The Role of Membrane-Type 1 Matrix Metalloproteinase–Substrate Interactions in Pathogenesis
Abstract
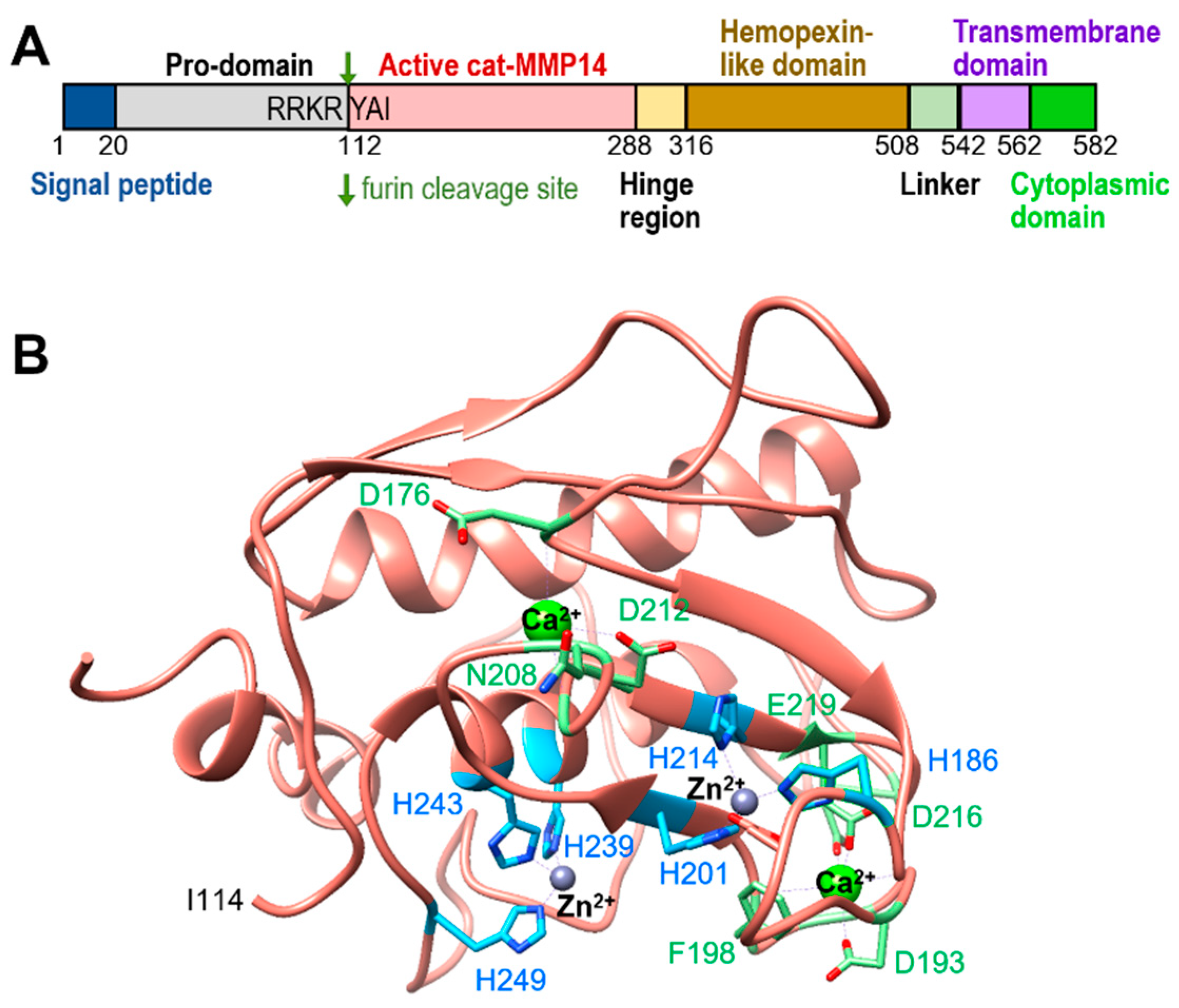
1. Introduction
2. Substrates of MT1-MMP
2.1. ECM Substrates
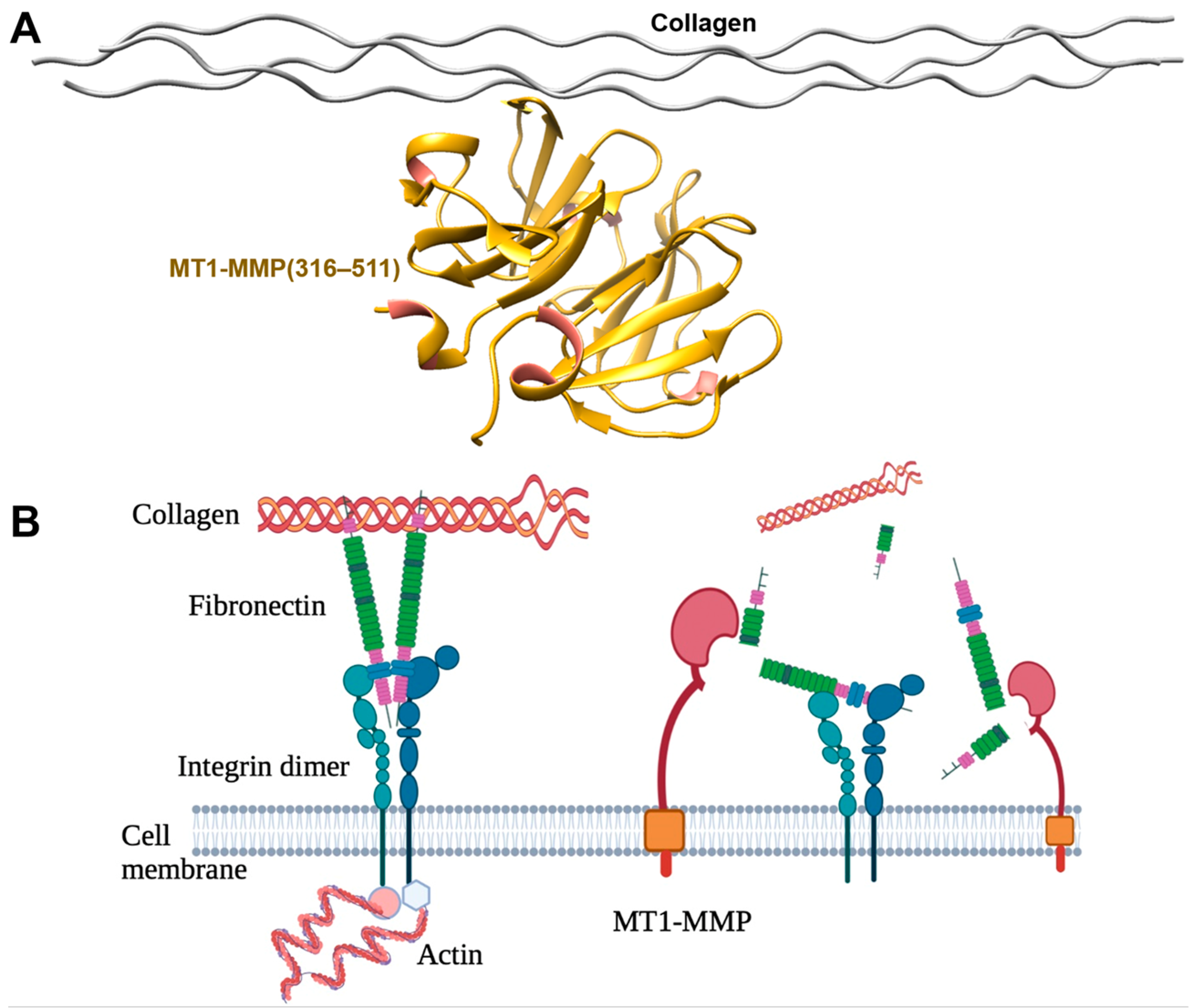
2.1.1. Collagen
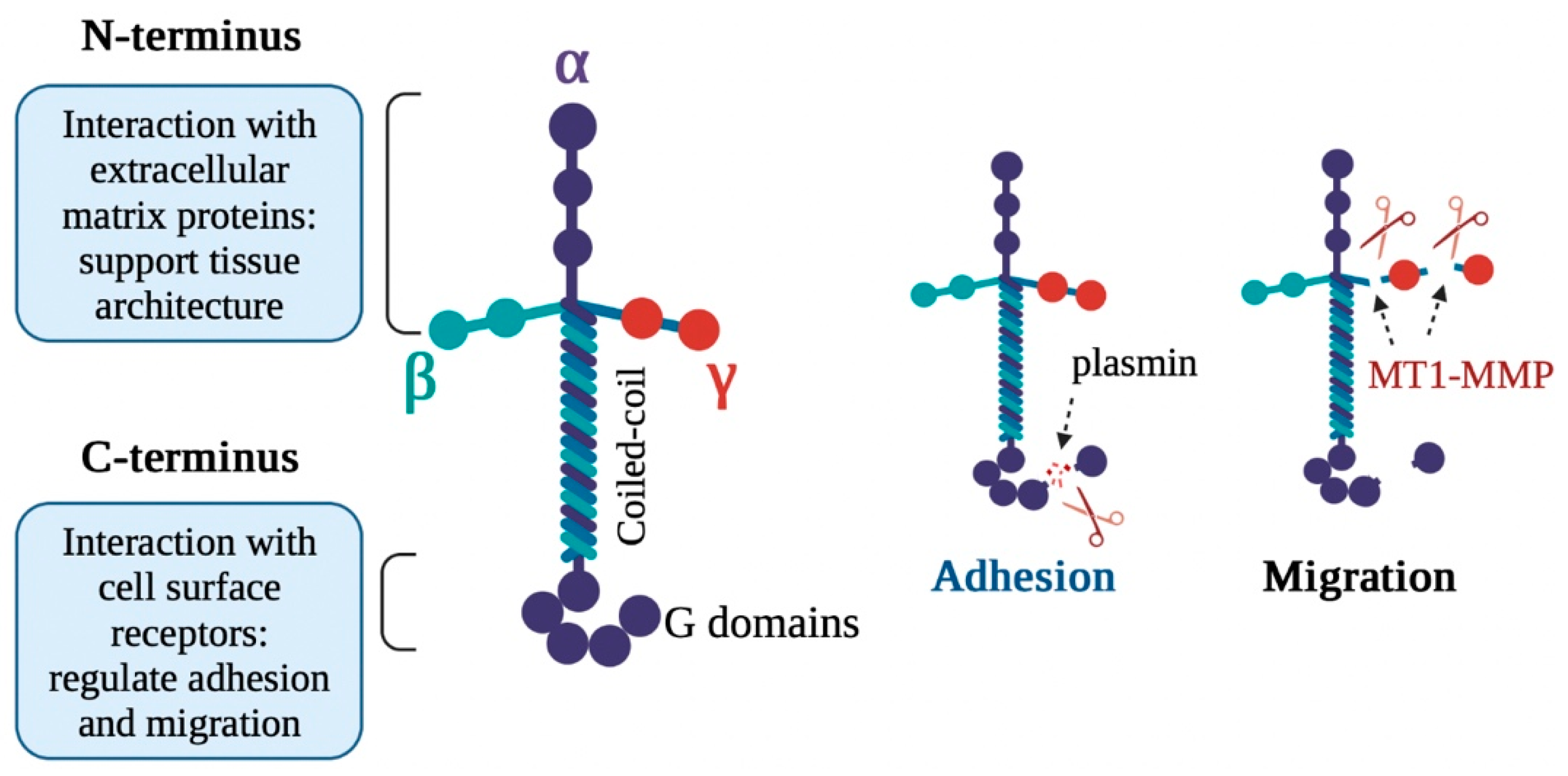
2.1.2. Fibronectin
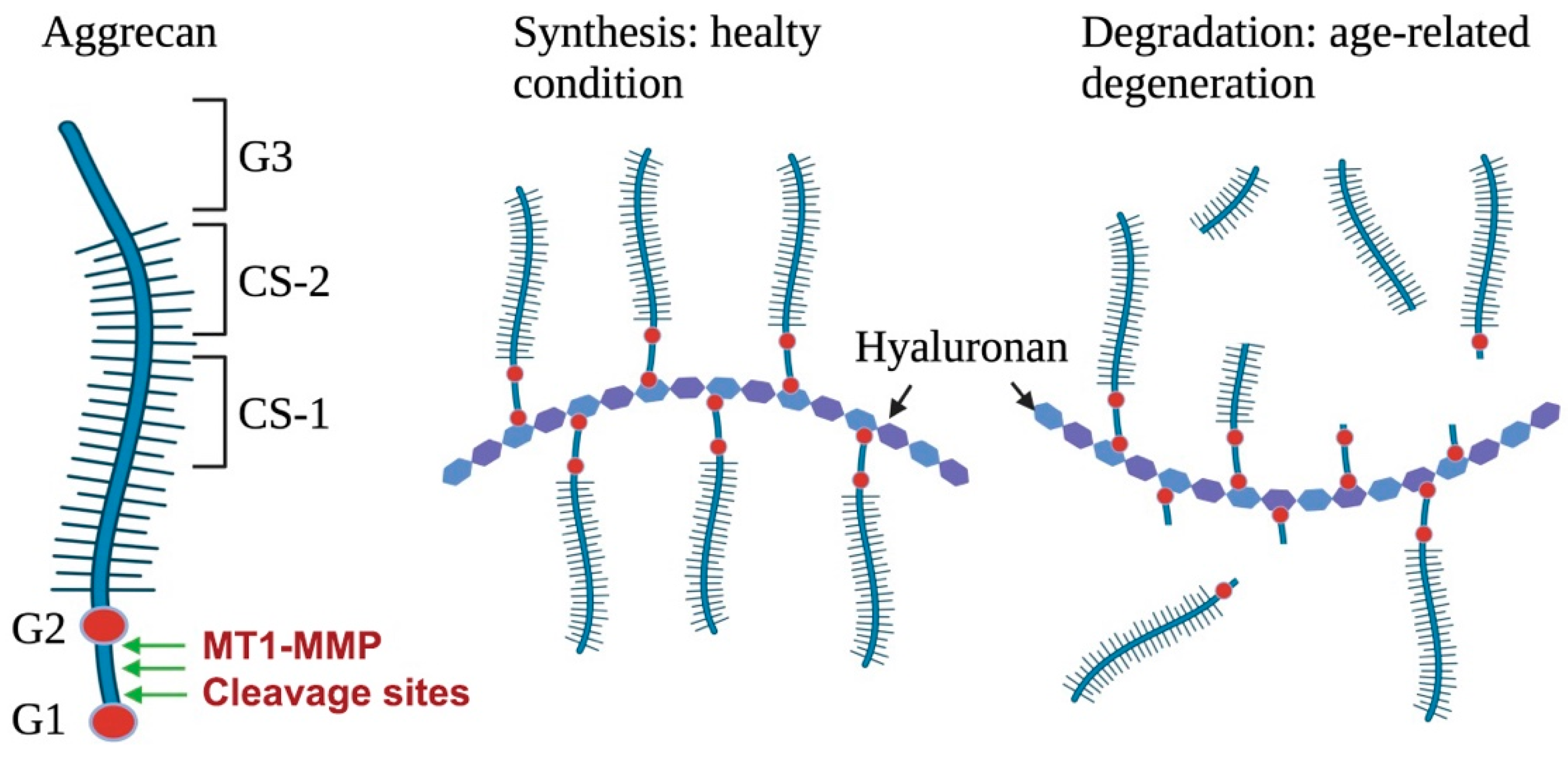
2.1.3. Aggrecan
2.1.4. Laminin
2.1.5. Other ECM Substrates
2.2. Non-ECM Substrates
2.2.1. pro-MMP-2 and pro-MMP-13
2.2.2. Vascular Endothelial Growth Factor Receptor-1 and Fibroblast Growth Factor Receptor-2
2.3. Other Non-ECM Substrates
3. Summary
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Khokha, R.; Murthy, A.; Weiss, A. Metalloproteinases and their natural inhibitors in inflammation and immunity. Nat. Rev. Immunol. 2013, 13, 649–665. [Google Scholar] [CrossRef]
- Page-McCaw, A.; Ewald, A.J.; Werb, Z. Matrix metalloproteinases and the regulation of tissue remodelling. Nat. Rev. Mol. Cell Biol. 2007, 8, 221–233. [Google Scholar] [CrossRef]
- Hadler-Olsen, E.; Fadnes, B.; Sylte, I.; Uhlin-Hansen, L.; Winberg, J.O. Regulation of matrix metalloproteinase activity in health and disease. FEBS J. 2011, 278, 28–45. [Google Scholar] [CrossRef] [PubMed]
- Chakraborti, S.; Mandal, M.; Das, S.; Mandal, A.; Chakraborti, T. Regulation of matrix metalloproteinases: An overview. Mol. Cell Biochem. 2003, 253, 269–285. [Google Scholar] [CrossRef]
- Curry Jr, T.E.; Osteen, K.G. The matrix metalloproteinase system: Changes, regulation, and impact throughout the ovarian and uterine reproductive cycle. Endocr. Rev. 2003, 24, 428–465. [Google Scholar] [PubMed]
- López-Boado, Y.S.; Wilson, C.L.; Hooper, L.V.; Gordon, J.I.; Hultgren, S.J.; Parks, W.C. Bacterial exposure induces and activates matrilysin in mucosal epithelial cells. J. Cell Biol. 2000, 148, 1305–1315. [Google Scholar] [CrossRef] [PubMed]
- Arroyo, A.; Genis, L.; Gonzalo, P.; Matias-Roman, S.; Pollan, A.; Galvez, B. Matrix metalloproteinases: New routes to the use of MT1-MMP as a therapeutic target in angiogenesis-related disease. Curr. Pharm. Des. 2007, 13, 1787–1802. [Google Scholar] [CrossRef]
- Cui, N.; Hu, M.; Khalil, R.A. Biochemical and Biological Attributes of Matrix Metalloproteinases. Prog. Mol. Biol. Transl. Sci. 2017, 147, 1–73. [Google Scholar] [PubMed]
- Itoh, Y. Membrane-type matrix metalloproteinases: Their functions and regulations. Matrix Biol. 2015, 44–46, 207–223. [Google Scholar] [CrossRef]
- Qiang, L.; Cao, H.; Chen, J.; Weller, S.G.; Krueger, E.W.; Zhang, L.; Razidlo, G.L.; McNiven, M.A. Pancreatic tumor cell metastasis is restricted by MT1-MMP binding protein MTCBP-1. J. Cell Biol. 2019, 218, 317–332. [Google Scholar] [CrossRef]
- Schmuck, R.B.; Lippens, E.; Wulsten, D.; Garske, D.S.; Stronisch, A.; Pratschke, J.; Sauer, I.M.; Duda, G.N.; Bahra, M.; Cipitria, A. Role of extracellular matrix structural components and tissue mechanics in the development of postoperative pancreatic fistula. J. Biomech. 2021, 128, 110714. [Google Scholar] [CrossRef]
- King, M.W. Extracellular Matrix: Glycosaminoglycans and Proteoglycans. In Integrative Medical Biochemistry Examination and Board Review; McGraw-Hill Education: New York, NY, USA, 2014. [Google Scholar]
- Jalali, S.; del Pozo, M.A.; Chen, K.; Miao, H.; Li, Y.; Schwartz, M.A.; Shyy, J.Y.; Chien, S. Integrin-mediated mechanotransduction requires its dynamic interaction with specific extracellular matrix (ECM) ligands. Proc. Natl. Acad. Sci. USA 2001, 98, 1042–1046. [Google Scholar] [CrossRef] [PubMed]
- De Arcangelis, A.; Georges-Labouesse, E. Integrin and ECM functions: Roles in vertebrate development. Trends Genet. 2000, 16, 389–395. [Google Scholar] [CrossRef] [PubMed]
- Tarca, A.L.; Draghici, S.; Khatri, P.; Hassan, S.S.; Mittal, P.; Kim, J.S.; Kim, C.J.; Kusanovic, J.P.; Romero, R. A novel signaling pathway impact analysis. Bioinformatics 2009, 25, 75–82. [Google Scholar] [CrossRef] [PubMed]
