Dissection of Antibody Responses of Gam-COVID-Vac-Vaccinated Subjects Suggests Involvement of Epitopes Outside RBD in SARS-CoV-2 Neutralization
Abstract
1. Introduction
2. Results
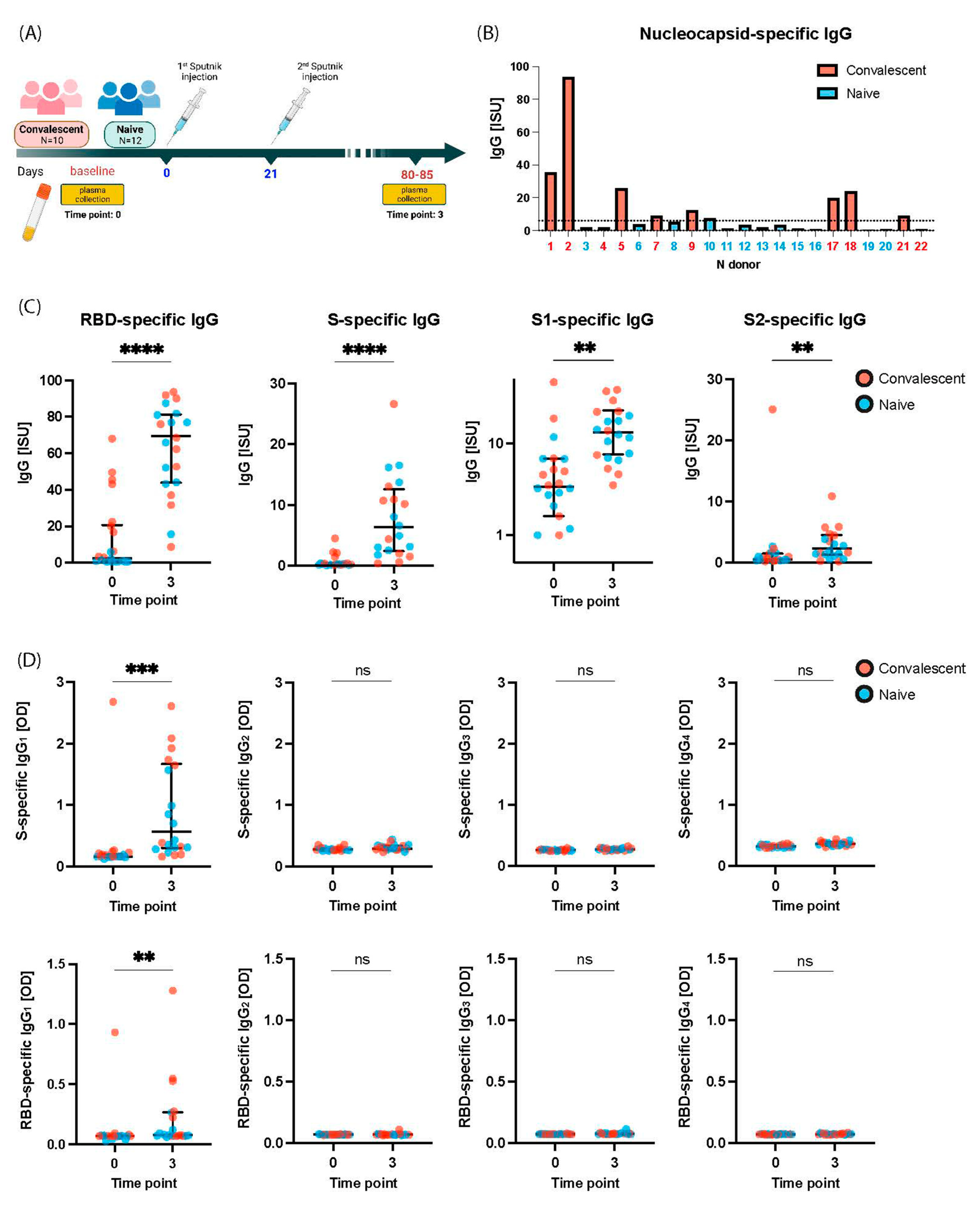
2.1. Gam-COVID-Vac-Vaccinated Subjects Include COVID-19 Naïve and Convalescent Subjects
2.2. Gam-COVID-Vac Vaccination Induces Broad IgG Responses to S Including S1, RBD, and S2
2.3. Gam-COVID-Vac Vaccination Induces Mainly an IgG1 Subclass Response to S and RBD
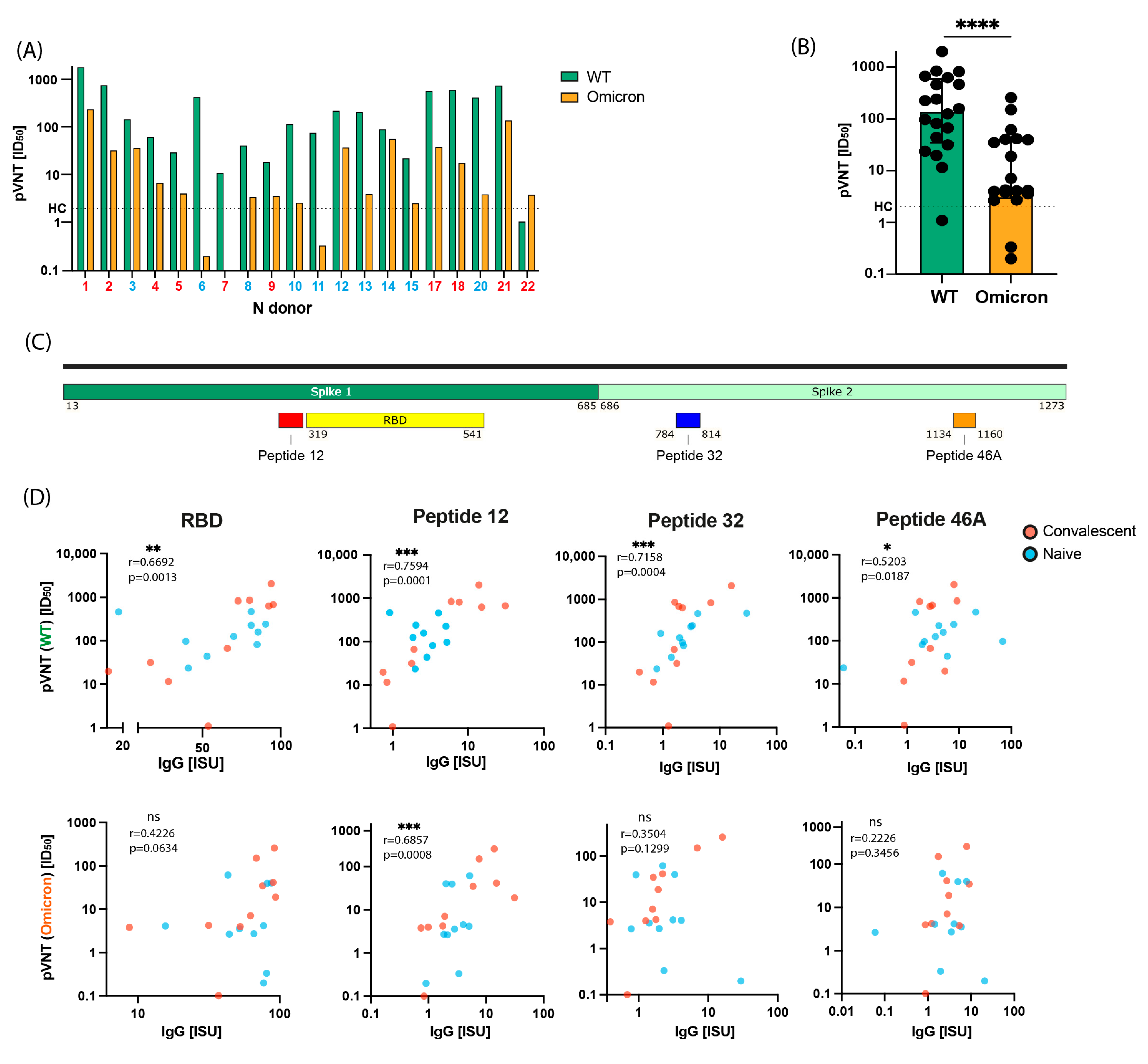
2.4. Induction of Virus-Neutralizing Antibodies and Antibodies Inhibiting the RBD-ACE2 Interaction by Gam-COVID-Vac Vaccination
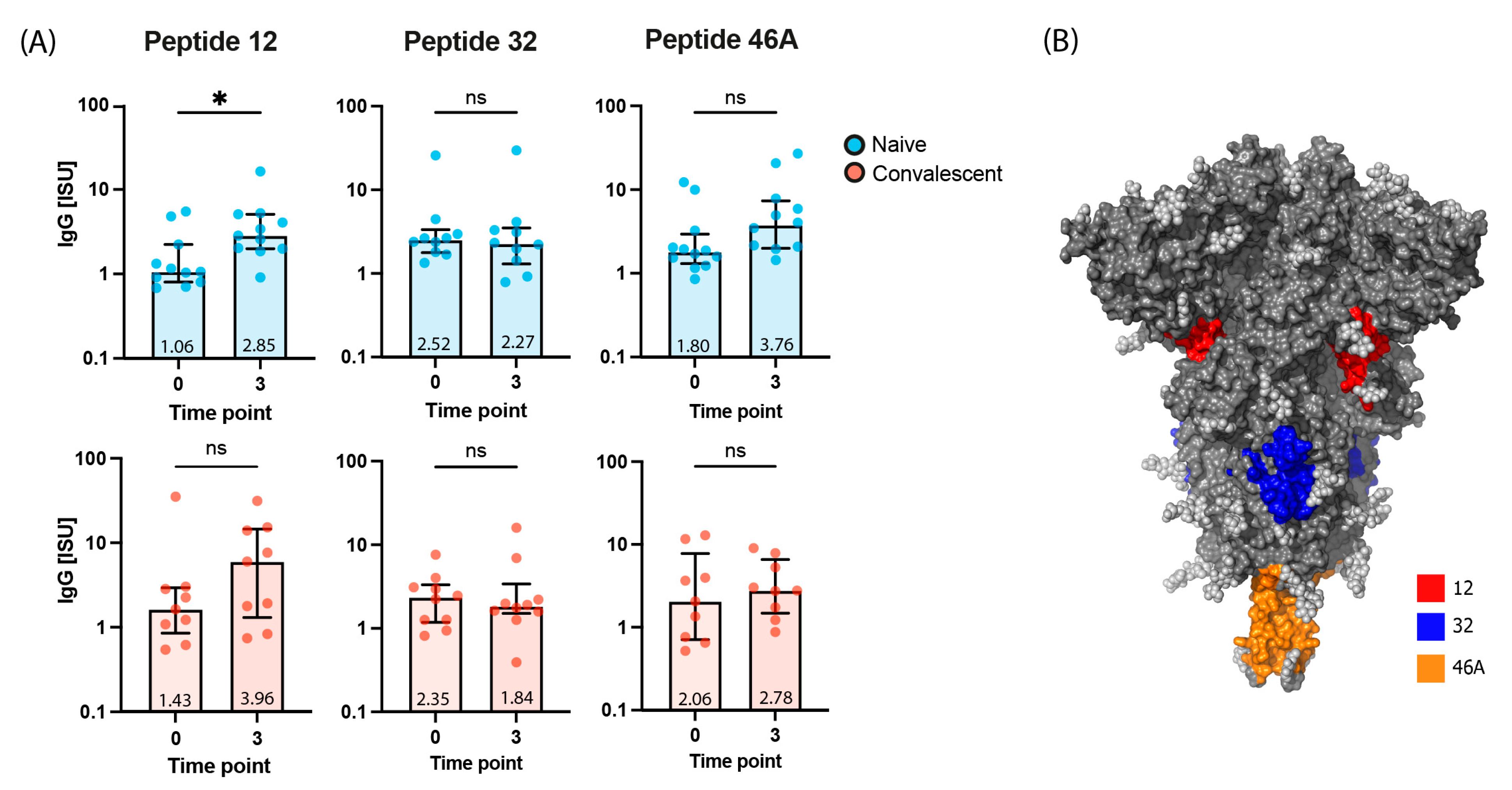
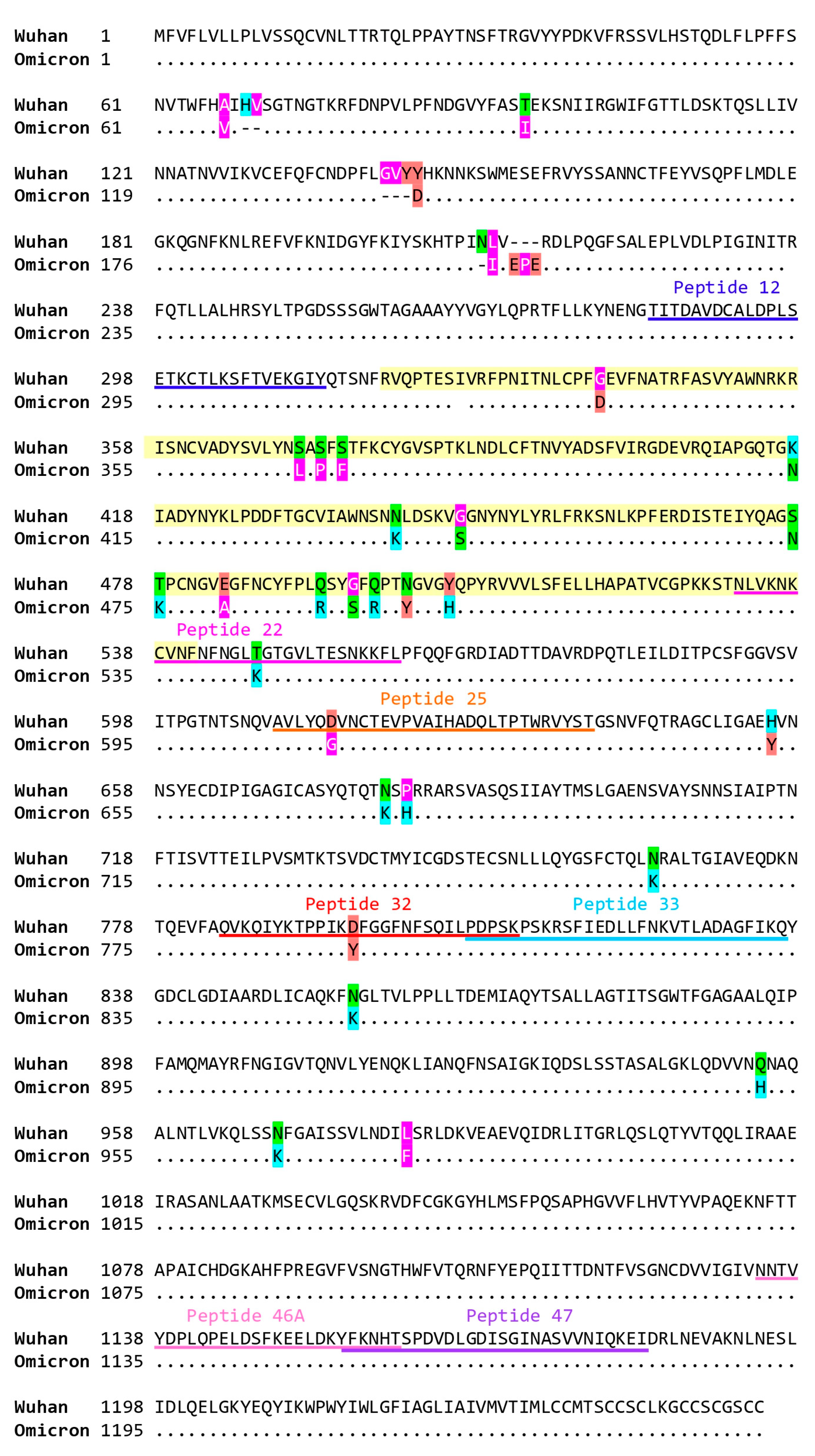
2.5. Virus Neutralization in Gam-COVID-Vac-Vaccinated Subjects Is Strongly Correlated with IgG Responses to RBD and to the S-Derived Peptide 12
3. Discussion
4. Materials and Methods
4.1. Study Population and Ethics Statement
4.2. Detection of SARS-CoV-2-Specific Antibody Responses
4.3. Far UV Circular Dichroism (CD) Spectra
4.4. Pseudo-Virus-Based Virus Neutralization Assay (pVNT)
4.5. Molecular Interaction Assay (MIA)
4.6. Visualization of Peptides in the Spike Protein Structure
4.7. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Rodriguez-Coira, J.; Sokolowska, M. SARS-CoV-2 candidate vaccines—Composition, mechanisms of action and stages of clinical development. Allergy 2021, 76, 1922–1924. [Google Scholar] [CrossRef] [PubMed]
- Sharma, A.; Ahmad, F.I.; Lal, S.K. COVID-19: A Review on the Novel Coronavirus Disease Evolution, Transmission, Detection, Control and Prevention. Viruses 2021, 13, 202. [Google Scholar] [CrossRef] [PubMed]
- Arshad, B.; Iqbal, T.; Bhatti, K.P. Review-Insights into Off-Label therapeutic strategies against mild and severe COVID-19 infection. Pak. J. Pharm. Sci. 2021, 34, 1469–1484. [Google Scholar] [CrossRef] [PubMed]
- Zaman, W.; Saqib, S.; Ullah, F. COVID-19: Phylogenetic approaches may help in finding resources for natural cure. Phytother. Res. 2020, 34, 2783–2785. [Google Scholar] [CrossRef]
- Rubio-Casillas, A.; Redwan, E.M.; Uversky, V.N. SARS-CoV-2 Intermittent Virulence as a Result of Natural Selection. COVID 2022, 2, 1089–1101. [Google Scholar] [CrossRef]
