-Kapłon, A. SIRT1-SIRT7 Expression in Patients with Lymphoproliferative Disorders Undergoing Hematopoietic Stem Cell Mobilization. Cancers 2022, 14, 1213. [Google Scholar] [CrossRef]
- Edatt, L.; Aswini, P.; Grace, R.R.; Vishnu, R.; Sharath, S.S.; Sameer, K.V.B. Role of Sirtuins in Tumor Angiogenesis. Front. Oncol. 2020, 9, 1516. [Google Scholar] [CrossRef]
- Fiorentino, F.; Mai, A.; Rotili, D. Emerging Therapeutic Potential of SIRT6 Modulators. J. Med. Chem. 2021, 64, 9732–9758. [Google Scholar] [CrossRef]
- Sociali, G.; Grozio, A.; Caffa, I.; Schuster, S.; Becherini, P.; Damonte, P.; Sturla, L.; Fresia, C.; Passalacqua, M.; Mazzola, F.; et al. SIRT6 deacetylase activity regulates NAMPT activity and NAD(P)(H) pools in cancer cells. FASEB J. 2018, 33, 3704–3717. [Google Scholar] [CrossRef] [PubMed]
- Kobayashi, C.I.; Suda, T. Regulation of reactive oxygen species in stem cells and cancer stem cells. J. Cell. Physiol. 2011, 227, 421–430. [Google Scholar] [CrossRef] [PubMed]
- Morris, G.; Gevezova, M.; Sarafian, V.; Maes, M. Redox regulation of the immune response. Cell. Mol. Immunol. 2022, 19, 1079–1101. [Google Scholar] [CrossRef]
- Ali, M.H.; Schlidt, S.A.; Chandel, N.S.; Hynes, K.L.; Schumacker, P.T.; Gewertz, B.L. Endothelial permeability and IL-6 production during hypoxia: Role of ROS in signal transduction. Am. J. Physiol. 1999, 277, L1057–L1065. [Google Scholar] [CrossRef] [PubMed]
- Lu, X.; Kang, Y. Hypoxia and Hypoxia-Inducible Factors: Master Regulators of Metastasis. Clin. Cancer Res. 2010, 16, 5928–5935. [Google Scholar] [CrossRef]
- Wu, L.; Yu, H.; Zhao, Y.; Zhang, C.; Wang, J.; Yue, X.; Yang, Q.; Hu, W. HIF-2α mediates hypoxia-induced LIF expression in human colorectal cancer cells. Oncotarget 2015, 6, 4406–4417. [Google Scholar] [CrossRef] [PubMed]
- Houtkooper, R.H.; Pirinen, E.; Auwerx, J. Sirtuins as regulators of metabolism and healthspan. Nat. Rev. Mol. Cell Biol. 2012, 13, 225–238. [Google Scholar] [CrossRef]
- Klein, M.A.; Denu, J.M. Biological and catalytic functions of sirtuin 6 as targets for small-molecule modulators. J. Biol. Chem. 2020, 295, 11021–11041. [Google Scholar] [CrossRef]
- Krishna Mohan, S.; Krishna Mohan, P.; Krishna, R. Solution structure of CXCL5–a novel chemokine and adipokine implicated in inflammation and obesity. PLoS ONE 2014, 9, e93228. [Google Scholar]
- Min, Y.; Xiaokun, M.; Dudu, J.; Lijing, W.; Qing, Z.; Jiangmin, Z. CXC chemokine ligand 5 (CXCL5) disrupted the permeability of human brain microvascular endothelial cells via regulating p38 signal. Microbiol. Immunol. 2021, 65, 40–47. [Google Scholar]
- Yoon, K.-A.; Cho, H.-S.; Shin, H.-I.; Cho, J.-Y. Differential Regulation of CXCL5 by FGF2 in Osteoblastic and Endothelial Niche Cells Supports Hematopoietic Stem Cell Migration. Stem Cells Dev. 2012, 21, 3391–3402. [Google Scholar] [CrossRef]
- Murugaesu, N.; Iravani, M.; Van Weverwijk, A.; Ivetic, A.; Johnson, D.A.; Antonopoulos, A. An In Vivo Functional Screen Identifies ST6GalNAc2 Sialyltransferase as a Breast Cancer Metastasis Suppressor. Cancer Discov. 2014, 4, 304–317. [Google Scholar] [CrossRef] [PubMed]
- Wetzler, M.; Talpaz, M.; Lowe, D.G.; Baiocchi, G.; Gutterman, J.U.; Kurzrock, R. Constitutive expression of leukemia inhibitory factor RNA by human bone marrow stromal cells and modulation by IL-1, TNF-alpha, and TGF-beta. Exp. Hematol. 1991, 19, 347–351. [Google Scholar] [PubMed]
- Shahgaldi, S.; Kahmini, F.R. A comprehensive review of Sirtuins: With a major focus on redox homeostasis and metabolism. Life Sci. 2021, 282, 119803. [Google Scholar] [CrossRef] [PubMed]
