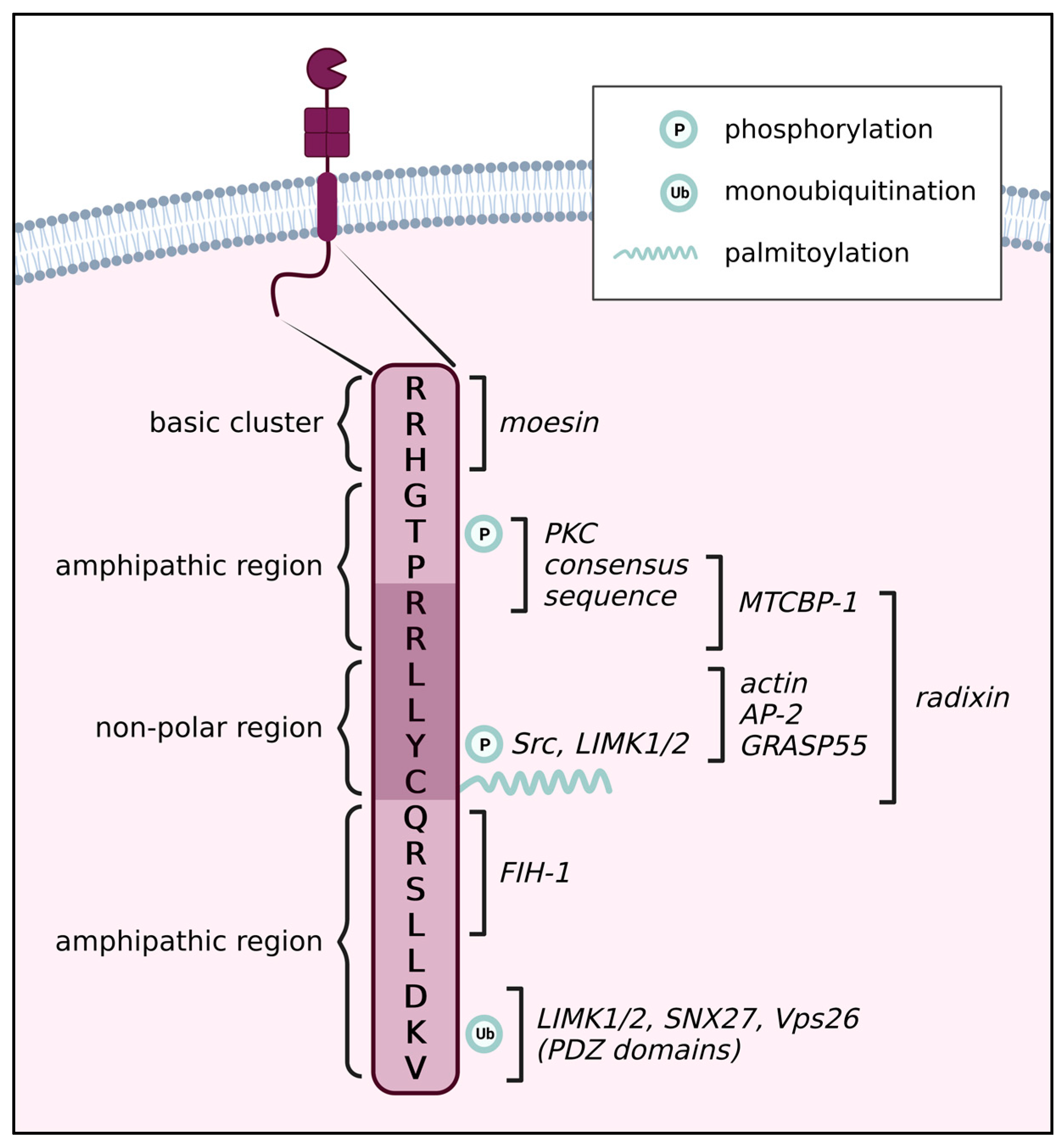
Cytoplasmic Tail of MT1-MMP: A Hub of MT1-MMP Regulation and Function
Abstract
1. Introduction
2. Post-Translational Modifications
2.1. The Role of the CT on the Regulation of O-Linked Glycosylation
2.2. The Role of the CT in MT1-MMP Activation via Prodomain Cleavage
2.3. The Role of the CT in MT1-MMP Autoprocessing
2.4. Palmitoylation of the CT
2.5. Phosphorylation of the CT
2.5.1. Thr567
2.5.2. Tyr573
2.6. Ubiquitination of the CT
3. The Role of the CT in Homodimerization
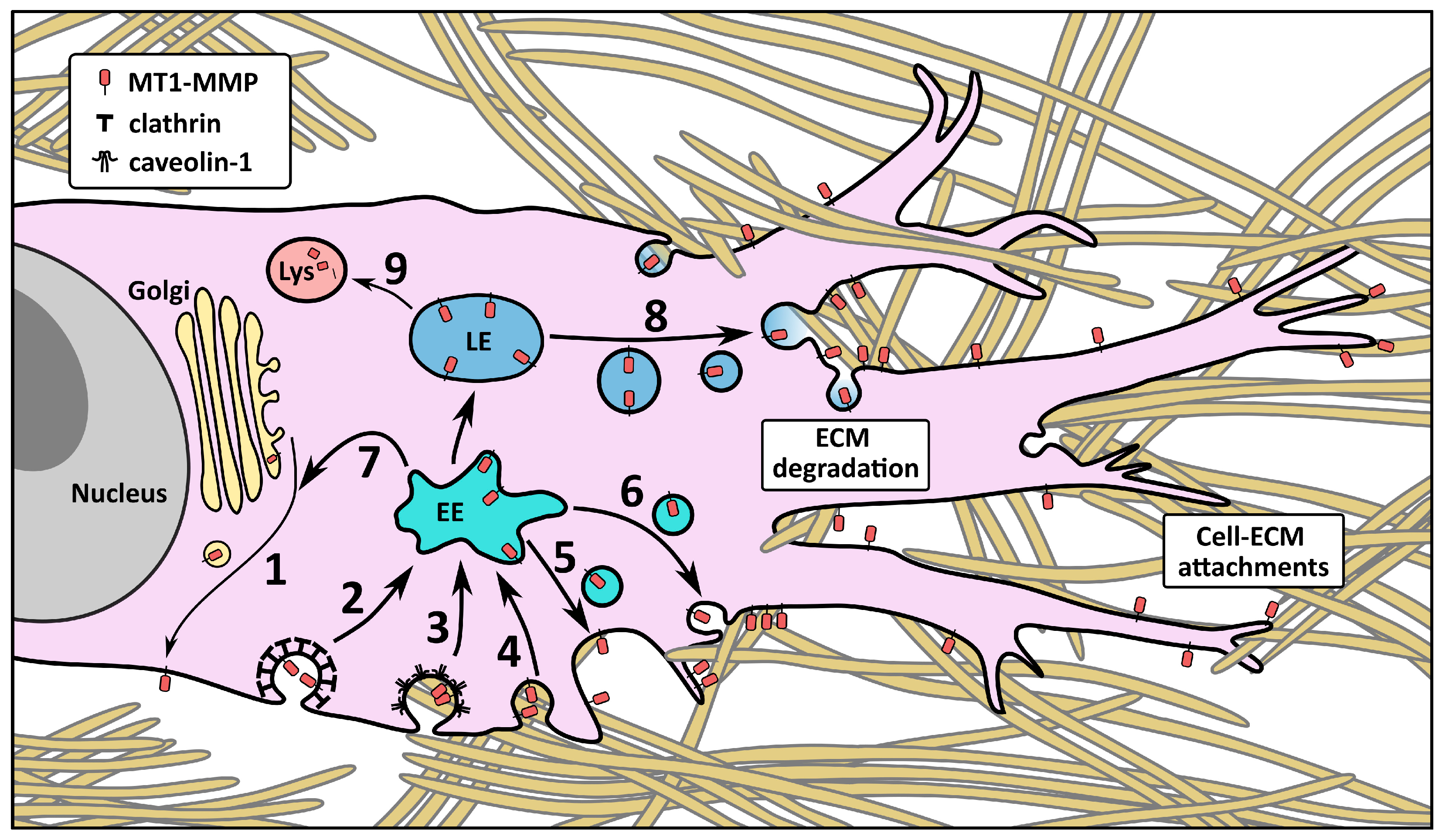
4. The Influence of the CT on MT1-MMP Trafficking and Localization
4.1. Membrane Localization and Endocytosis
4.2. Intracellular Trafficking and Recycling
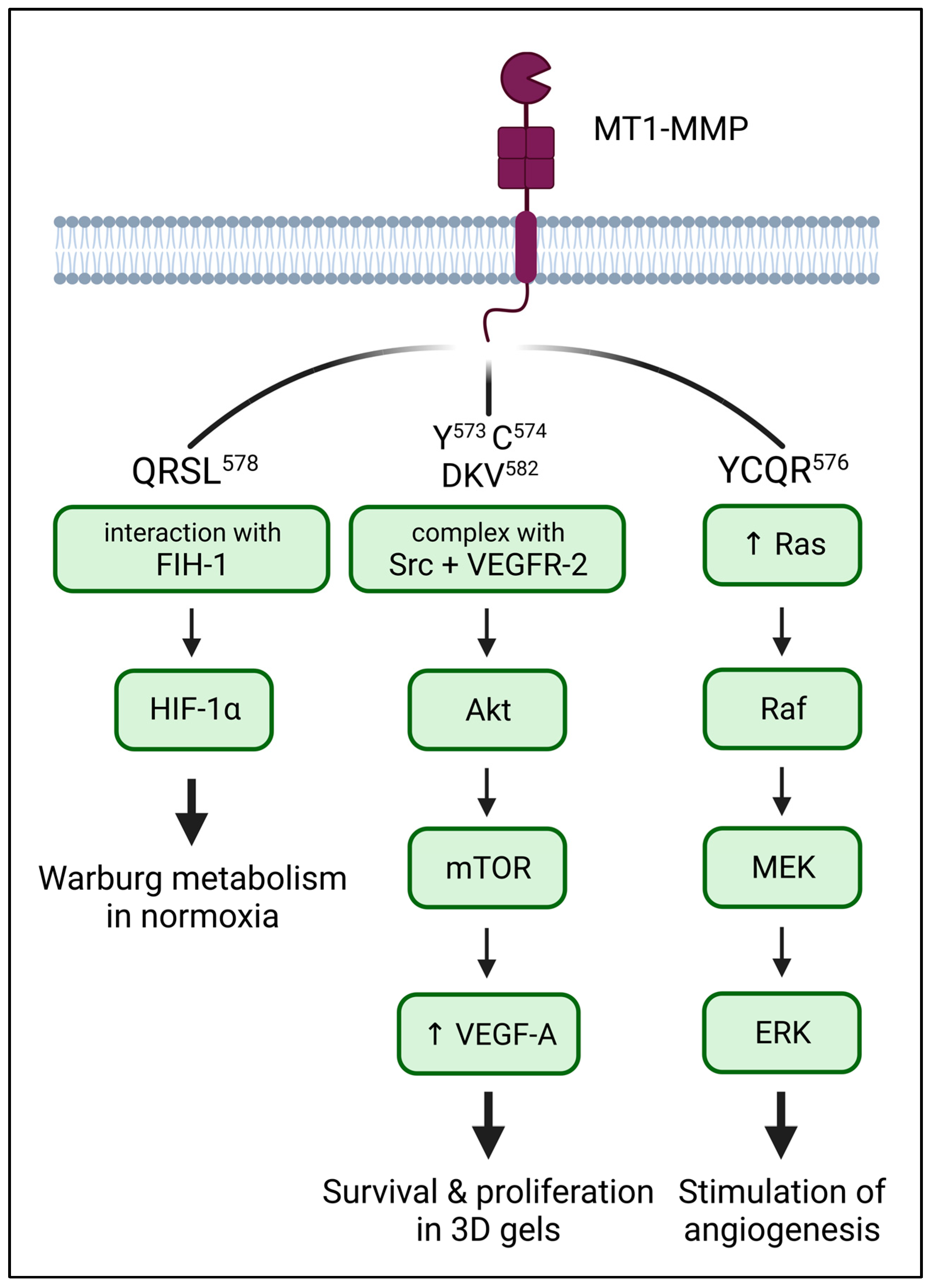
5. The Role of the CT in Downstream Signalling of MT1-MMP
5.1. HIF-1α: Metabolism
5.2. ERK: MAPK Signaling
5.3. VEGF: Stimulation of Angiogenesis
6. The Importance of the CT for Cell Adhesion
7. Regulation of Invasiveness through the CT
8. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Linder, S.; Wiesner, C.; Himmel, M. Degrading Devices: Invadosomes in Proteolytic Cell Invasion. Annu. Rev. Cell Dev. Biol. 2011, 27, 185–211. [Google Scholar] [CrossRef] [PubMed]
- Gimona, M.; Buccione, R. Adhesions That Mediate Invasion. Int. J. Biochem. Cell Biol. 2006, 38, 1875–1892. [Google Scholar] [CrossRef] [PubMed]
- Lizárraga, F.; Poincloux, R.; Romão, M.; Montagnac, G.; le Dez, G.; Bonne, I.; Rigaill, G.; Raposo, G.; Chavrier, P. Diaphanous-Related Formins Are Required for Invadopodia Formation and Invasion of Breast Tumor Cells. Cancer Res. 2009, 69, 2792–2800. [Google Scholar] [CrossRef]
- Tolde, O.; Rösel, D.; Veselý, P.; Folk, P.; Brábek, J. The Structure of Invadopodia in a Complex 3D Environment. Eur. J. Cell Biol. 2010, 89, 674–680. [Google Scholar] [CrossRef] [PubMed]
- Artym, V.V.; Swatkoski, S.; Matsumoto, K.; Campbell, C.B.; Petrie, R.J.; Dimitriadis, E.K.; Li, X.; Mueller, S.C.; Bugge, T.H.; Gucek, M.; et al. Dense Fibrillar Collagen Is a Potent Inducer of Invadopodia via a Specific Signaling Network. J. Cell Biol. 2015, 208, 331–350. [Google Scholar] [CrossRef] [PubMed]
- Dalecká, M.; Sabó, J.; Backová, L.; Rösel, D.; Brábek, J.; Benda, A.; Tolde, O. Invadopodia Structure in 3D Environment Resolved by Near-Infrared Branding Protocol Combining Correlative Confocal and FIB-SEM Microscopy. Int. J. Mol. Sci. 2021, 22, 7805. [Google Scholar] [CrossRef]
- Artym, V.V.; Zhang, Y.; Seillier-Moiseiwitsch, F.; Yamada, K.M.; Mueller, S.C. Dynamic Interactions of Cortactin and Membrane Type 1 Matrix Metalloproteinase at Invadopodia: Defining the Stages of Invadopodia Formation and Function. Cancer Res. 2006, 66, 3034–3043. [Google Scholar] [CrossRef]
- Pedersen, N.M.; Wenzel, E.M.; Wang, L.; Antoine, S.; Chavrier, P.; Stenmark, H.; Raiborg, C. Protrudin-Mediated ER-Endosome Contact Sites Promote MT1-MMP Exocytosis and Cell Invasion. J. Cell Biol. 2020, 219, e202003063. [Google Scholar] [CrossRef] [PubMed]
- Nagase, H.; Visse, R.; Murphy, G. Structure and Function of Matrix Metalloproteinases and TIMPs. Cardiovasc. Res. 2006, 69, 562–573. [Google Scholar] [CrossRef]
- Marcink, T.C.; Simoncic, J.A.; An, B.; Knapinska, A.M.; Fulcher, Y.G.; Akkaladevi, N.; Fields, G.B.; van Doren, S.R. MT1-MMP Binds Membranes by Opposite Tips of Its β Propeller to Position It for Pericellular Proteolysis. Structure 2019, 27, 281–292.e6. [Google Scholar] [CrossRef]
- Cao, J.; Sato, H.; Takino, T.; Seiki, M. The C-Terminal Region of Membrane Type Matrix Metalloproteinase Is a Functional Transmembrane Domain Required for pro-Gelatinase A Activation. J. Biol. Chem. 1995, 270, 801–805. [Google Scholar] [CrossRef] [PubMed]
- Sato, H.; Takino, T.; Okada, Y.; Cao, J.; Shinagawa, A.; Yamamoto, E.; Seiki, M. A Matrix Metalloproteinase Expressed on the Surface of Invasive Cancer Cells. Nature 1994, 370, 61–65. [Google Scholar] [CrossRef] [PubMed]
- Gifford, V.; Itoh, Y. MT1-MMP-Dependent Cell Migration: Proteolytic and Non-Proteolytic Mechanisms. Biochem. Soc. Trans. 2019, 47, 811–826. [Google Scholar] [CrossRef] [PubMed]
- Knapinska, A.M.; Fields, G.B. The Expanding Role of MT1-MMP in Cancer Progression. Pharmaceuticals 2019, 12, 77. [Google Scholar] [CrossRef]
- Itoh, Y. Membrane-Type Matrix Metalloproteinases: Their Functions and Regulations. Matrix Biol. 2015, 44–46, 207–223. [Google Scholar] [CrossRef] [PubMed]
- Terawaki, S.; Kitano, K.; Aoyama, M.; Mori, T.; Hakoshima, T. MT1-MMP Recognition by ERM Proteins and Its Implication in CD44 Shedding. Genes Cells 2015, 20, 847–859. [Google Scholar] [CrossRef]
- Wu, Y.I.; Munshi, H.G.; Sen, R.; Snipas, S.J.; Salvesen, G.S.; Fridman, R.; Stack, M.S. Glycosylation Broadens the Substrate Profile of Membrane Type 1 Matrix Metalloproteinase. J. Biol. Chem. 2004, 279, 8278–8289. [Google Scholar] [CrossRef]