- Logunov, D.Y.; Dolzhikova, I.V.; Zubkova, O.V. Safety and immunogenicity of an rAd26 and rAd5 vector-based heterologous prime-boost COVID-19 vaccine in two formulations: Two open, non-randomised phase 1/2 studies from Russia. Lancet 2020, 396, 887–897, Correction in Lancet 2021, 397, 98. [Google Scholar] [CrossRef]
- Folegatti, P.M.; Ewer, K.J.; Aley, P.K. Safety and immunogenicity of the ChAdOx1 nCoV-19 vaccine against SARS-CoV-2: A preliminary report of a phase 1/2, single-blind, randomised controlled trial. Lancet 2020, 396, 467–478, Correction in Lancet 2020, 396, 1884. [Google Scholar] [CrossRef]
- Logunov, D.Y.; Dolzhikova, I.V.; Shcheblyakov, D.V. Safety and efficacy of an rAd26 and rAd5 vector-based heterologous prime-boost COVID-19 vaccine: An interim analysis of a randomised controlled phase 3 trial in Russia. Lancet 2021, 397, 671–681, Correction in Lancet 2021, 397, 670. [Google Scholar] [CrossRef]
- Jackson, C.B.; Farzan, M.; Chen, B.; Choe, H. Mechanisms of SARS-CoV-2 entry into cells. Nat. Rev. Mol. Cell Biol. 2022, 23, 3–20. [Google Scholar] [CrossRef]
- Heinz, F.X.; Stiasny, K. Distinguishing features of current COVID-19 vaccines: Knowns and unknowns of antigen presentation and modes of action. NPJ Vaccines 2021, 6, 104. [Google Scholar] [CrossRef]
- Lim, H.X.; Masomian, M.; Khalid, K.; Kumar, A.U.; MacAry, P.A.; Poh, C.L. Identification of B-Cell Epitopes for Eliciting Neutralizing Antibodies against the SARS-CoV-2 Spike Protein through Bioinformatics and Monoclonal Antibody Targeting. Int. J. Mol. Sci. 2022, 23, 4341. [Google Scholar] [CrossRef]
- Moroy, G.; Tuffery, P. Peptide-Based Strategies Against SARS-CoV-2 Attack: An Updated In Silico Perspective. Front. Drug. Discov. 2022, 2, 899477. [Google Scholar] [CrossRef]
- Gattinger, P.; Niespodziana, K.; Stiasny, K. Neutralization of SARS-CoV-2 requires antibodies against conformational receptor-binding domain epitopes. Allergy 2022, 77, 230–242. [Google Scholar] [CrossRef]
- Niu, L.; Wittrock, K.N.; Clabaugh, G.C. A Structural Landscape of Neutralizing Antibodies Against SARS-CoV-2 Receptor Binding Domain. Front. Immunol. 2021, 12, 647934. [Google Scholar] [CrossRef]
- Shan, S.; Mok, C.K.; Zhang, S. A Potent and Protective Human Neutralizing Antibody Against SARS-CoV-2 Variants. Front. Immunol. 2021, 12, 766821. [Google Scholar] [CrossRef]