- Bentzinger, C.F.; Wang, Y.X.; von Maltzahn, J.; Soleimani, V.D.; Yin, H.; Rudnicki, M.A. Fibronectin regulates Wnt7a signaling and satellite cell expansion. Cell Stem Cell 2013, 12, 75–87. [Google Scholar] [CrossRef]
- Agrawal, V.; Tottey, S.; Johnson, S.A.; Freund, J.M.; Siu, B.F.; Badylak, S.F. Recruitment of progenitor cells by an extracellular matrix cryptic peptide in a mouse model of digit amputation. Tissue Eng. Part A 2011, 17, 2435–2443. [Google Scholar] [CrossRef]
- Bowers, S.L.; Baudino, T.A. Laying the groundwork for growth: Cell-cell and cell-ECM interactions in cardiovascular development. Birth Defects Res. C Embryo Today 2010, 90, 1–7. [Google Scholar] [CrossRef]
- Ohuchi, E.; Imai, K.; Fujii, Y.; Sato, H.; Seiki, M.; Okada, Y. Membrane type 1 matrix metalloproteinase digests interstitial collagens and other extracellular matrix macromolecules. J. Biol. Chem. 1997, 272, 2446–2451. [Google Scholar] [CrossRef]
- Zhytnik, L.; Maasalu, K.; Pashenko, A.; Khmyzov, S.; Reimann, E.; Prans, E.; Koks, S.; Martson, A. COL1A1/2 Pathogenic Variants and Phenotype Characteristics in Ukrainian Osteogenesis Imperfecta Patients. Front. Genet. 2019, 10, 722. [Google Scholar] [CrossRef]
- Steiner, R.D.; Basel, D. COL1A1/2 Osteogenesis Imperfecta. In GeneReviews.((R.)); Adam, M.P., Mirzaa, G.M., Pagon, R.A., Wallace, S.E., Bean, L.J.H., Gripp, K.W., Amemiya, A., Eds.; University of Washington: Seattle, WA, USA, 1993. [Google Scholar]
- Chung, L.; Dinakarpandian, D.; Yoshida, N.; Lauer-Fields, J.L.; Fields, G.B.; Visse, R.; Nagase, H. Collagenase unwinds triple-helical collagen prior to peptide bond hydrolysis. EMBO J. 2004, 23, 3020–3030. [Google Scholar] [CrossRef] [PubMed]
- Szabova, L.; Chrysovergis, K.; Yamada, S.S.; Holmbeck, K. MT1-MMP is required for efficient tumor dissemination in experimental metastatic disease. Oncogene 2008, 27, 3274–3281. [Google Scholar] [CrossRef]
- Orgel, J.P.; Irving, T.C.; Miller, A.; Wess, T.J. Microfibrillar structure of type I collagen in situ. Proc. Natl. Acad. Sci. USA 2006, 103, 9001–9005. [Google Scholar] [CrossRef]
- Salsas-Escat, R.; Stultz, C.M. Conformational selection and collagenolysis in type III collagen. Proteins 2010, 78, 325–335. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Y.; Marcink, T.C.; Sanganna Gari, R.R.; Marsh, B.P.; King, G.M.; Stawikowska, R.; Fields, G.B.; Van Doren, S.R. Transient collagen triple helix binding to a key metalloproteinase in invasion and development. Structure 2015, 23, 257–269. [Google Scholar] [CrossRef] [PubMed]
- Hotary, K.; Allen, E.; Punturieri, A.; Yana, I.; Weiss, S.J. Regulation of cell invasion and morphogenesis in a three-dimensional type I collagen matrix by membrane-type matrix metalloproteinases 1, 2, and 3. J. Cell Biol. 2000, 149, 1309–1323. [Google Scholar] [CrossRef]
- Zigrino, P.; Brinckmann, J.; Niehoff, A.; Lu, Y.; Giebeler, N.; Eckes, B.; Kadler, K.E.; Mauch, C. Fibroblast-Derived MMP-14 Regulates Collagen Homeostasis in Adult Skin. J. Investig. Dermatol. 2016, 136, 1575–1583. [Google Scholar] [CrossRef] [PubMed]
- Holmbeck, K.; Bianco, P.; Caterina, J.; Yamada, S.; Kromer, M.; Kuznetsov, S.A.; Mankani, M.; Robey, P.G.; Poole, A.R.; Pidoux, I.; et al. MT1-MMP-deficient mice develop dwarfism, osteopenia, arthritis, and connective tissue disease due to inadequate collagen turnover. Cell 1999, 99, 81–92. [Google Scholar] [CrossRef] [PubMed]
- Hiraoka, N.; Allen, E.; Apel, I.J.; Gyetko, M.R.; Weiss, S.J. Matrix metalloproteinases regulate neovascularization by acting as pericellular fibrinolysins. Cell 1998, 95, 365–377. [Google Scholar] [CrossRef]
- Cole, W.G. Collagen genes: Mutations affecting collagen structure and expression. Prog. Nucleic Acid Res. Mol. Biol. 1994, 47, 29–80. [Google Scholar]
- Shoulders, M.D.; Raines, R.T. Collagen structure and stability. Annu. Rev. Biochem. 2009, 78, 929–958. [Google Scholar] [CrossRef]