- Ludwig, T.; Theissen, S.M.; Morton, M.J.; Caplan, M.J. The Cytoplasmic Tail Dileucine Motif LL572 Determines the Glycosylation Pattern of Membrane-Type 1 Matrix Metalloproteinase. J. Biol. Chem. 2008, 283, 35410–35418. [Google Scholar] [CrossRef] [PubMed]
- Roghi, C.; Jones, L.; Gratian, M.; English, W.R.; Murphy, G. Golgi Reassembly Stacking Protein 55 Interacts with Membrane-Type (MT) 1-Matrix Metalloprotease (MMP) and Furin and Plays a Role in the Activation of the MT1-MMP Zymogen. FEBS J. 2010, 277, 3158–3175. [Google Scholar] [CrossRef]
- Xiang, Y.; Zhang, X.; Nix, D.B.; Katoh, T.; Aoki, K.; Tiemeyer, M.; Wang, Y. Regulation of Protein Glycosylation and Sorting by the Golgi Matrix Proteins GRASP55/65. Nat. Commun. 2013, 4, 1659. [Google Scholar] [CrossRef]
- Pothukuchi, P.; Agliarulo, I.; Pirozzi, M.; Rizzo, R.; Russo, D.; Turacchio, G.; Nüchel, J.; Yang, J.; Gehin, C.; Capolupo, L.; et al. GRASP55 Regulates Intra-Golgi Localization of Glycosylation Enzymes to Control Glycosphingolipid Biosynthesis. EMBO J. 2021, 40, e107766. [Google Scholar] [CrossRef]
- Stein, M.F.; Blume, K.; Heilingloh, C.S.; Kummer, M.; Biesinger, B.; Sticht, H.; Steinkasserer, A. CD83 and GRASP55 Interact in Human Dendritic Cells. Biochem. Biophys. Res. Commun. 2015, 459, 42–48. [Google Scholar] [CrossRef] [PubMed]
- Yana, I.; Weiss, S.J. Regulation of Membrane Type-1 Matrix Metalloproteinase Activity by Proprotein Convertases. Mol. Biol. Cell 2000, 11, 2387–2401. [Google Scholar] [CrossRef] [PubMed]
- Remacle, A.G.; Rozanov, D.V.; Fugere, M.; Day, R.; Strongin, A.Y. Furin Regulates the Intracellular Activation and the Uptake Rate of Cell Surface-Associated MT1-MMP. Oncogene 2006, 25, 5648–5655. [Google Scholar] [CrossRef]
- Golubkov, V.S.; Cieplak, P.; Chekanov, A.V.; Ratnikov, B.I.; Aleshin, A.E.; Golubkova, N.V.; Postnova, T.I.; Radichev, I.A.; Rozanov, D.V.; Zhu, W.; et al. Internal Cleavages of the Autoinhibitory Prodomain Are Required for Membrane Type 1 Matrix Metalloproteinase Activation, Although Furin Cleavage Alone Generates Inactive Proteinase. J. Biol. Chem. 2010, 285, 27726–27736. [Google Scholar] [CrossRef] [PubMed]
- Cepeda, M.A.; Pelling, J.J.; Evered, C.L.; Leong, H.S.; Damjanovski, S. The Cytoplasmic Domain of MT1-MMP Is Dispensable for Migration Augmentation but Necessary to Mediate Viability of MCF-7 Breast Cancer Cells. Exp. Cell Res. 2017, 350, 169–183. [Google Scholar] [CrossRef] [PubMed]
- Lehti, K.; Lohi, J.; Valtanen, H.; Keski-Oja, J. Proteolytic Processing of Membrane-Type-1 Matrix Metalloproteinase Is Associated with Gelatinase A Activation at the Cell Surface. Biochem. J. 1998, 334, 345–353. [Google Scholar] [CrossRef]
- Stanton, H.; Gavrilovic, J.; Atkinson, S.J.; D’Ortho, M.P.; Yamada, K.M.; Zardi, L.; Murphy, G. The Activation of ProMMP-2 (Gelatinase A) by HT1080 Fibrosarcoma Cells Is Promoted by Culture on a Fibronectin Substrate and Is Concomitant with an Increase in Processing of MT1-MMP (MMP-14) to a 45 KDa Form. J. Cell Sci. 1998, 111, 2789–2798. [Google Scholar] [CrossRef]
- Annabi, B.; Lachambre, M.; Bousquet-Gagnon, N.P.; Pagé, M.; Gingras, D.; Béliveau, R. Localization of Membrane-Type 1 Matrix Metalloproteinase in Caveolae Membrane Domains. Biochem. J. 2001, 353, 547–553. [Google Scholar] [CrossRef]
- Toth, M.; Hernandez-Barrantes, S.; Osenkowski, P.; Margarida Bernardo, M.; Gervasi, D.C.; Shimura, Y.; Meroueh, O.; Kotra, L.P.; Gálvez, B.G.; Arroyo, A.G.; et al. Complex Pattern of Membrane Type 1 Matrix Metalloproteinase Shedding. Regulation by Autocatalytic Cells Surface Inactivation of Active Enzyme. J. Biol. Chem. 2002, 277, 26340–26350. [Google Scholar] [CrossRef]
- Lehti, K.; Lohi, J.; Juntunen, M.M.; Pei, D.; Keski-Oja, J. Oligomerization through Hemopexin and Cytoplasmic Domains Regulates the Activity and Turnover of Membrane-Type 1 Matrix Metalloproteinase. J. Biol. Chem. 2002, 277, 8440–8448. [Google Scholar] [CrossRef]
- Fehon, R.G.; McClatchey, A.I.; Bretscher, A. Organizing the Cell Cortex: The Role of ERM Proteins. Nat. Rev. Mol. Cell Biol. 2010, 11, 276–287. [Google Scholar] [CrossRef] [PubMed]
- Suárez, H.; López-Martín, S.; Toribio, V.; Zamai, M.; Hernández-Riquer, M.V.; Genís, L.; Arroyo, A.G.; Yáñez-Mó, M. Regulation of MT1-MMP Activity through Its Association with ERMs. Cells 2020, 9, 348. [Google Scholar] [CrossRef] [PubMed]
- Song, I.W.; Li, W.R.; Chen, L.Y.; Shen, L.F.; Liu, K.M.; Yen, J.J.Y.; Chen, Y.J.; Chen, Y.J.; Kraus, V.B.; Wu, J.Y.; et al. Palmitoyl Acyltransferase, Zdhhc13, Facilitates Bone Mass Acquisition by Regulating Postnatal Epiphyseal Development and Endochondral Ossification: A Mouse Model. PLoS ONE 2014, 9, e92194. [Google Scholar] [CrossRef] [PubMed]
- Anilkumar, N.; Uekita, T.; Couchman, J.R.; Nagase, H.; Seiki, M.; Itoh, Y. Palmitoylation at Cys 574 Is Essential for MT1-MMP to Promote Cell Migration. FASEB J. 2005, 19, 1326–1328. [Google Scholar] [CrossRef] [PubMed]