- Bajpai, P.; Singh, V.; Chandele, A. Broadly Neutralizing Antibodies to SARS-CoV-2 Provide Novel Insights Into the Neutralization of Variants and Other Human Coronaviruses. Front. Cell. Infect. Microbiol. 2022, 12, 928279. [Google Scholar] [CrossRef]
- Vishwakarma, P.; Yadav, N.; Rizvi, Z.A. Severe Acute Respiratory Syndrome Coronavirus 2 Spike Protein Based Novel Epitopes Induce Potent Immune Responses in vivo and Inhibit Viral Replication in vitro. Front. Immunol. 2021, 12, 613045. [Google Scholar] [CrossRef]
- Garrett, M.E.; Galloway, J.G.; Wolf, C. Comprehensive characterization of the antibody responses to SARS-CoV-2 Spike protein finds additional vaccine-induced epitopes beyond those for mild infection. eLife 2022, 11, e73490. [Google Scholar] [CrossRef]
- Peng, M.; Dou, X.; Zhang, X. Protective antigenic epitopes revealed by immunosignatures after three doses of inactivated SARS-CoV-2 vaccine. Front. Immunol. 2022, 13, 938378. [Google Scholar] [CrossRef]
- Polvere, I.; Parrella, A.; Zerillo, L. Humoral Immune Response Diversity to Different COVID-19 Vaccines: Implications for the “Green Pass” Policy. Front. Immunol. 2022, 13, 833085. [Google Scholar] [CrossRef]
- Tukhvatulin, A.I.; Dolzhikova, I.V.; Shcheblyakov, D.V. An open, non-randomised, phase 1/2 trial on the safety, tolerability, and immunogenicity of single-dose vaccine “Sputnik Light” for prevention of coronavirus infection in healthy adults. Lancet Reg. Health Eur. 2021, 11, 100241. [Google Scholar] [CrossRef] [PubMed]
- Gonzalez, S.; Olszevicki, S.; Salazar, M. Effectiveness of the first component of Gam-COVID-Vac (Sputnik V) on reduction of SARS-CoV-2 confirmed infections, hospitalisations and mortality in patients aged 60–79: A retrospective cohort study in Argentina. EClinicalMedicine 2021, 40, 101126. [Google Scholar] [CrossRef] [PubMed]
- Gushchin, V.A.; Dolzhikova, I.V.; Shchetinin, A.M. Neutralizing Activity of Sera from Sputnik V-Vaccinated People against Variants of Concern (VOC: B.1.1.7, B.1.351, P.1, B.1.617.2, B.1.617.3) and Moscow Endemic SARS-CoV-2 Variants. Vaccines 2021, 9, 779. [Google Scholar] [CrossRef] [PubMed]
- Voko, Z.; Kiss, Z.; Surjan, G. Nationwide effectiveness of five SARS-CoV-2 vaccines in Hungary-the HUN-VE study. Clin. Microbiol. Infect. 2022, 28, 398–404. [Google Scholar] [CrossRef] [PubMed]
- Jeewandara, C.; Fernando, S.; Pushpakumara, P.D. Immune responses following the first dose of the Sputnik V (Gam-COVID-Vac). Sci. Rep. 2022, 12, 1727. [Google Scholar] [CrossRef]