- Maurer, L.M.; Ma, W.; Mosher, D.F. Dynamic structure of plasma fibronectin. Crit. Rev. Biochem. Mol. Biol. 2015, 51, 213–227. [Google Scholar] [CrossRef] [PubMed]
- Grinnell, F. Fibronectin and wound healing. J. Cell Biochem. 1984, 26, 107–116. [Google Scholar] [CrossRef]
- Ramos Gde, O.; Bernardi, L.; Lauxen, I.; Sant’Ana Filho, M.; Horwitz, A.R.; Lamers, M.L. Fibronectin Modulates Cell Adhesion and Signaling to Promote Single Cell Migration of Highly Invasive Oral Squamous Cell Carcinoma. PLoS ONE 2016, 11, e0151338. [Google Scholar] [CrossRef]
- Romberger, D.J. Fibronectin. Int. J. Biochem. Cell Biol. 1997, 29, 939–943. [Google Scholar] [CrossRef]
- Miyamoto, S.; Katz, B.Z.; Lafrenie, R.M.; Yamada, K.M. Fibronectin and integrins in cell adhesion, signaling, and morphogenesis. Ann. N. Y. Acad. Sci. 1998, 857, 119–129. [Google Scholar] [CrossRef] [PubMed]
- Niland, S.; Eble, J.A. Hold on or Cut? Integrin- and MMP-Mediated Cell-Matrix Interactions in the Tumor Microenvironment. Int. J. Mol. Sci. 2020, 22, 238. [Google Scholar] [CrossRef]
- George, E.L.; Baldwin, H.S.; Hynes, R.O. Fibronectins are essential for heart and blood vessel morphogenesis but are dispensable for initial specification of precursor cells. Blood 1997, 90, 3073–3081. [Google Scholar] [CrossRef] [PubMed]
- Chen, W.T.; Chen, J.M.; Parsons, S.J.; Parsons, J.T. Local degradation of fibronectin at sites of expression of the transforming gene product pp60src. Nature 1985, 316, 156–158. [Google Scholar] [CrossRef]
- Shi, F.; Sottile, J. MT1-MMP regulates the turnover and endocytosis of extracellular matrix fibronectin. J. Cell Sci. 2011, 124 Pt 23, 4039–4050. [Google Scholar] [CrossRef]
- Zhang, X.; Chen, C.T.; Bhargava, M.; Torzilli, P.A. A Comparative Study of Fibronectin Cleavage by MMP-1, -3, -13, and -14. Cartilage 2012, 3, 267–277. [Google Scholar] [CrossRef]
- Araki, Y.; Mimura, T. Matrix Metalloproteinase Gene Activation Resulting from Disordred Epigenetic Mechanisms in Rheumatoid Arthritis. Int. J. Mol. Sci. 2017, 18, 905. [Google Scholar] [CrossRef]
- Miller, M.C.; Manning, H.B.; Jain, A.; Troeberg, L.; Dudhia, J.; Essex, D.; Sandison, A.; Seiki, M.; Nanchahal, J.; Nagase, H.; et al. Membrane type 1 matrix metalloproteinase is a crucial promoter of synovial invasion in human rheumatoid arthritis. Arthritis Rheum 2009, 60, 686–697. [Google Scholar] [CrossRef] [PubMed]
- Gleghorn, J.P.; Jones, A.R.; Flannery, C.R.; Bonassar, L.J. Boundary mode lubrication of articular cartilage by recombinant human lubricin. J. Orthop. Res. 2009, 27, 771–777. [Google Scholar] [CrossRef]
- Wang, M.; Liu, C.; Thormann, E.; Dedinaite, A. Hyaluronan and phospholipid association in biolubrication. Biomacromolecules 2013, 14, 4198–4206. [Google Scholar] [CrossRef]
- Fosang, A.J.; Last, K.; Neame, P.J.; Murphy, G.; Knauper, V.; Tschesche, H.; Hughes, C.E.; Caterson, B.; Hardingham, T.E. Neutrophil collagenase (MMP-8) cleaves at the aggrecanase site E373-A374 in the interglobular domain of cartilage aggrecan. Biochem. J. 1994, 304 Pt 2, 347–351. [Google Scholar] [CrossRef] [PubMed]
- Buttner, F.H.; Hughes, C.E.; Margerie, D.; Lichte, A.; Tschesche, H.; Caterson, B.; Bartnik, E. Membrane type 1 matrix metalloproteinase (MT1-MMP) cleaves the recombinant aggrecan substrate rAgg1mut at the ‘aggrecanase’ and the MMP sites. Characterization of MT1-MMP catabolic activities on the interglobular domain of aggrecan. Biochem. J. 1998, 333 Pt 1, 159–165. [Google Scholar] [CrossRef]
- Pratta, M.A.; Yao, W.; Decicco, C.; Tortorella, M.D.; Liu, R.Q.; Copeland, R.A.; Magolda, R.; Newton, R.C.; Trzaskos, J.M.; Arner, E.C. Aggrecan protects cartilage collagen from proteolytic cleavage. J. Biol. Chem. 2003, 278, 45539–45545. [Google Scholar] [CrossRef]