- Moss, N.M.; Wu, Y.I.; Liu, Y.; Munshi, H.G.; Stack, M.S. Modulation of the Membrane Type 1 Matrix Metalloproteinase Cytoplasmic Tail Enhances Tumor Cell Invasion and Proliferation in Three-Dimensional Collagen Matrices. J. Biol. Chem. 2009, 284, 19791–19799. [Google Scholar] [CrossRef]
- Williams, K.C.; Coppolino, M.G. Phosphorylation of Membrane Type 1-Matrix Metalloproteinase (MT1-MMP) and Its Vesicle-Associated Membrane Protein 7 (VAMP7)-Dependent Trafficking Facilitate Cell Invasion and Migration. J. Biol. Chem. 2011, 286, 43405–43416. [Google Scholar] [CrossRef]
- Rosse, C.; Lodillinsky, C.; Fuhrmann, L.; Nourieh, M.; Monteiro, P.; Irondelle, M.; Lagoutte, E.; Vacher, S.; Waharte, F.; Paul-Gilloteaux, P.; et al. Control of MT1-MMP Transport by Atypical PKC during Breast-Cancer Progression. Proc. Natl. Acad. Sci. USA 2014, 111, E1872–E1879. [Google Scholar] [CrossRef]
- Moro, L.; Dolce, L.; Cabodi, S.; Bergatto, E.; Erba, E.B.; Smeriglio, M.; Turco, E.; Retta, S.F.; Giuffrida, M.G.; Venturino, M.; et al. Integrin-Induced Epidermal Growth Factor (EGF) Receptor Activation Requires c-Src and P130Cas and Leads to Phosphorylation of Specific EGF Receptor Tyrosines. J. Biol. Chem. 2002, 277, 9405–9414. [Google Scholar] [CrossRef]
- Grafinger, O.R.; Gorshtein, G.; Stirling, T.; Brasher, M.I.; Coppolino, M.G. Β1 Integrin-Mediated Signaling Regulates MT1-MMP Phosphorylation to Promote Tumor Cell Invasion. J. Cell Sci. 2020, 133, jcs239152. [Google Scholar] [CrossRef]
- Zhang, X.A.; Bontrager, A.L.; Hemler, M.E. Transmembrane-4 Superfamily Proteins Associate with Activated Protein Kinase C (PKC) and Link PKC to Specific Beta(1) Integrins. J. Biol. Chem. 2001, 276, 25005–25013. [Google Scholar] [CrossRef] [PubMed]
- Termini, C.M.; Gillette, J.M. Tetraspanins Function as Regulators of Cellular Signaling. Front. Cell Dev. Biol. 2017, 5, 34. [Google Scholar] [CrossRef] [PubMed]
- Tavsan, Z.; Kayali, H.A. Protein Kinase C Regulates the Complex between Cell Membrane Molecules in Ovarian Cancer. Process Biochem. 2020, 92, 182–189. [Google Scholar] [CrossRef]
- Schröder, H.M.; Hoffmann, S.C.; Hecker, M.; Korff, T.; Ludwig, T. The Tetraspanin Network Modulates MT1-MMP Cell Surface Trafficking. Int. J. Biochem. Cell Biol. 2013, 45, 1133–1144. [Google Scholar] [CrossRef] [PubMed]
- Yañez-Mó, M.; Barreiro, O.; Gonzalo, P.; Batista, A.; Megías, D.; Genís, L.; Sachs, N.; Sala-Valdés, M.; Alonso, M.A.; Montoya, M.C.; et al. MT1-MMP Collagenolytic Activity Is Regulated through Association with Tetraspanin CD151 in Primary Endothelial Cells. Blood 2008, 112, 3217–3226. [Google Scholar] [CrossRef] [PubMed]
- Nyalendo, C.; Michaud, M.; Beaulieu, E.; Roghi, C.; Murphy, G.; Gingras, D.; Béliveau, R. Src-Dependent Phosphorylation of Membrane Type I Matrix Metalloproteinase on Cytoplasmic Tyrosine 573: ROLE IN ENDOTHELIAL AND TUMOR CELL MIGRATION. J. Biol. Chem. 2007, 282, 15690–15699. [Google Scholar] [CrossRef]
- Moss, N.M.; Liu, Y.; Johnson, J.J.; Debiase, P.; Jones, J.; Hudson, L.G.; Munshi, H.G.; Stack, M.S. Epidermal Growth Factor Receptor-Mediated Membrane Type 1 Matrix Metalloproteinase Endocytosis Regulates the Transition between Invasive versus Expansive Growth of Ovarian Carcinoma Cells in Three-Dimensional Collagen. Mol. Cancer Res. 2009, 7, 809–820. [Google Scholar] [CrossRef]
- Lagoutte, E.; Villeneuve, C.; Lafanechère, L.; Wells, C.M.; Jones, G.E.; Chavrier, P.; Rossé, C. LIMK Regulates Tumor-Cell Invasion and Matrix Degradation Through Tyrosine Phosphorylation of MT1-MMP. Sci. Rep. 2016, 6, 24925. [Google Scholar] [CrossRef]
- Gingras, D.; Michaud, M.; di Tomasso, G.; Béliveau, E.; Nyalendo, C.; Béliveau, R. Sphingosine-1-Phosphate Induces the Association of Membrane-Type 1 Matrix Metalloproteinase with P130Cas in Endothelial Cells. FEBS Lett. 2008, 582, 399–404. [Google Scholar] [CrossRef]
- Komander, D.; Rape, M. The Ubiquitin Code. Annu. Rev. Biochem. 2012, 81, 203–229. [Google Scholar] [CrossRef]
- Eisenach, P.A.; de Sampaio, P.C.; Murphy, G.; Roghi, C. Membrane Type 1 Matrix Metalloproteinase (MT1-MMP) Ubiquitination at Lys581 Increases Cellular Invasion through Type I Collagen. J. Biol. Chem. 2012, 287, 11533–11545. [Google Scholar] [CrossRef] [PubMed]
- Rozanov, D.V.; Deryugina, E.I.; Ratnikov, B.I.; Monosov, E.Z.; Marchenko, G.N.; Quigley, J.P.; Strongin, A.Y. Mutation Analysis of Membrane Type-1 Matrix Metalloproteinase (MT1-MMP): The Role of the Cytoplasmic Tail Cys574, the Active Site Glu 240, and Furin Cleavage Motifs in Oligomerization, Processing, and Self-Proteolysis of MT1-MMP Expressed in Breast Carcin. J. Biol. Chem. 2001, 276, 25705–25714. [Google Scholar] [CrossRef] [PubMed]
- Itoh, Y.; Ito, N.; Nagase, H.; Evans, R.D.; Bird, S.A.; Seiki, M. Cell Surface Collagenolysis Requires Homodimerization of the Membrane-Bound Collagenase MT1-MMP. Mol. Biol. Cell 2006, 17, 5390–5399. [Google Scholar] [CrossRef]
- Itoh, Y.; Palmisano, R.; Anilkumar, N.; Nagase, H.; Miyawaki, A.; Seiki, M. Dimerization of MT1-MMP during Cellular Invasion Detected by Fluorescence Resonance Energy Transfer. Biochem. J. 2011, 440, 319–326. [Google Scholar] [CrossRef]