- Ikegame, S.; Siddiquey, M.N.A.; Hung, C.T. Neutralizing activity of Sputnik V vaccine sera against SARS-CoV-2 variants. Nat. Commun. 2021, 12, 4598. [Google Scholar] [CrossRef]
- Gushchin, V.A.; Tsyganova, E.V.; Ogarkova, D.A. Sputnik V protection from COVID-19 in people living with HIV under antiretroviral therapy. EClinicalMedicine 2022, 46, 101360. [Google Scholar] [CrossRef]
- Shamji, M.H.; Valenta, R.; Jardetzky, T. The role of allergen-specific IgE, IgG and IgA in allergic disease. Allergy 2021, 76, 3627–3641. [Google Scholar] [CrossRef]
- Eckl-Dorna, J.; Weber, M.; Stanek, V. Two years of treatment with the recombinant grass pollen allergy vaccine BM32 induces a continuously increasing allergen-specific IgG4 response. EBioMedicine 2019, 50, 421–432. [Google Scholar] [CrossRef]
- Shamji, M.H.; Kappen, J.H.; Akdis, M. Biomarkers for monitoring clinical efficacy of allergen immunotherapy for allergic rhinoconjunctivitis and allergic asthma: An EAACI Position Paper. Allergy 2017, 72, 1156–1173. [Google Scholar] [CrossRef]
- Tortorici, M.A.; Beltramello, M.; Lempp, F.A. Ultrapotent human antibodies protect against SARS-CoV-2 challenge via multiple mechanisms. Science 2020, 370, 950–957. [Google Scholar] [CrossRef]
- Cia, G.; Pucci, F.; Rooman, M. Analysis of the Neutralizing Activity of Antibodies Targeting Open or Closed SARS-CoV-2 Spike Protein Conformations. Int. J. Mol. Sci. 2022, 23, 2078. [Google Scholar] [CrossRef]
- Meng, B.; Datir, R.; Choi, J. SARS-CoV-2 spike N-terminal domain modulates TMPRSS2-dependent viral entry and fusogenicity. Cell Rep. 2022, 40, 111220. [Google Scholar] [CrossRef]
- Resch, Y.; Weghofer, M.; Seiberler, S. Molecular characterization of Der p 10: A diagnostic marker for broad sensitization in house dust mite allergy. Clin. Exp. Allergy 2011, 41, 1468–1477. [Google Scholar] [CrossRef]
- Byazrova, M.; Yusubalieva, G.; Spiridonova, A. Pattern of circulating SARS-CoV-2-specific antibody-secreting and memory B-cell generation in patients with acute COVID-19. Clin. Transl. Immunol. 2021, 10, e1245. [Google Scholar] [CrossRef]
- Gattinger, P.; Borochova, K.; Dorofeeva, Y. Antibodies in serum of convalescent patients following mild COVID-19 do not always prevent virus-receptor binding. Allergy 2021, 76, 878–883. [Google Scholar] [CrossRef]