- Little, C.B.; Hughes, C.E.; Curtis, C.L.; Janusz, M.J.; Bohne, R.; Wang-Weigand, S.; Taiwo, Y.O.; Mitchell, P.G.; Otterness, I.G.; Flannery, C.R.; et al. Matrix metalloproteinases are involved in C-terminal and interglobular domain processing of cartilage aggrecan in late stage cartilage degradation. Matrix Biol. 2002, 21, 271–288. [Google Scholar] [PubMed]
- Goldring, S.R.; Goldring, M.B. The role of cytokines in cartilage matrix degeneration in osteoarthritis. Clin. Orthop. Relat. Res. 2004, 427, S27–S36. [Google Scholar] [CrossRef]
- Takahashi, M.; Fujikawa, K.; Angammana, R.; Shibata, S. An in situ hybridization study of MMP-2, -9, -13, -14, TIMP-1, and -2 mRNA in fetal mouse mandibular condylar cartilage as compared with limb bud cartilage. Gene Expr. Patterns 2019, 32, 1–11. [Google Scholar] [CrossRef]
- Tetlow, L.C.; Adlam, D.J.; Woolley, D.E. Matrix metalloproteinase and proinflammatory cytokine production by chondrocytes of human osteoarthritic cartilage: Associations with degenerative changes. Arthritis Rheum 2001, 44, 585–594. [Google Scholar] [CrossRef]
- Aumailley, M. The laminin family. Cell Adhes. Migr. 2013, 7, 48–55. [Google Scholar] [CrossRef]
- Miner, J.H.; Patton, B.L.; Lentz, S.I.; Gilbert, D.J.; Snider, W.D.; Jenkins, N.A.; Copeland, N.G.; Sanes, J.R. The laminin alpha chains: Expression, developmental transitions, and chromosomal locations of alpha1-5, identification of heterotrimeric laminins 8-11, and cloning of a novel alpha3 isoform. J. Cell Biol. 1997, 137, 685–701. [Google Scholar] [CrossRef]
- Kleinman, H.K.; Cannon, F.B.; Laurie, G.W.; Hassell, J.R.; Aumailley, M.; Terranova, V.P.; Martin, G.R.; DuBois-Dalcq, M. Biological activities of laminin. J. Cell Biochem. 1985, 27, 317–325. [Google Scholar] [CrossRef] [PubMed]
- Deutzmann, R.; Aumailley, M.; Wiedemann, H.; Pysny, W.; Timpl, R.; Edgar, D. Cell adhesion, spreading and neurite stimulation by laminin fragment E8 depends on maintenance of secondary and tertiary structure in its rod and globular domain. Eur. J. Biochem. 1990, 191, 513–522. [Google Scholar] [CrossRef]
- Rousselle, P.; Golbik, R.; van der Rest, M.; Aumailley, M. Structural requirement for cell adhesion to kalinin (laminin-5). J. Biol. Chem. 1995, 270, 13766–13770. [Google Scholar] [CrossRef] [PubMed]
- Jones, J.C.; Hopkinson, S.B.; Goldfinger, L.E. Structure and assembly of hemidesmosomes. Bioessays 1998, 20, 488–494. [Google Scholar] [CrossRef]
- Legan, P.K.; Collins, J.E.; Garrod, D.R. The molecular biology of desmosomes and hemidesmosomes: “what’s in a name”? Bioessays 1992, 14, 385–393. [Google Scholar] [CrossRef]
- Galliano, M.F.; Aberdam, D.; Aguzzi, A.; Ortonne, J.P.; Meneguzzi, G. Cloning and complete primary structure of the mouse laminin alpha 3 chain. Distinct expression pattern of the laminin alpha 3A and alpha 3B chain isoforms. J. Biol. Chem. 1995, 270, 21820–21826. [Google Scholar] [CrossRef] [PubMed]
- Ryan, M.C.; Tizard, R.; VanDevanter, D.R.; Carter, W.G. Cloning of the LamA3 gene encoding the alpha 3 chain of the adhesive ligand epiligrin. Expression in wound repair. J. Biol. Chem. 1994, 269, 22779–22787. [Google Scholar] [CrossRef] [PubMed]
- Goldfinger, L.E.; Stack, M.S.; Jones, J.C. Processing of laminin-5 and its functional consequences: Role of plasmin and tissue-type plasminogen activator. J. Cell Biol. 1998, 141, 255–265. [Google Scholar] [CrossRef]
- Marinkovich, M.P.; Lunstrum, G.P.; Burgeson, R.E. The anchoring filament protein kalinin is synthesized and secreted as a high molecular weight precursor. J. Biol. Chem. 1992, 267, 17900–17906. [Google Scholar] [CrossRef] [PubMed]
- Giannelli, G.; Falk-Marzillier, J.; Schiraldi, O.; Stetler-Stevenson, W.G.; Quaranta, V. Induction of cell migration by matrix metalloprotease-2 cleavage of laminin-5. Science 1997, 277, 225–228. [Google Scholar] [CrossRef]
- Bair, E.L.; Chen, M.L.; McDaniel, K.; Sekiguchi, K.; Cress, A.E.; Nagle, R.B.; Bowden, G.T. Membrane type 1 matrix metalloprotease cleaves laminin-10 and promotes prostate cancer cell migration. Neoplasia 2005, 7, 380–389. [Google Scholar] [CrossRef]