- Itoh, Y.; Takamura, A.; Ito, N.; Maru, Y.; Sato, H.; Suenaga, N.; Aoki, T.; Seiki, M. Homophilic Complex Formation of MT1-MMP Facilitates ProMMP-2 Activation on the Cell Surface and Promotes Tumor Cell Invasion. EMBO J. 2001, 20, 4782. [Google Scholar] [CrossRef] [PubMed]
- Itoh, Y.; Ito, N.; Nagase, H.; Seiki, M. The Second Dimer Interface of MT1-MMP, the Transmembrane Domain, Is Essential for ProMMP-2 Activation on the Cell Surface. J. Biol. Chem. 2008, 283, 13053. [Google Scholar] [CrossRef] [PubMed]
- Tochowicz, A.; Goettig, P.; Evans, R.; Visse, R.; Shitomi, Y.; Palmisano, R.; Ito, N.; Richter, K.; Maskos, K.; Franke, D.; et al. The Dimer Interface of the Membrane Type 1 Matrix Metalloproteinase Hemopexin Domain: Crystal Structure and Biological Functions. J. Biol. Chem. 2011, 286, 7587–7600. [Google Scholar] [CrossRef]
- Fogarasi, M.; Dima, S. The Catalytic Domain Mediates Homomultimerization of MT1-MMP and the Prodomain Interferes with MT1-MMP Oligomeric Complex Assembly. Biomolecules 2022, 12, 1145. [Google Scholar] [CrossRef]
- Uekita, T.; Itoh, Y.; Yana, I.; Ohno, H.; Seiki, M. Cytoplasmic Tail-Dependent Internalization of Membrane-Type 1 Matrix Metalloproteinase Is Important for Its Invasion-Promoting Activity. J. Cell Biol. 2001, 155, 1345–1356. [Google Scholar] [CrossRef] [PubMed]
- Planchon, D.; Rios Morris, E.; Genest, M.; Comunale, F.; Vacher, S.; Bièche, I.; Denisov, E.V.; Tashireva, L.A.; Perelmuter, V.M.; Linder, S.; et al. MT1-MMP Targeting to Endolysosomes Is Mediated by Upregulation of Flotillins. J. Cell Sci. 2018, 131, jcs218925. [Google Scholar] [CrossRef]
- Remacle, A.; Murphy, G.; Roghi, C. Membrane Type I-Matrix Metalloproteinase (MT1-MMP) Is Internalised by Two Different Pathways and Is Recycled to the Cell Surface. J. Cell Sci. 2003, 116, 3905–3916. [Google Scholar] [CrossRef] [PubMed]
- Yamaguchi, H.; Takeo, Y.; Yoshida, S.; Kouchi, Z.; Nakamura, Y.; Fukami, K. Lipid Rafts and Caveolin-1 Are Required for Invadopodia Formation and Extracellular Matrix Degradation by Human Breast Cancer Cells. Cancer Res. 2009, 69, 8594–8602. [Google Scholar] [CrossRef] [PubMed]
- Poincloux, R.; Lizarraga, F.; Chavrier, P. Matrix Invasion by Tumour Cells: A Focus on MT1-MMP Trafficking to Invadopodia. J. Cell Sci. 2009, 122, 3015–3024. [Google Scholar] [CrossRef] [PubMed]
- Wolf, K.; Friedl, P. Functional Imaging of Pericellular Proteolysis in Cancer Cell Invasion. Biochimie 2005, 87, 315–320. [Google Scholar] [CrossRef]
- Jiang, A.; Lehti, K.; Wang, X.; Weiss, S.J.; Keski-Oja, J.; Pei, D. Regulation of Membrane-Type Matrix Metalloproteinase 1 Activity by Dynamin-Mediated Endocytosis. Proc. Natl. Acad. Sci. USA 2001, 98, 13693–13698. [Google Scholar] [CrossRef]
- Rozanov, D.V.; Deryugina, E.I.; Monosov, E.Z.; Marchenko, N.D.; Strongin, A.Y. Aberrant, Persistent Inclusion into Lipid Rafts Limits the Tumorigenic Function of Membrane Type-1 Matrix Metalloproteinase in Malignant Cells. Exp. Cell Res. 2004, 293, 81–95. [Google Scholar] [CrossRef]
- Kaksonen, M.; Roux, A. Mechanisms of Clathrin-Mediated Endocytosis. Nat. Rev. Mol. Cell Biol. 2018, 19, 313–326. [Google Scholar] [CrossRef]
- Matthaeus, C.; Taraska, J.W. Energy and Dynamics of Caveolae Trafficking. Front. Cell Dev. Biol. 2021, 8, 1842. [Google Scholar] [CrossRef]
- Cheng, J.P.X.; Nichols, B.J. Caveolae: One Function or Many? Trends Cell Biol. 2016, 26, 177–189. [Google Scholar] [CrossRef]
- Atkinson, S.J.; English, J.L.; Holway, N.; Murphy, G. Cellular Cholesterol Regulates MT1 MMP Dependent Activation of MMP 2 via MEK-1 in HT1080 Fibrosarcoma Cells. FEBS Lett. 2004, 566, 65–70. [Google Scholar] [CrossRef]
- Yang, H.; Guan, L.; Li, S.; Jiang, Y.; Xiong, N.; Li, L.; Wu, C.; Zeng, H.; Liu, Y. Mechanosensitive Caveolin-1 Activation-Induced PI3K/Akt/MTOR Signaling Pathway Promotes Breast Cancer Motility, Invadopodia Formation and Metastasis In Vivo. Oncotarget 2016, 7, 16227–16247. [Google Scholar] [CrossRef]
- Gálvez, B.G.; Matías-Román, S.; Yáñez-Mó, M.; Vicente-Manzanares, M.; Sánchez-Madrid, F.; Arroyo, A.G. Caveolae Are a Novel Pathway for Membrane-Type 1 Matrix Metalloproteinase Traffic in Human Endothelial Cells. Mol. Biol. Cell 2004, 15, 678–687. [Google Scholar] [CrossRef] [PubMed]
- Gingras, D.; Béliveau, R. Emerging Concepts in the Regulation of Membrane-Type 1 Matrix Metalloproteinase Activity. Biochim. Biophys. Acta (BBA)-Mol. Cell Res. 2010, 1803, 142–150. [Google Scholar] [CrossRef]
- Couet, J.; Li, S.; Okamoto, T.; Ikezu, T.; Lisanti, M.P. Identification of Peptide and Protein Ligands for the Caveolin-Scaffolding Domain: IMPLICATIONS FOR THE INTERACTION OF CAVEOLIN WITH CAVEOLAE-ASSOCIATED PROTEINS. J. Biol. Chem. 1997, 272, 6525–6533. [Google Scholar] [CrossRef] [PubMed]
- Collins, B.M.; Davis, M.J.; Hancock, J.F.; Parton, R.G. Structure-Based Reassessment of the Caveolin Signaling Model: Do Caveolae Regulate Signaling Through Caveolin-Protein Interactions? Dev. Cell 2012, 23, 11. [Google Scholar] [CrossRef]