- Gattinger, P.; Tulaeva, I.; Borochova, K. Omicron: A SARS-CoV-2 variant of real concern. Allergy 2022, 77, 1616–1620. [Google Scholar] [CrossRef]
- Gattinger, P.; Ohradanova-Repic, A.; Valenta, R. Importance, Applications and Features of Assays Measuring SARS-CoV-2 Neutralizing Antibodies. Int. J. Mol. Sci. 2023, 24, 5352. [Google Scholar] [CrossRef]
- Gattinger, P.; Kratzer, B.; Tulaeva, I.; Niespodziana, K.; Ohradanova-Repic, A.; Gebetsberger, L.; Borochova, K.; Garner-Spitzer, E.; Trapin, D.; Hofer, G.; et al. Vaccine based on folded receptor binding domain-PreS fusion protein with potential to induce sterilizing immunity to SARS-CoV-2 variants. Allergy 2022, 77, 2431–2445. [Google Scholar] [CrossRef]
| Subjects | Sex | Age | Polymerase Chain Reaction (PCR)-Confirmed COVID-19 Symptoms (before Vaccination) | PCR-Confirmed COVID-19 Symptoms (Days after Vaccination) | Nucleocapsid-Specific IgG at Baseline (ISAC Standardized Units (ISU)) | RBD-Specific IgG at Baseline (ISU) | |
|---|---|---|---|---|---|---|---|
| Convalescent | 1 | M | 61 | −70 to −61 | no | 35.6 | 20.1 |
| 2 | M | 69 | −53 to −46 | no | 93.6 | 49.6 | |
| 4 | F | 60 | no | no | 1.9 | 16.8 | |
| 5 | M | 60 | no | no | 25.6 | 6.2 | |
| 7 | F | 66 | no | no | 9.2 | 3.4 | |
| 9 | M | 66 | no | no | 12.6 | 2.9 | |
| 17 | M | 42 | −86 to −78 | no | 19.8 | 43.0 | |
| 18 | M | 61 | −120 to −110 | no | 23.8 | 22.5 | |
| 21 | F | 53 | −88 to −71 | no | 8.9 | 68.0 | |
| 22 | F | 53 | no | 110 to 128 | 0.8 | 45.5 | |
| Naive | 3 | F | 62 | no | no | 2.0 | 0.7 |
| 6 | M | 70 | no | no | 3.8 | 0.8 | |
| 8 | F | 54 | no | no | 5.3 | 5.9 | |
| 10 | F | 43 | no | no | 7.7 | 0.5 | |
| 11 | F | 61 | no | no | 1.2 | 2.1 | |
| 12 | F | 52 | no | no | 3.5 | 1.0 | |
| 13 | F | 25 | no | no | 1.8 | 0.9 | |
| 14 | F | 46 | no | no | 3.4 | 0.7 | |
| 15 | F | 43 | no | no | 1.0 | 1.5 | |
| 16 | F | 51 | no | no | 0.8 | 0.5 | |
| 19 | F | 66 | no | no | 0.4 | 0.5 | |
| 20 | F | 62 | no | no | 0.8 | 0.9 |
| Subject ID | Time Point | S (Folded) [ISU] | S1 (Folded) [ISU] | S2 (Folded) [ISU] | RBD (Folded) [ISU] | N-Protein [ISU] | Inhibition of RBD-ACE2 [%] | pVNT (Wuhan) [ID50] | pVNT (Omicron) [ID50] |
|---|---|---|---|---|---|---|---|---|---|
| 1 | 0 | 1.434 | 18.549 | 0.799 | 20.124 | 35.639 | −6.67 | 37.78 | 3.6 |