- Pan, H.; Guang-Hui, L.; Liu, X.; Li, J.; Wang, L.; Wu, J.; Weizhou, Z.; Zhang, W.; Ren, R.; Zhang, W.; et al. SIRT6 safeguards human mesenchymal stem cells from oxidative stress by coactivating NRF2. Cell Res. 2016, 26, 190–205. [Google Scholar] [CrossRef]
- Greiten, L.E.; Zhang, B.; Roos, C.M.; Hagler, M.; Jahns, F.-P.; Miller, J.D. Sirtuin 6 Protects Against Oxidative Stress and Vascular Dysfunction in Mice. Front. Physiol. 2021, 12, 753501. [Google Scholar] [CrossRef]
- Butler, J.M.; Nolan, D.J.; Vertes, E.L.; Varnum-Finney, B.; Kobayashi, H.; Hooper, A.T.; Seandel, M.; Shido, K.; White, I.A.; Kobayashi, M.; et al. Endothelial Cells Are Essential for the Self-Renewal and Repopulation of Notch-Dependent Hematopoietic Stem Cells. Cell Stem Cell 2010, 6, 251–264. [Google Scholar] [CrossRef]





| Samples | IL-6 | IL-8 | MCP-1 | IL-1b | IL-2 | IL-5 | PDGF-BB | IP-10 |
|---|---|---|---|---|---|---|---|---|
| S1 | 3167 | 110,236 | 8097 | 43 | 102 | 196 | 832 | 1222 |
| S2 | 23,941 | 358,257 | 40,761 | 34 | 122 | 444 | 716 | 63,475 |
| S2 vs. S1 (fold) | 7.56 | 3.25 | 5.03 | 0.79 | 1.19 | 2.26 | 0.86 | 51.96 |
| Gene Description | Fold Change |
|---|---|
| GSR, gluthatione reductase (NADPH) | 1.40 |
| IDHI, isocitrate dehydrogenase | 1.27 |
| PGD, 6-phosphogluconate dehydrogenase | 1.01 |
| G6PD, glucose-6 phosphate deydrogenase | 0.74 |
| GPX1, glutathione peroxidase 1 | 1.31 |
| GPX1P1, glutathione peroxidase, pseudogene 1 | 1.34 |
| GPX3, glutathione peroxidase 3 | 0.76 |
| GPX4, glutathione peroxidase 4 | 1.21 |
| GPX7, glutathione peroxidase 7 | 2.68 |
| GPX8, glutathione peroxidase 8 | 1.44 |
| RRM1, ribonucleoside-diphosphate reductase subunit M1 | 2.10 |
| RRM2, ribonucleoside-diphosphate reductase subunit M2 | 3.56 |
| OPLAH, 5 oxoprolinase (ATP-hydrolysing) | 0.71 |
| GCLM, glutamate-cysteine ligase regulatory subunit | 0.91 |
| GCLC, glutamate-cisteine ligase (6.3.2.2) | 0.99 |
| GSS, glutathione synthase (6.3.2.3) | 1.06 |
| SLC6A9, glycin transporter (Glyt 1) | 0.70 |
| SLC1A1, glutamate transporter | 1.51 |
| SLC7A11, cystine glutamate transporter | 1.40 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Antonelli, A.; Scarpa, E.S.; Bruzzone, S.; Astigiano, C.; Piacente, F.; Bruschi, M.; Fraternale, A.; Di Buduo, C.A.; Balduini, A.; Magnani, M. Anoxia Rapidly Induces Changes in Expression of a Large and Diverse Set of Genes in Endothelial Cells. Int. J. Mol. Sci. 2023, 24, 5157. https://doi.org/10.3390/ijms24065157
Antonelli A, Scarpa ES, Bruzzone S, Astigiano C, Piacente F, Bruschi M, Fraternale A, Di Buduo CA, Balduini A, Magnani M. Anoxia Rapidly Induces Changes in Expression of a Large and Diverse Set of Genes in Endothelial Cells. International Journal of Molecular Sciences. 2023; 24(6):5157. https://doi.org/10.3390/ijms24065157
Chicago/Turabian StyleAntonelli, Antonella, Emanuele Salvatore Scarpa, Santina Bruzzone, Cecilia Astigiano, Francesco Piacente, Michela Bruschi, Alessandra Fraternale, Christian A. Di Buduo, Alessandra Balduini, and Mauro Magnani. 2023. "Anoxia Rapidly Induces Changes in Expression of a Large and Diverse Set of Genes in Endothelial Cells" International Journal of Molecular Sciences 24, no. 6: 5157. https://doi.org/10.3390/ijms24065157
APA StyleAntonelli, A., Scarpa, E. S., Bruzzone, S., Astigiano, C., Piacente, F., Bruschi, M., Fraternale, A., Di Buduo, C. A., Balduini, A., & Magnani, M. (2023). Anoxia Rapidly Induces Changes in Expression of a Large and Diverse Set of Genes in Endothelial Cells. International Journal of Molecular Sciences, 24(6), 5157. https://doi.org/10.3390/ijms24065157