- Labrecque, L.; Nyalendo, C.; Langlois, S.; Durocher, Y.; Roghi, C.; Murphy, G.; Gingras, D.; Béliveau, R. Src-Mediated Tyrosine Phosphorylation of Caveolin-1 Induces Its Association with Membrane Type 1 Matrix Metalloproteinase. J. Biol. Chem. 2004, 279, 52132–52140. [Google Scholar] [CrossRef]
- Gauthier-Rouvière, C.; Bodin, S.; Comunale, F.; Planchon, D. Flotillin Membrane Domains in Cancer. Cancer Metastasis Rev. 2020, 39, 361–374. [Google Scholar] [CrossRef] [PubMed]
- Wiesner, C.; el Azzouzi, K.; Linder, S. A Specific Subset of RabGTPases Controls Cell Surface Exposure of MT1-MMP, Extracellular Matrix Degradation and Three-Dimensional Invasion of Macrophages. J. Cell Sci. 2013, 126, 2820–2833. [Google Scholar] [CrossRef]
- Frittoli, E.; Palamidessi, A.; Marighetti, P.; Confalonieri, S.; Bianchi, F.; Malinverno, C.; Mazzaro, G.; Viale, G.; Martin-Padura, G.; Garré, M.; et al. A RAB5/RAB4 Recycling Circuitry Induces a Proteolytic Invasive Program and Promotes Tumor Dissemination. J. Cell Biol. 2014, 206, 307–328. [Google Scholar] [CrossRef]
- Colombero, C.; Remy, D.; Antoine-Bally, S.; Macé, A.S.; Monteiro, P.; ElKhatib, N.; Fournier, M.; Dahmani, A.; Montaudon, E.; Montagnac, G.; et al. MTOR Repression in Response to Amino Acid Starvation Promotes ECM Degradation Through MT1-MMP Endocytosis Arrest. Adv. Sci. 2021, 8, e2101614. [Google Scholar] [CrossRef]
- Loskutov, Y.V.; Kozyulina, P.Y.; Kozyreva, V.K.; Ice, R.J.; Jones, B.C.; Roston, T.J.; Smolkin, M.B.; Ivanov, A.V.; Wysolmerski, R.B.; Pugacheva, E.N. NEDD9/Arf6-Dependent Endocytic Trafficking of Matrix Metalloproteinase 14: A Novel Mechanism for Blocking Mesenchymal Cell Invasion and Metastasis of Breast Cancer. Oncogene 2015, 34, 3662–3675. [Google Scholar] [CrossRef] [PubMed]
- Chevalier, C.; Collin, G.; Descamps, S.; Touaitahuata, H.; Simon, V.; Reymond, N.; Fernandez, L.; Milhiet, P.E.; Georget, V.; Urbach, S.; et al. TOM1L1 Drives Membrane Delivery of MT1-MMP to Promote ERBB2-Induced Breast Cancer Cell Invasion. Nat. Commun. 2016, 7, 10765. [Google Scholar] [CrossRef] [PubMed]
- Steffen, A.; le Dez, G.; Poincloux, R.; Recchi, C.; Nassoy, P.; Rottner, K.; Galli, T.; Chavrier, P. MT1-MMP-Dependent Invasion Is Regulated by TI-VAMP/VAMP7. Curr. Biol. 2008, 18, 926–931. [Google Scholar] [CrossRef] [PubMed]
- Williams, K.C.; McNeilly, R.E.; Coppolino, M.G. SNAP23, Syntaxin4, and Vesicle-Associated Membrane Protein 7 (VAMP7) Mediate Trafficking of Membrane Type 1-Matrix Metalloproteinase (MT1-MMP) during Invadopodium Formation and Tumor Cell Invasion. Mol. Biol. Cell 2014, 25, 2061–2070. [Google Scholar] [CrossRef] [PubMed]
- Miyagawa, T.; Hasegawa, K.; Aoki, Y.; Watanabe, T.; Otagiri, Y.; Arasaki, K.; Wakana, Y.; Asano, K.; Tanaka, M.; Yamaguchi, H.; et al. MT1-MMP Recruits the ER-Golgi SNARE Bet1 for EfficientMT1-MMP Transport to the Plasmamembrane. J. Cell Biol. 2019, 218, 3355–3371. [Google Scholar] [CrossRef]
- Wang, X.; Ma, D.; Keski-Oja, J.; Pei, D. Co-Recycling of MT1-MMP and MT3-MMP through the Trans-Golgi Network: Identification of DKV582 as a Recycling Signal. J. Biol. Chem. 2004, 279, 9331–9336. [Google Scholar] [CrossRef]
- Bravo-Cordero, J.J.; Marrero-Diaz, R.; Megías, D.; Genís, L.; García-Grande, A.; García, M.A.; Arroyo, A.G.; Montoya, M.C. MT1-MMP Proinvasive Activity Is Regulated by a Novel Rab8-Dependent Exocytic Pathway. EMBO J. 2007, 26, 1499–1510. [Google Scholar] [CrossRef]
- Castro-Castro, A.; Marchesin, V.; Monteiro, P.; Lodillinsky, C.; Rossé, C.; Chavrier, P. Cellular and Molecular Mechanisms of MT1-MMP-Dependent Cancer Cell Invasion. Annu. Rev. Cell Dev. Biol. 2016, 32, 555–576. [Google Scholar] [CrossRef]
- Marchesin, V.; Castro-Castro, A.; Lodillinsky, C.; Castagnino, A.; Cyrta, J.; Bonsang-Kitzis, H.; Fuhrmann, L.; Irondelle, M.; Infante, E.; Montagnac, G.; et al. ARF6-JIP3/4 Regulate Endosomal Tubules for MT1-MMP Exocytosis in Cancer Invasion. J. Cell Biol. 2015, 211, 339–358. [Google Scholar] [CrossRef] [PubMed]
- Monteiro, P.; Rossé, C.; Castro-Castro, A.; Irondelle, M.; Lagoutte, E.; Paul-Gilloteaux, P.; Desnos, C.; Formstecher, E.; Darchen, F.; Perrais, D.; et al. Endosomal WASH and Exocyst Complexes Control Exocytosis of MT1-MMP at Invadopodia. J. Cell Biol. 2013, 203, 1063–1079. [Google Scholar] [CrossRef]
- Sakurai-Yageta, M.; Recchi, C.; le Dez, G.; Sibarita, J.B.; Daviet, L.; Camonis, J.; D’Souza-Schorey, C.; Chavrier, P. The Interaction of IQGAP1 with the Exocyst Complex Is Required for Tumor Cell Invasion Downstream of Cdc42 and RhoA. J. Cell Biol. 2008, 181, 985–998. [Google Scholar] [CrossRef] [PubMed]
- Clancy, J.W.; Sedgwick, A.; Rosse, C.; Muralidharan-Chari, V.; Raposo, G.; Method, M.; Chavrier, P.; D’Souza-Schorey, C. Regulated Delivery of Molecular Cargo to Invasive Tumour-Derived Microvesicles. Nat. Commun. 2015, 6, 6919. [Google Scholar] [CrossRef]