| 3 | 13.031 | 36.751 | 10.887 | 91.877 | 12.119 | 99.36 | 2014.01 | 258.8 | |
| 2 | 0 | 2.239 | 5.195 | 0.983 | 49.559 | 93.621 | 2.05 | 80.46 | 1.49 |
| 3 | 0.384 | 21.944 | 4.731 | 75.926 | 42.004 | 94.92 | 845.2 | 34.81 | |
| 3 | 0 | 0.153 | 0.977 | 0.391 | 0.693 | 1.979 | 3.38 | 0.91 | 0.52 |
| 3 | 16.509 | 19.899 | 1.959 | 81.847 | 1.263 | 37.07 | 158.2 | 39.41 | |
| 4 | 0 | 0.126 | 4.534 | 0.424 | 16.791 | 1.891 | 2.01 | 0.01 | 0.34 |
| 3 | 10.17 | 13.711 | 1.685 | 62.309 | 0.866 | 5.07 | 66.89 | 7.12 | |
| 5 | 0 | 0.214 | 0.857 | 1.501 | 6.184 | 25.591 | 1.21 | 0.01 | 1.22 |
| 3 | 4.364 | 5.291 | 3.503 | 31.586 | 26.784 | 5.26 | 31.43 | 4.25 | |
| 6 | 0 | 0.295 | 6.772 | 0.469 | 0.784 | 3.793 | −1.22 | 2.5 | 0.23 |
| 3 | 3.045 | 13.996 | 2.777 | 76.984 | 6.099 | −26.45 | 468.64 | 0.2 | |
| 7 | 0 | 0.22 | 4.933 | 0.546 | 3.362 | 9.165 | 1.83 | 0.72 | 0.12 |
| 3 | 2.006 | 7.43 | 1.611 | 37.039 | 2.497 | 12.04 | 11.57 | 0.1 | |
| 8 | 0 | 0.161 | 2.737 | 0.382 | 5.921 | 5.293 | 7.24 | 0.07 | 2.45 |
| 3 | 6.624 | 12.456 | 1.465 | 52.134 | 0.889 | −6.91 | 43.82 | 3.575 | |
| 9 | 0 | 0.12 | 3.664 | 0.237 | 2.907 | 12.566 | −1.25 | 0.01 | 1.12 |
| 3 | 0.59 | 3.497 | 0.219 | 8.714 | 2.664 | 23.59 | 19.68 | 3.797 | |
| 10 | 0 | 0.07 | 3.209 | 0.497 | 0.486 | 7.663 | −12.19 | 0.08 | 0.17 |
| 3 | 8.034 | 10.476 | 1.369 | 65.884 | 1.248 | 8.6 | 125.56 | 2.713 | |
| 11 | 0 | 0.308 | 2.071 | 0.371 | 2.141 | 1.206 | 13.39 | 2.81 | 0.54 |
| 3 | 13.721 | 17.25 | 3.061 | 81.083 | 1.476 | −10.69 | 81.45 | 0.3323 | |
| 12 | 0 | 0.287 | 1.171 | 2.658 | 1.017 | 3.53 | −2.96 | 4.91 | 1.03 |
| 3 | 16.199 | 17.508 | 3.85 | 87.431 | 3.791 | −4.49 | 241.16 | 39.98 | |
| 13 | 0 | 0.414 | 3.224 | 0.38 | 0.929 | 1.836 | 0.01 | 0.91 | 0.06 |
| 3 | 4.931 | 11.493 | 1.107 | 76.989 | 0.716 | −14.85 | 226.12 | 4.169 | |
| 14 | 0 | 0.103 | 6.739 | 1.6 | 0.651 | 3.365 | 2.36 | 2.43 | 0.15 |
| 3 | 2.533 | 7.719 | 1.5 | 43.182 | 1.09 | −1.21 | 96.93 | 61.77 | |
| 15 | 0 | 0.136 | 2.887 | 1.739 | 1.5 | 1.004 | −17.01 | 1.39 | 2.34 |
| 3 | 3.139 | 6.521 | 0.626 | 44.147 | 0.431 | −2.66 | 23.53 | 2.67 | |
| 16 | 0 | 0.179 | 1.581 | 0.541 | 0.535 | 0.786 | −7.95 | 1.27 | 2.77 |
| 3 | 6.161 | 11.499 | 3.866 | 70.174 | 8.907 | −8.25 | 506.21 | 24.31 | |
| 17 | 0 | 4.492 | 6.83 | 2.311 | 43.009 | 19.826 | −13.41 | 78.81 | 2.58 |
| 3 | 26.607 | 29.214 | 4.489 | 90.084 | 6.7 | 35.58 | 631.3 | 41.34 | |