- Gialeli, C.; Theocharis, A.D.; Karamanos, N.K. Roles of matrix metalloproteinases in cancer progression and their pharmacological targeting. FEBS J. 2011, 278, 16–27. [Google Scholar] [CrossRef] [PubMed]
- Ohtake, Y.; Tojo, H.; Seiki, M. Multifunctional roles of MT1-MMP in myofiber formation and morphostatic maintenance of skeletal muscle. J. Cell Sci. 2006, 119 Pt 18, 3822–3832. [Google Scholar] [CrossRef]
- Wewer, U.M.; Engvall, E. Merosin/laminin-2 and muscular dystrophy. Neuromuscul. Disord. 1996, 6, 409–418. [Google Scholar] [CrossRef] [PubMed]
- Koshikawa, N.; Giannelli, G.; Cirulli, V.; Miyazaki, K.; Quaranta, V. Role of cell surface metalloprotease MT1-MMP in epithelial cell migration over laminin-5. J. Cell Biol. 2000, 148, 615–624. [Google Scholar] [CrossRef]
- Bernfield, M.; Götte, M.; Park, P.W.; Reizes, O.; Fitzgerald, M.L.; Lincecum, J.; Zako, M. Functions of cell surface heparan sulfate proteoglycans. Annu. Rev. Biochem. 1999, 68, 729–777. [Google Scholar] [CrossRef]
- Szatmari, T.; Dobra, K. The role of syndecan-1 in cellular signaling and its effects on heparan sulfate biosynthesis in mesenchymal tumors. Front. Oncol. 2013, 3, 310. [Google Scholar] [CrossRef]
- Nikolova, V.; Koo, C.Y.; Ibrahim, S.A.; Wang, Z.; Spillmann, D.; Dreier, R.; Kelsch, R.; Fischgrabe, J.; Smollich, M.; Rossi, L.H.; et al. Differential roles for membrane-bound and soluble syndecan-1 (CD138) in breast cancer progression. Carcinogenesis 2009, 30, 397–407. [Google Scholar] [CrossRef]
- Endo, K.; Takino, T.; Miyamori, H.; Kinsen, H.; Yoshizaki, T.; Furukawa, M.; Sato, H. Cleavage of syndecan-1 by membrane type matrix metalloproteinase-1 stimulates cell migration. J. Biol. Chem. 2003, 278, 40764–40770. [Google Scholar] [CrossRef]
- Li, Q.; Park, P.W.; Wilson, C.L.; Parks, W.C. Matrilysin shedding of syndecan-1 regulates chemokine mobilization and transepithelial efflux of neutrophils in acute lung injury. Cell 2002, 111, 635–646. [Google Scholar] [CrossRef] [PubMed]
- Nadanaka, S.; Bai, Y.; Kitagawa, H. Cleavage of Syndecan-1 Promotes the Proliferation of the Basal-Like Breast Cancer Cell Line BT-549 Via Akt SUMOylation. Front. Cell Dev. Biol. 2021, 9, 659428. [Google Scholar] [CrossRef]
- Knudson, C.B.; Knudson, W. Cartilage Proteoglycans; Seminars in Cell & Developmental Biology; Elsevier: Amsterdam, The Netherlands, 2001; pp. 69–78. [Google Scholar]
- Ying, S.; Shiraishi, A.; Kao, C.W.-C.; Converse, R.L.; Funderburgh, J.L.; Swiergiel, J.; Roth, M.R.; Conrad, G.W.; Kao, W.W.-Y. Characterization and expression of the mouse lumican gene. J. Biol. Chem. 1997, 272, 30306–30313. [Google Scholar] [CrossRef]
- Blochberger, T.C.; Vergnes, J.P.; Hempel, J.; Hassell, J.R. cDNA to chick lumican (corneal keratan sulfate proteoglycan) reveals homology to the small interstitial proteoglycan gene family and expression in muscle and intestine. J. Biol. Chem. 1992, 267, 347–352. [Google Scholar] [CrossRef] [PubMed]
- Yoshioka, N.; Inoue, H.; Nakanishi, K.; Oka, K.; Yutsudo, M.; Yamashita, A.; Hakura, A.; Nojima, H. Isolation of transformation suppressor genes by cDNA subtraction: Lumican suppresses transformation induced by v-src and vK-ras. J. Virol. 2000, 74, 1008–1013. [Google Scholar] [CrossRef]
- Pietraszek, K.; Chatron-Colliet, A.; Brezillon, S.; Perreau, C.; Jakubiak-Augustyn, A.; Krotkiewski, H.; Maquart, F.X.; Wegrowski, Y. Lumican: A new inhibitor of matrix metalloproteinase-14 activity. FEBS Lett. 2014, 588, 4319–4324. [Google Scholar] [CrossRef]
- Li, Y.; Aoki, T.; Mori, Y.; Ahmad, M.; Miyamori, H.; Takino, T.; Sato, H. Cleavage of lumican by membrane-type matrix metalloproteinase-1 abrogates this proteoglycan-mediated suppression of tumor cell colony formation in soft agar. Cancer Res. 2004, 64, 7058–7064. [Google Scholar] [CrossRef]
- Yana, I.; Weiss, S.J. Regulation of membrane type-1 matrix metalloproteinase activation by proprotein convertases. Mol. Biol. Cell 2000, 11, 2387–2401. [Google Scholar] [CrossRef] [PubMed]