- Sneeggen, M.; Pedersen, N.M.; Campsteijn, C.; Haugsten, E.M.; Stenmark, H.; Schink, K.O. WDFY2 Restrains Matrix Metalloproteinase Secretion and Cell Invasion by Controlling VAMP3-Dependent Recycling. Nat. Commun. 2019, 10, 2850. [Google Scholar] [CrossRef] [PubMed]
- Kean, M.J.; Williams, K.C.; Skalski, M.; Myers, D.; Burtnik, A.; Foster, D.; Coppolino, M.G. VAMP3, Syntaxin-13 and SNAP23 Are Involved in Secretion of Matrix Metalloproteinases, Degradation of the Extracellular Matrix and Cell Invasion. J. Cell Sci. 2009, 122, 4089–4098. [Google Scholar] [CrossRef]
- Seaman, M.N.J. The Retromer Complex: From Genesis to Revelations. Trends Biochem. Sci. 2021, 46, 608–620. [Google Scholar] [CrossRef]
- Sharma, P.; Parveen, S.; Shah, L.V.; Mukherjee, M.; Kalaidzidis, Y.; Kozielski, A.J.; Rosato, R.; Chang, J.C.; Datta, S. SNX27-Retromer Assembly Recycles MT1-MMP to Invadopodia and Promotes Breast Cancer Metastasis. J. Cell Biol. 2020, 219, e201812098. [Google Scholar] [CrossRef]
- Hutagalung, A.H.; Novick, P.J. Role of Rab GTPases in Membrane Traffic and Cell Physiology. Physiol. Rev. 2011, 91, 119–149. [Google Scholar] [CrossRef]
- Harbour, M.E.; Breusegem, S.Y.; Seaman, M.N.J. Recruitment of the Endosomal WASH Complex Is Mediated by the Extended ‘Tail’ of Fam21 Binding to the Retromer Protein Vps35. Biochem. J. 2012, 442, 209–220. [Google Scholar] [CrossRef]
- Rojas, R.; van Vlijmen, T.; Mardones, G.A.; Prabhu, Y.; Rojas, A.L.; Mohammed, S.; Heck, A.J.R.; Raposo, G.; van der Sluijs, P.; Bonifacino, J.S. Regulation of Retromer Recruitment to Endosomes by Sequential Action of Rab5 and Rab7. J. Cell Biol. 2008, 183, 513–526. [Google Scholar] [CrossRef]
- Liberti, M.V.; Locasale, J.W. The Warburg Effect: How Does It Benefit Cancer Cells? Trends Biochem. Sci. 2016, 41, 211–218. [Google Scholar] [CrossRef]
- Kelly, B.; O’Neill, L.A.J. Metabolic Reprogramming in Macrophages and Dendritic Cells in Innate Immunity. Cell Res. 2015, 25, 771–784. [Google Scholar] [CrossRef]
- Sakamoto, T.; Seiki, M. Cytoplasmic Tail of MT1-MMP Regulates Macrophage Motility Independently from Its Protease Activity. Genes Cells 2009, 14, 617–626. [Google Scholar] [CrossRef]
- Sakamoto, T.; Niiya, D.; Seiki, M. Targeting the Warburg Effect That Arises in Tumor Cells Expressing Membrane Type-1 Matrix Metalloproteinase. J. Biol. Chem. 2011, 286, 14691–14704. [Google Scholar] [CrossRef] [PubMed]
- Annabi, B.; Lee, Y.; Turcotte, S.; Naud, E.; Desrosiers, R.R.; Champagne, M.; Eliopoulos, N.; Galipeau, J.; Béliveau, R. Hypoxia Promotes Murine Bone-Marrow-Derived Stromal Cell Migration and Tube Formation. Stem Cells 2003, 21, 337–347. [Google Scholar] [CrossRef] [PubMed]
- Proulx-Bonneau, S.; Guezguez, A.; Annabi, B. A Concerted HIF-1α/MT1-MMP Signalling Axis Regulates the Expression of the 3BP2 Adaptor Protein in Hypoxic Mesenchymal Stromal Cells. PLoS ONE 2011, 6, e21511. [Google Scholar] [CrossRef]
- Zhang, W.; Liu, H.T. MAPK Signal Pathways in the Regulation of Cell Proliferation in Mammalian Cells. Cell Res. 2002, 12, 9–18. [Google Scholar] [CrossRef] [PubMed]
- Katz, M.; Amit, I.; Yarden, Y. Regulation of MAPKs by Growth Factors and Receptor Tyrosine Kinases. Biochim. Biophys. Acta BBA-Mol. Cell Res. 2007, 1773, 1161–1176. [Google Scholar] [CrossRef]
- Soares-Silva, M.; Diniz, F.F.; Gomes, G.N.; Bahia, D. The Mitogen-Activated Protein Kinase (MAPK) Pathway: Role in Immune Evasion by Trypanosomatids. Front. Microbiol. 2016, 7, 183. [Google Scholar] [CrossRef]
- Gingras, D.; Bousquet-Gagnon, N.; Langlois, S.; Lachambre, M.P.; Annabi, B.; Béliveau, R. Activation of the Extracellular Signal-Regulated Protein Kinase (ERK) Cascade by Membrane-Type-1 Matrix Metalloproteinase (MT1-MMP). FEBS Lett. 2001, 507, 231–236. [Google Scholar] [CrossRef]
- Takino, T.; Miyamori, H.; Watanabe, Y.; Yoshioka, K.; Seiki, M.; Sato, H. Membrane Type 1 Matrix Metalloproteinase Regulates Collagen-Dependent Mitogen-Activated Protein/Extracellular Signal-Related Kinase Activation and Cell Migration. Cancer Res. 2004, 64, 1044–1049. [Google Scholar] [CrossRef]
- D’Alessio, S.; Ferrari, G.; Cinnante, K.; Scheerer, W.; Galloway, A.C.; Roses, D.F.; Rozanov, D.V.; Remacle, A.G.; Oh, E.S.; Shiryaev, S.A.; et al. Tissue Inhibitor of Metalloproteinases-2 Binding to Membrane-Type 1 Matrix Metalloproteinase Induces MAPK Activation and Cell Growth by a Non-Proteolytic Mechanism. J. Biol. Chem. 2008, 283, 87–99. [Google Scholar] [CrossRef] [PubMed]
- Sounni, N.E.; Rozanov, D.V.; Remacle, A.G.; Golubkov, V.S.; Noel, A.; Strongin, A.Y. Timp-2 Binding with Cellular MT1-MMP Stimulates Invasion-Promoting MEK/ERK Signaling in Cancer Cells. Int. J. Cancer. 2010, 126, 1067. [Google Scholar] [CrossRef] [PubMed]
- Takino, T.; Tsuge, H.; Ozawa, T.; Sato, H. MT1-MMP Promotes Cell Growth and ERK Activation through c-Src and Paxillin in Three-Dimensional Collagen Matrix. Biochem. Biophys. Res. Commun. 2010, 396, 1042–1047. [Google Scholar] [CrossRef] [PubMed]