| 18 | 0 | 2.111 | 3.474 | 0.741 | 22.527 | 23.754 | 8.37 | 65.39 | 3.1 |
| 3 | 10.915 | 38.111 | 5.796 | 93.621 | 10.836 | 23.64 | 673.21 | 18.92 | |
| 19 | 0 | 0.311 | 1.259 | 0.3 | 0.466 | 0.374 | 7.24 | 13.14 | 6.87 |
| 3 | 12.446 | 35.571 | 15.249 | 78.687 | 11.299 | 99.6 | 780.4 | 188.4 | |
| 20 | 0 | 0.32 | 11.647 | 1.021 | 0.92 | 0.819 | 4.95 | 3.93 | 2.6 |
| 3 | 1.763 | 6.95 | 0.584 | 15.657 | 0.552 | −3.68 | 460.98 | 4.1 | |
| 21 | 0 | 0.35 | 46.049 | 25.096 | 68.013 | 8.907 | 98.49 | 1224.01 | 109.6 |
| 3 | 10.709 | 22.227 | 5.884 | 68.567 | 0.104 | 98.97 | 823.67 | 150.6 | |
| 22 | 0 | 0.385 | 1.607 | 0.281 | 45.468 | 0.828 | 4.46 | 0.08 | 0.5 |
| 3 | 1.5 | 4.581 | 0.3 | 52.691 | 0.834 | −3.33 | 1.09 | 4 |
| Peptide | Amino Acid Sequence | Molecular Weight [Da] | No. of Amino Acids |
|---|---|---|---|
| 1 | PLVSSQCVNLTTRTQLPPAYTNSFTRGVYY | 3377.8 | 30 |
| 2 | RGVYYPDKVFRSSVLHSTQDLFLPFFSNVT | 3520.9 | 30 |
| 3 | FSNVTWFHAIHVSGTNGTKRFDNPVLPFND | 3418.7 | 30 |
| 4 | LPFNDGVYFASTEKSNIIRGWIFGTTLDS | 3249.6 | 29 |
| 5 | TLDSKTQSLLIVNNATNVVIKVCEFQFCND | 3357.8 | 30 |
| 6 | QFCNDPFLGVYYHKNNKSWMESEFRVYSSA | 3648.0 | 30 |
| 7 | VYSSANNCTFEYVSQPFLMDLEGKQGNFKN | 3431.8 | 30 |
| 8 | GNFKNLREFVFKNIDGYFKIYSKHTPINLV | 3603.1 | 30 |
| 9 | PINLVRDLPQGFSALEPLVDLPIGINITR | 3171.7 | 29 |
| 10 | NITRFQTLLALHRSYLTPGDSSSGWTAGAA | 3192.5 | 30 |
| 11 | TAGAAAYYVGYLQPRTFLLKYNENGTITDA | 3282.6 | 30 |
| 12 | TITDAVDCALDPLSETKCTLKSFTVEKGIY | 3262.7 | 30 |
| 13 | EKGIYQTSNFRVQPTESIVRFPNITNLC | 3255.7 | 28 |
| 14 | FNATRFASVYAWNRKRISNCVADYS | 2940.2 | 25 |
| 15 | VADYSVLYNSASFSTFKCYGVSPTK | 2735.0 | 25 |
| 16 | VSPTKLNDLCFTNVYADSFVIRGDEVRQIA | 3371.8 | 30 |
| 17 | VRQIAPGQTGKIADYNYKLPDDFTGCVIAW | 3340.8 | 30 |
| 18 | CVIAWNSNNLDSKVGGNYNYLYRLFRKSNL | 3522.9 | 30 |
| 19 | KPFERDISTEIYQAGSTPCNGVEGF | 2746.0 | 25 |
| 20 | GVEGFNCYFPLQSYGFQPTNGVGYQPYRVV | 3387.7 | 30 |
| 21 | PYRVVVLSFELLHAPATVCGPKKSTNLVKN | 3281.9 | 30 |
| 22 | NLVKNKCVNFNFNGLTGTGVLTESNKKFL | 3200.7 | 29 |
| 23 | PFQQFGRDIADTTDAVRDPQTLEILDIT | 3176.4 | 28 |
| 24 | ILDITPCSFGGVSVITPGTNTSNQVAVLY | 2967.3 | 29 |
| 25 | AVLYQDVNCTEVPVAIHADQLTPTWRVYST | 3390.8 | 30 |
| 26 | RVYSTGSNVFQTRAGCLIGAEHVNNSYECD | 3291.5 | 30 |
| 27 | SYECDIPIGAGICASYQTQTNSPRRARSVA | 3215.5 | 30 |
| 28 | TMSLGAENSVAYSNNSIANNSIAIPTNFTI | 3115.4 | 30 |