- Maskos, K. Crystal structures of MMPs in complex with physiological and pharmacological inhibitors. Biochimie. 2005, 87, 249–263. [Google Scholar] [CrossRef]
- Van Wart, H.E.; Birkedal-Hansen, H. The cysteine switch: A principle of regulation of metalloproteinase activity with potential applicability to the entire matrix metalloproteinase gene family. Proc. Natl. Acad. Sci. USA 1990, 87, 5578–5582. [Google Scholar] [CrossRef] [PubMed]
- Jozic, D.; Bourenkov, G.; Lim, N.H.; Visse, R.; Nagase, H.; Bode, W.; Maskos, K. X-ray structure of human proMMP-1: New insights into procollagenase activation and collagen binding. J. Biol. Chem. 2005, 280, 9578–9585. [Google Scholar] [CrossRef] [PubMed]
- Nagase, H. Activation mechanisms of matrix metalloproteinases. Biol. Chem. 1997, 378, 151–160. [Google Scholar] [PubMed]
- Busti, C.; Falcinelli, E.; Momi, S.; Gresele, P. Matrix metalloproteinases and peripheral arterial disease. Intern. Emerg. Med. 2010, 5, 13–25. [Google Scholar] [CrossRef]
- Itoh, T.; Tanioka, M.; Yoshida, H.; Yoshioka, T.; Nishimoto, H.; Itohara, S. Reduced angiogenesis and tumor progression in gelatinase A-deficient mice. Cancer Res. 1998, 58, 1048–1051. [Google Scholar]
- Nagase, H. Cell surface activation of progelatinase A (proMMP-2) and cell migration. Cell Res. 1998, 8, 179–186. [Google Scholar] [CrossRef]
- Strongin, A.Y.; Collier, I.; Bannikov, G.; Marmer, B.L.; Grant, G.A.; Goldberg, G.I. Mechanism of cell surface activation of 72-kDa type IV collagenase. Isolation of the activated form of the membrane metalloprotease. J. Biol. Chem. 1995, 270, 5331–5338. [Google Scholar] [CrossRef]
- Fernandez-Catalan, C.; Bode, W.; Huber, R.; Turk, D.; Calvete, J.J.; Lichte, A.; Tschesche, H.; Maskos, K. Crystal structure of the complex formed by the membrane type 1-matrix metalloproteinase with the tissue inhibitor of metalloproteinases-2, the soluble progelatinase A receptor. EMBO J. 1998, 17, 5238–5248. [Google Scholar] [CrossRef]
- Bernardo, M.M.; Fridman, R. TIMP-2 (tissue inhibitor of metalloproteinase-2) regulates MMP-2 (matrix metalloproteinase-2) activity in the extracellular environment after pro-MMP-2 activation by MT1 (membrane type 1)-MMP. Biochem. J. 2003, 374, 739–745. [Google Scholar] [CrossRef]
- English, J.L.; Kassiri, Z.; Koskivirta, I.; Atkinson, S.J.; Di Grappa, M.; Soloway, P.D.; Nagase, H.; Vuorio, E.; Murphy, G.; Khokha, R. Individual Timp deficiencies differentially impact pro-MMP-2 activation. J. Biol. Chem. 2006, 281, 10337–10346. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Z.; Apte, S.S.; Soininen, R.; Cao, R.; Baaklini, G.Y.; Rauser, R.W.; Wang, J.; Cao, Y.; Tryggvason, K. Impaired endochondral ossification and angiogenesis in mice deficient in membrane-type matrix metalloproteinase I. Proc. Natl. Acad. Sci. USA 2000, 97, 4052–4057. [Google Scholar] [CrossRef]
- Knauper, V.; Will, H.; Lopez-Otin, C.; Smith, B.; Atkinson, S.J.; Stanton, H.; Hembry, R.M.; Murphy, G. Cellular mechanisms for human procollagenase-3 (MMP-13) activation. Evidence that MT1-MMP (MMP-14) and gelatinase a (MMP-2) are able to generate active enzyme. J. Biol. Chem. 1996, 271, 17124–17131. [Google Scholar]
- Schneider, F.; Sukhova, G.K.; Aikawa, M.; Canner, J.; Gerdes, N.; Tang, S.M.; Shi, G.P.; Apte, S.S.; Libby, P. Matrix-metalloproteinase-14 deficiency in bone-marrow-derived cells promotes collagen accumulation in mouse atherosclerotic plaques. Circulation 2008, 117, 931–939. [Google Scholar] [CrossRef]
- Hu, Q.; Ecker, M. Overview of MMP-13 as a Promising Target for the Treatment of Osteoarthritis. Int. J. Mol. Sci. 2021, 22, 1742. [Google Scholar] [CrossRef]
- Wang, Z.; Juttermann, R.; Soloway, P.D. TIMP-2 is required for efficient activation of proMMP-2 in vivo. J. Biol. Chem. 2000, 275, 26411–26415. [Google Scholar] [CrossRef] [PubMed]