- Tanimura, S.; Asato, K.; Fujishiro, S.H.; Kohno, M. Specific Blockade of the ERK Pathway Inhibits the Invasiveness of Tumor Cells: Down-Regulation of Matrix Metalloproteinase-3/-9/-14 and CD44. Biochem. Biophys. Res. Commun. 2003, 304, 801–806. [Google Scholar] [CrossRef] [PubMed]
- Deryugina, E.I.; Ratnikov, B.I.; Postnova, T.I.; Rozanov, D.V.; Strongin, A.Y. Processing of Integrin Alpha(v) Subunit by Membrane Type 1 Matrix Metalloproteinase Stimulates Migration of Breast Carcinoma Cells on Vitronectin and Enhances Tyrosine Phosphorylation of Focal Adhesion Kinase. J. Biol. Chem. 2002, 277, 9749–9756. [Google Scholar] [CrossRef]
- Sounni, N.E.; Devy, L.; Hajitou, A.; Frankenne, F.; Munaut, C.; Gilles, C.; Deroanne, C.; Thompson, E.W.; Foidart, J.M.; Noel, A. MT1-MMP Expression Promotes Tumor Growth and Angiogenesis through an up-Regulation of Vascular Endothelial Growth Factor Expression. FASEB J. 2002, 16, 555–564. [Google Scholar] [CrossRef]
- Sounni, N.E.; Roghi, C.; Chabottaux, V.; Janssen, M.; Munaut, C.; Maquoi, E.; Galvez, B.G.; Gilles, C.; Frankenne, F.; Murphy, G.; et al. Up-Regulation of Vascular Endothelial Growth Factor-A by Active Membrane-Type 1 Matrix Metalloproteinase through Activation of Src-Tyrosine Kinases. J. Biol. Chem. 2004, 279, 13564–13574. [Google Scholar] [CrossRef]
- Eisenach, P.A.; Roghi, C.; Fogarasi, M.; Murphy, G.; English, W.R. MT1-MMP Regulates VEGF-A Expression through a Complex with VEGFR-2 and Src. J. Cell Sci. 2010, 123, 4182–4193. [Google Scholar] [CrossRef]
- Funahashi, Y.; Shawber, C.J.; Sharma, A.; Kanamaru, E.; Choi, Y.K.; Kitajewski, J. Notch Modulates VEGF Action in Endothelial Cells by Inducing Matrix Metalloprotease Activity. Vasc. Cell 2011, 3, 2. [Google Scholar] [CrossRef]
- Itoh, Y.; Seiki, M. MT1-MMP: A Potent Modifier of Pericellular Microenvironment. J. Cell Physiol. 2006, 206, 1–8. [Google Scholar] [CrossRef]
- Gonzalo, P.; Moreno, V.; Gálvez, B.G.; Arroyo, A.G. MT1-MMP and Integrins: Hand-to-Hand in Cell Communication. Biofactors 2010, 36, 248–254. [Google Scholar] [CrossRef] [PubMed]
- Gálvez, B.G.; Matías-Román, S.; Yáñez-Mó, M.; Sánchez-Madrid, F.; Arroyo, A.G. ECM Regulates MT1-MMP Localization with Beta1 or Alphavbeta3 Integrins at Distinct Cell Compartments Modulating Its Internalization and Activity on Human Endothelial Cells. J. Cell Biol. 2002, 159, 509–521. [Google Scholar] [CrossRef]
- Goodison, S.; Urquidi, V.; Tarin, D. CD44 Cell Adhesion Molecules. Mol. Pathol. 1999, 52, 189–196. [Google Scholar] [CrossRef]
- Mori, H.; Tomari, T.; Koshikawa, N.; Kajita, M.; Itoh, Y.; Sato, H.; Tojo, H.; Yana, I.; Seiki, M. CD44 Directs Membrane-Type 1 Matrix Metalloproteinase to Lamellipodia by Associating with Its Hemopexin-like Domain. EMBO J. 2002, 21, 3949–3959. [Google Scholar] [CrossRef]
- Suenaga, N.; Mori, H.; Itoh, Y.; Seiki, M. CD44 Binding through the Hemopexin-like Domain Is Critical for Its Shedding by Membrane-Type 1 Matrix Metalloproteinase. Oncogene 2005, 24, 859–868. [Google Scholar] [CrossRef]
- Kajita, M.; Itoh, Y.; Chiba, T.; Mori, H.; Okada, A.; Kinoh, H.; Seiki, M. Membrane-Type 1 Matrix Metalloproteinase Cleaves CD44 and Promotes Cell Migration. J. Cell Biol. 2001, 153, 893–904. [Google Scholar] [CrossRef] [PubMed]
- Mori, T.; Kitano, K.; Terawaki, S.I.; Maesaki, R.; Fukami, Y.; Hakoshima, T. Structural Basis for CD44 Recognition by ERM Proteins. J. Biol. Chem. 2008, 283, 29602. [Google Scholar] [CrossRef]
- Janoštiak, R.; Tolde, O.; Brůhová, Z.; Novotný, M.; Hanks, S.K.; Rösel, D.; Brábek, J. Tyrosine Phosphorylation within the SH3 Domain Regulates CAS Subcellular Localization, Cell Migration, and Invasiveness. Mol. Biol. Cell 2011, 22, 4256. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; McNiven, M.A. Invasive Matrix Degradation at Focal Adhesions Occurs via Protease Recruitment by a FAK-P130Cas Complex. J. Cell Biol. 2012, 196, 375–385. [Google Scholar] [CrossRef] [PubMed]
- Yang, J.; Kasberg, W.C.; Celo, A.; Liang, Z.; Quispe, K.; Sharon Stack, M. Post-Translational Modification of the Membrane Type 1 Matrix Metalloproteinase (MT1-MMP) Cytoplasmic Tail Impacts Ovarian Cancer Multicellular Aggregate Dynamics. J. Biol. Chem. 2017, 292, 13111–13121. [Google Scholar] [CrossRef]
- Bruney, L.; Liu, Y.; Grisoli, A.; Ravosa, M.J.; Stack, M.S. Integrin-Linked Kinase Activity Modulates the pro-Metastatic Behavior of Ovarian Cancer Cells. Oncotarget 2016, 7, 21968. [Google Scholar] [CrossRef] [PubMed]
- Wickström, S.A.; Lange, A.; Montanez, E.; Fässler, R. The ILK/PINCH/Parvin Complex: The Kinase Is Dead, Long Live the Pseudokinase! EMBO J. 2010, 29, 281–291. [Google Scholar] [CrossRef] [PubMed]
- Lange, A.; Wickström, S.A.; Jakobson, M.; Zent, R.; Sainio, K.; Fässler, R. Integrin-Linked Kinase Is an Adaptor with Essential Functions during Mouse Development. Nature 2009, 461, 1002–1006. [Google Scholar] [CrossRef] [PubMed]