| 30 | TSVDCTMYICGDSTECSNLLLQYGSFCTQL | 3296.7 | 30 |
| 31 | FCTQLNRALTGIAVEQDKNTQEVFAQVKQI | 3393.8 | 30 |
| 32 | QVKQIYKTPPIKDFGGFNFSQILPDPSK | 3193.6 | 28 |
| 33 | PDPSKPSKRSFIEDLLFNKVTLADAGFIKQ | 3362.8 | 30 |
| 34 | GFIKQYGDCLGDIAARDLICAQKFNGLTVL | 3243.7 | 30 |
| 35 | TDEMIAQYTSALLAGTITSGW | 2229.4 | 21 |
| 36 | ITSGWTFGAGAALQIPFAMQMAYRFNGIGV | 3176.7 | 30 |
| 37 | NGIGVTQNVLYENQKLIANQFNSAIGKIQD | 3290.6 | 30 |
| 38 | GKIQDSLSSTASALGKLQDVVNQNAQALNT | 3072.3 | 30 |
| 39 | LNTLVKQLSSNFGAISSVLNDILSRLDK | 3046.5 | 28 |
| 40 | LDKVEAEVQIDRLITGRLQSLQTYVTQQ | 3245.6 | 28 |
| 41 | YVTQQLIRAAEIRASANLAATKMSECVL | 3051.5 | 28 |
| 42 | CVLGQSKRVDFCGKGYHLMSFPQSAPH | 2993.4 | 27 |
| 43 | PHGVVFLHVTYVPAQEKNFTTAPAICHDGK | 3277.7 | 30 |
| 44 | CHDGKAHFPREGVFVSNGTHWFVTQRNFYE | 3566.9 | 30 |
| 45 | RNFYEPQIITTDNTFVSGNC | 2319.5 | 20 |
| 46 | NNTVYDPLQPELDSFKEELDKYFKNHT | 3285.5 | 27 |
| 47 | FKNHTSPDVDLGDISGINASVVNIQKEI | 3011.3 | 28 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Byazrova, M.; Gattinger, P.; Astakhova, E.; Hofer, G.; Khaitov, M.; Filatov, A.; Valenta, R. Dissection of Antibody Responses of Gam-COVID-Vac-Vaccinated Subjects Suggests Involvement of Epitopes Outside RBD in SARS-CoV-2 Neutralization. Int. J. Mol. Sci. 2023, 24, 5104. https://doi.org/10.3390/ijms24065104
Byazrova M, Gattinger P, Astakhova E, Hofer G, Khaitov M, Filatov A, Valenta R. Dissection of Antibody Responses of Gam-COVID-Vac-Vaccinated Subjects Suggests Involvement of Epitopes Outside RBD in SARS-CoV-2 Neutralization. International Journal of Molecular Sciences. 2023; 24(6):5104. https://doi.org/10.3390/ijms24065104
Chicago/Turabian StyleByazrova, Maria, Pia Gattinger, Ekaterina Astakhova, Gerhard Hofer, Musa Khaitov, Alexander Filatov, and Rudolf Valenta. 2023. "Dissection of Antibody Responses of Gam-COVID-Vac-Vaccinated Subjects Suggests Involvement of Epitopes Outside RBD in SARS-CoV-2 Neutralization" International Journal of Molecular Sciences 24, no. 6: 5104. https://doi.org/10.3390/ijms24065104
APA StyleByazrova, M., Gattinger, P., Astakhova, E., Hofer, G., Khaitov, M., Filatov, A., & Valenta, R. (2023). Dissection of Antibody Responses of Gam-COVID-Vac-Vaccinated Subjects Suggests Involvement of Epitopes Outside RBD in SARS-CoV-2 Neutralization. International Journal of Molecular Sciences, 24(6), 5104. https://doi.org/10.3390/ijms24065104