- Caterina, J.J.; Yamada, S.; Caterina, N.C.; Longenecker, G.; Holmback, K.; Shi, J.; Yermovsky, A.E.; Engler, J.A.; Birkedal-Hansen, H. Inactivating mutation of the mouse tissue inhibitor of metalloproteinases-2(Timp-2) gene alters proMMP-2 activation. J. Biol. Chem. 2000, 275, 26416–26422. [Google Scholar] [CrossRef]
- Ferrara, N.; Houck, K.; Jakeman, L.; Leung, D.W. Molecular and biological properties of the vascular endothelial growth factor family of proteins. Endocr. Rev. 1992, 13, 18–32. [Google Scholar] [CrossRef]
- Chintalgattu, V.; Nair, D.M.; Katwa, L.C. Cardiac myofibroblasts: A novel source of vascular endothelial growth factor (VEGF) and its receptors Flt-1 and KDR. J. Mol. Cell Cardiol. 2003, 35, 277–286. [Google Scholar] [CrossRef] [PubMed]
- Ferrara, N.; Carver-Moore, K.; Chen, H.; Dowd, M.; Lu, L.; O’Shea, K.S.; Powell-Braxton, L.; Hillan, K.J.; Moore, M.W. Heterozygous embryonic lethality induced by targeted inactivation of the VEGF gene. Nature 1996, 380, 439–442. [Google Scholar] [CrossRef]
- Hattori, K.; Dias, S.; Heissig, B.; Hackett, N.R.; Lyden, D.; Tateno, M.; Hicklin, D.J.; Zhu, Z.; Witte, L.; Crystal, R.G.; et al. Vascular endothelial growth factor and angiopoietin-1 stimulate postnatal hematopoiesis by recruitment of vasculogenic and hematopoietic stem cells. J. Exp. Med. 2001, 193, 1005–1014. [Google Scholar] [CrossRef] [PubMed]
- Han, K.Y.; Dugas-Ford, J.; Lee, H.; Chang, J.H.; Azar, D.T. MMP14 Cleavage of VEGFR1 in the Cornea Leads to a VEGF-Trap Antiangiogenic Effect. Investig. Ophthalmol. Vis. Sci. 2015, 56, 5450–5456. [Google Scholar] [CrossRef]
- Han, K.Y.; Chang, J.H.; Lee, H.; Azar, D.T. Proangiogenic Interactions of Vascular Endothelial MMP14 With VEGF Receptor 1 in VEGFA-Mediated Corneal Angiogenesis. Investig. Ophthalmol. Vis. Sci. 2016, 57, 3313–3322. [Google Scholar] [CrossRef]
- Beenken, A.; Mohammadi, M. The FGF family: Biology, pathophysiology and therapy. Nat. Rev. Drug. Discov. 2009, 8, 235–253. [Google Scholar] [CrossRef]
- Chang, J.-H.; Huang, Y.-H.; Cunningham, C.M.; Han, K.-Y.; Chang, M.; Seiki, M.; Zhou, Z.; Azar, D.T. Matrix metalloproteinase 14 modulates signal transduction and angiogenesis in the cornea. Surv. Ophthalmol. 2016, 61, 478–497. [Google Scholar]
- Chan, K.M.; Wong, H.L.; Jin, G.; Liu, B.; Cao, R.; Cao, Y.; Lehti, K.; Tryggvason, K.; Zhou, Z. MT1-MMP inactivates ADAM9 to regulate FGFR2 signaling and calvarial osteogenesis. Dev. Cell 2012, 22, 1176–1190. [Google Scholar] [CrossRef] [PubMed]
- Cukier, A.M.; Therond, P.; Didichenko, S.A.; Guillas, I.; Chapman, M.J.; Wright, S.D.; Kontush, A. Structure-function relationships in reconstituted HDL: Focus on antioxidative activity and cholesterol efflux capacity. Biochim. Biophys. Acta (BBA.)-Mol. Cell Biol. Lipids 2017, 1862, 890–900. [Google Scholar] [CrossRef] [PubMed]
- Yu, X.-H.; Zhang, D.-W.; Zheng, X.-L.; Tang, C.-K. Cholesterol transport system: An integrated cholesterol transport model involved in atherosclerosis. Prog. Lipid Res. 2019, 73, 65–91. [Google Scholar]
- Walldius, G.; Jungner, I. Apolipoprotein B and apolipoprotein A-I: Risk indicators of coronary heart disease and targets for lipid-modifying therapy. J. Intern. Med. 2004, 255, 188–205. [Google Scholar] [CrossRef]
- Schaefer, E.J.; Heaton, W.H.; Wetzel, M.G.; Brewer, H.B., Jr. Plasma apolipoprotein A-1 absence associated with a marked reduction of high density lipoproteins and premature coronary artery disease. Arteriosclerosis 1982, 2, 16–26. [Google Scholar] [CrossRef]
- Hwang, I.K.; Park, S.M.; Kim, S.Y.; Lee, S.T. A proteomic approach to identify substrates of matrix metalloproteinase-14 in human plasma. Biochim. Biophys. Acta 2004, 1702, 79–87. [Google Scholar] [CrossRef] [PubMed]
- Park, J.H.; Park, S.M.; Park, K.H.; Cho, K.H.; Lee, S.T. Analysis of apolipoprotein A-I as a substrate for matrix metalloproteinase-14. Biochem. Biophys. Res. Commun. 2011, 409, 58–63. [Google Scholar] [CrossRef] [PubMed]