- Fukuda, K.; Gupta, S.; Chen, K.; Wu, C.; Qin, J. The Pseudoactive Site of ILK Is Essential for Its Binding to Alpha-Parvin and Localization to Focal Adhesions. Mol. Cell 2009, 36, 819–830. [Google Scholar] [CrossRef]
- Wolf, K.; Mazo, I.; Leung, H.; Engelke, K.; von Andrian, U.H.; Deryugina, E.I.; Strongin, A.Y.; Bröcker, E.B.; Friedl, P. Compensation Mechanism in Tumor Cell Migration: Mesenchymal-Amoeboid Transition after Blocking of Pericellular Proteolysis. J. Cell Biol. 2003, 160, 267–277. [Google Scholar] [CrossRef]
- Rösel, D.; Brábek, J.; Tolde, O.; Mierke, C.T.; Zitterbart, D.P.; Raupach, C.; Bicanová, K.; Kollmannsberger, P.; Paňková, D.; Veselý, P.; et al. Up-Regulation of Rho/ROCK Signaling in Sarcoma Cells Drives Invasion and Increased Generation of Protrusive Forces. Mol. Cancer Res. 2008, 6, 1410–1420. [Google Scholar] [CrossRef]
- Clark, A.G.; Vignjevic, D.M. Modes of Cancer Cell Invasion and the Role of the Microenvironment. Curr. Opin. Cell Biol. 2015, 36, 13–22. [Google Scholar] [CrossRef]
- Cao, J.; Chiarelli, C.; Richman, O.; Zarrabi, K.; Kozarekar, P.; Zucker, S. Membrane Type 1 Matrix Metalloproteinase Induces Epithelial-to-Mesenchymal Transition in Prostate Cancer. J. Biol. Chem. 2008, 283, 6232–6240. [Google Scholar] [CrossRef]
- Yu, X.; Zech, T.; McDonald, L.; Gonzalez, E.G.; Li, A.; Macpherson, I.; Schwarz, J.P.; Spence, H.; Futó, K.; Timpson, P.; et al. N-WASP Coordinates the Delivery and F-Actin-Mediated Capture of MT1-MMP at Invasive Pseudopods. J. Cell Biol. 2012, 199, 527–544. [Google Scholar] [CrossRef]
- Sabeh, F.; Li, X.Y.; Saunders, T.L.; Rowe, R.G.; Weiss, S.J. Secreted versus Membrane-Anchored Collagenases: Relative Roles in Fibroblast-Dependent Collagenolysis and Invasion. J. Biol. Chem. 2009, 284, 23001–23011. [Google Scholar] [CrossRef]
- Cao, J.; Kozarekar, P.; Pavlaki, M.; Chiarelli, C.; Bahou, W.F.; Zucker, S. Distinct Roles for the Catalytic and Hemopexin Domains of Membrane Type 1-Matrix Metalloproteinase in Substrate Degradation and Cell Migration. J. Biol. Chem. 2004, 279, 14129–14139. [Google Scholar] [CrossRef]
- Li, X.-Y.; Ota, I.; Yana, I.; Sabeh, F.; Weiss, S.J. Molecular Dissection of the Structural Machinery Underlying the Tissue-Invasive Activity of Membrane Type-1 Matrix Metalloproteinase. Mol. Biol. Cell 2008, 19, 3221–3233. [Google Scholar] [CrossRef]
- Hotary, K.; Allen, E.; Punturieri, A.; Yana, I.; Weiss, S.J. Regulation of Cell Invasion and Morphogenesis in a Three-Dimensional Type I Collagen Matrix by Membrane-Type Matrix Metalloproteinases 1, 2, and 3. J. Cell Biol. 2000, 149, 1309–1323. [Google Scholar] [CrossRef]
- Pahwa, S.; Bhowmick, M.; Amar, S.; Cao, J.; Strongin, A.Y.; Fridman, R.; Weiss, S.J.; Fields, G.B. Characterization and Regulation of MT1-MMP Cell Surface-Associated Activity. Chem. Biol. Drug Des. 2019, 93, 1251–1264. [Google Scholar] [CrossRef]
- Nyalendo, C.; Sartelet, H.; Gingras, D.; Béliveau, R. Inhibition of Membrane-Type 1 Matrix Metalloproteinase Tyrosine Phosphorylation Blocks Tumor Progression in Mice. Anticancer Res. 2010, 30, 422. [Google Scholar]
- Uekita, T.; Gotoh, I.; Kinoshita, T.; Itoh, Y.; Sato, H.; Shiomi, T.; Okada, Y.; Seiki, M. Membrane-Type 1 Matrix Metalloproteinase Cytoplasmic Tail-Binding Protein-1 Is a New Member of the Cupin Superfamily. A Possible Multifunctional Protein Acting as an Invasion Suppressor down-Regulated in Tumors. J. Biol. Chem. 2004, 279, 12734–12743. [Google Scholar] [CrossRef] [PubMed]
- Qiang, L.; Cao, H.; Chen, J.; Weller, S.G.; Krueger, E.W.; Zhang, L.; Razidlo, G.L.; McNiven, M.A. Pancreatic Tumor Cell Metastasis Is Restricted by MT1-MMP Binding Protein MTCBP-1. J. Cell Biol. 2019, 218, 317–332. [Google Scholar] [CrossRef] [PubMed]
- Ferrari, R.; Martin, G.; Tagit, O.; Guichard, A.; Cambi, A.; Voituriez, R.; Vassilopoulos, S.; Chavrier, P. MT1-MMP Directs Force-Producing Proteolytic Contacts That Drive Tumor Cell Invasion. Nat. Commun. 2019, 10, 4886. [Google Scholar] [CrossRef] [PubMed]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Strouhalova, K.; Tolde, O.; Rosel, D.; Brábek, J. Cytoplasmic Tail of MT1-MMP: A Hub of MT1-MMP Regulation and Function. Int. J. Mol. Sci. 2023, 24, 5068. https://doi.org/10.3390/ijms24065068
Strouhalova K, Tolde O, Rosel D, Brábek J. Cytoplasmic Tail of MT1-MMP: A Hub of MT1-MMP Regulation and Function. International Journal of Molecular Sciences. 2023; 24(6):5068. https://doi.org/10.3390/ijms24065068
Chicago/Turabian StyleStrouhalova, Katerina, Ondřej Tolde, Daniel Rosel, and Jan Brábek. 2023. "Cytoplasmic Tail of MT1-MMP: A Hub of MT1-MMP Regulation and Function" International Journal of Molecular Sciences 24, no. 6: 5068. https://doi.org/10.3390/ijms24065068
APA StyleStrouhalova, K., Tolde, O., Rosel, D., & Brábek, J. (2023). Cytoplasmic Tail of MT1-MMP: A Hub of MT1-MMP Regulation and Function. International Journal of Molecular Sciences, 24(6), 5068. https://doi.org/10.3390/ijms24065068