- Xia, X.-D.; Alabi, A.; Wang, M.; Gu, H.-M.; Yang, R.Z.; Wang, G.-Q.; Zhang, D.-W. Membrane-type I matrix metalloproteinase (MT1-MMP), lipid metabolism, and therapeutic implications. J. Mol. Cell Biol. 2021, 13, 513–526. [Google Scholar] [CrossRef] [PubMed]
- Naor, D.; Sionov, R.V.; Ish-Shalom, D. CD44: Structure, function, and association with the malignant process. Adv. Cancer Res. 1997, 71, 241–319. [Google Scholar]
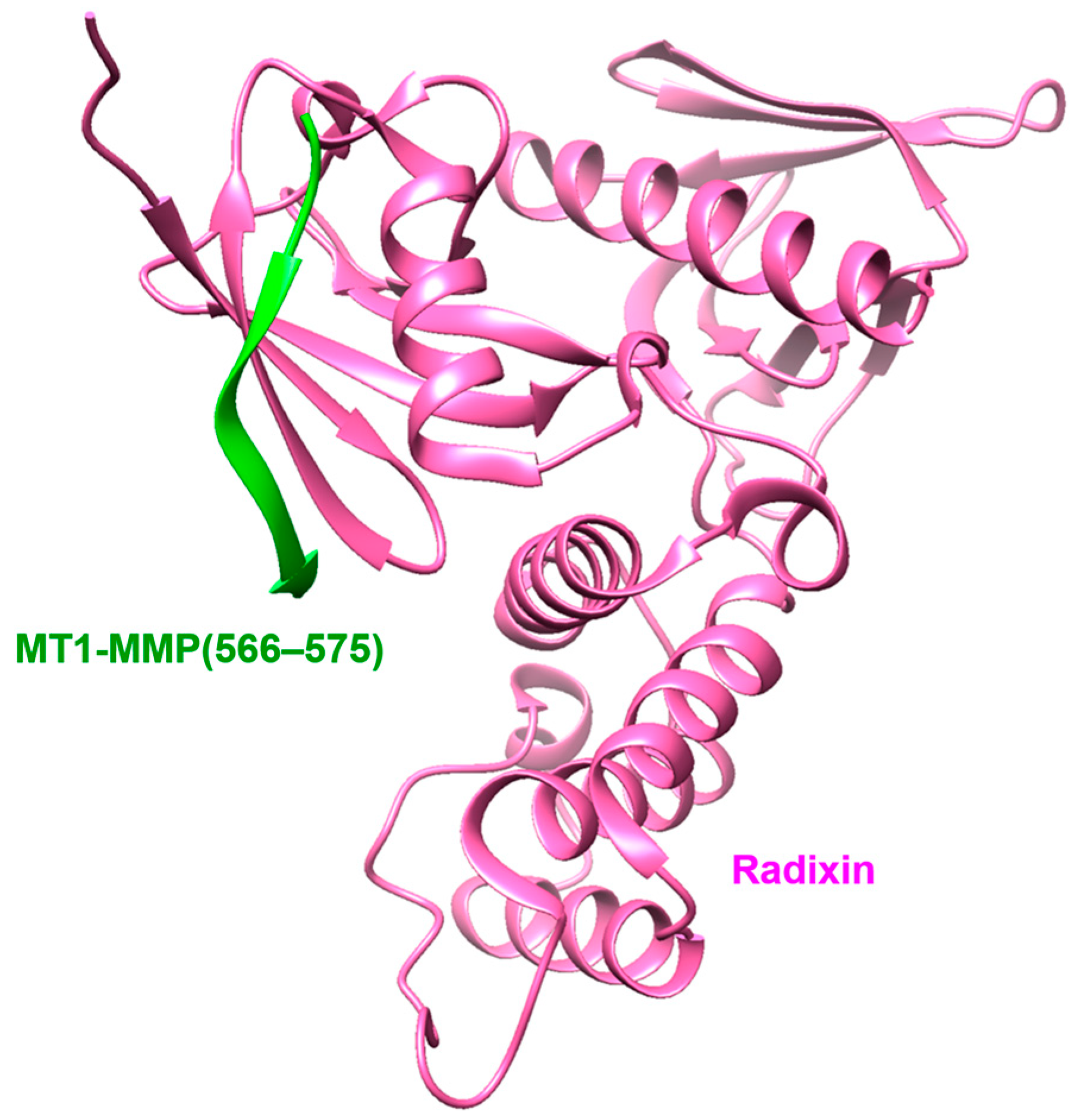
- Terawaki, S.; Kitano, K.; Aoyama, M.; Mori, T.; Hakoshima, T. MT1-MMP recognition by ERM proteins and its implication in CD44 shedding. Genes Cells 2015, 20, 847–859. [Google Scholar] [CrossRef]
- Haynes, B.F.; Hale, L.P.; Patton, K.L.; Martin, M.E.; McCallum, R.M. Measurement of an adhesion molecule as an indicator of inflammatory disease activity. Up-regulation of the receptor for hyaluronate (CD44) in rheumatoid arthritis. Arthritis Rheum 1991, 34, 1434–1443. [Google Scholar] [CrossRef]
- Goebeler, M.; Kaufmann, D.; Brocker, E.B.; Klein, C.E. Migration of highly aggressive melanoma cells on hyaluronic acid is associated with functional changes, increased turnover and shedding of CD44 receptors. J. Cell Sci. 1996, 109 Pt 7, 1957–1964. [Google Scholar] [CrossRef]
- Kajita, M.; Itoh, Y.; Chiba, T.; Mori, H.; Okada, A.; Kinoh, H.; Seiki, M. Membrane-type 1 matrix metalloproteinase cleaves CD44 and promotes cell migration. J. Cell Biol. 2001, 153, 893–904. [Google Scholar] [CrossRef]
- Zarrabi, K.; Dufour, A.; Li, J.; Kuscu, C.; Pulkoski-Gross, A.; Zhi, J.; Hu, Y.; Sampson, N.S.; Zucker, S.; Cao, J. Inhibition of matrix metalloproteinase 14 (MMP-14)-mediated cancer cell migration. J. Biol. Chem. 2011, 286, 33167–33177. [Google Scholar] [CrossRef]
- Kong, Y.Y.; Yoshida, H.; Sarosi, I.; Tan, H.L.; Timms, E.; Capparelli, C.; Morony, S.; Oliveira-dos-Santos, A.J.; Van, G.; Itie, A.; et al. OPGL is a key regulator of osteoclastogenesis, lymphocyte development and lymph-node organogenesis. Nature 1999, 397, 315–323. [Google Scholar] [CrossRef] [PubMed]
- Jones, D.H.; Kong, Y.Y.; Penninger, J.M. Role of RANKL and RANK in bone loss and arthritis. Ann. Rheum Dis. 2002, 61 (Suppl. 2), ii32–ii39. [Google Scholar] [CrossRef]
- Delgado-Calle, J.; Hancock, B.; Likine, E.F.; Sato, A.Y.; McAndrews, K.; Sanudo, C.; Bruzzaniti, A.; Riancho, J.A.; Tonra, J.R.; Bellido, T. MMP14 is a novel target of PTH signaling in osteocytes that controls resorption by regulating soluble RANKL production. FASEB J. 2018, 32, 2878–2890. [Google Scholar] [CrossRef] [PubMed]
- Hikita, A.; Yana, I.; Wakeyama, H.; Nakamura, M.; Kadono, Y.; Oshima, Y.; Nakamura, K.; Seiki, M.; Tanaka, S. Negative regulation of osteoclastogenesis by ectodomain shedding of receptor activator of NF-kappaB ligand. J. Biol. Chem. 2006, 281, 36846–36855. [Google Scholar] [CrossRef] [PubMed]
- Sabbota, A.L.; Kim, H.R.; Zhe, X.; Fridman, R.; Bonfil, R.D.; Cher, M.L. Shedding of RANKL by tumor-associated MT1-MMP activates Src-dependent prostate cancer cell migration. Cancer Res. 2010, 70, 5558–5566. [Google Scholar] [CrossRef]
- Chen, G.; Sircar, K.; Aprikian, A.; Potti, A.; Goltzman, D.; Rabbani, S.A. Expression of RANKL/RANK/OPG in primary and metastatic human prostate cancer as markers of disease stage and functional regulation. Cancer 2006, 107, 289–298. [Google Scholar] [CrossRef]
- Nanba, D.; Higashiyama, S. Dual intracellular signaling by proteolytic cleavage of membrane-anchored heparin-binding EGF-like growth factor. Cytokine Growth Factor Rev. 2004, 15, 13–19. [Google Scholar] [CrossRef]
- Tarbé, N.; Lösch, S.; Burtscher, H.; Jarsch, M.; Weidle, U.H. Identification of rat pancreatic carcinoma genes associated with lymphogenous metastasis. Anticancer Res. 2002, 22, 2015–2027. [Google Scholar]
- Suo, Z.; Risberg, B.; Karlsson, M.G.; Villman, K.; Skovlund, E.; Nesland, J.M. The expression of EGFR family ligands in breast carcinomas. Int. J. Surg. Pathol. 2002, 10, 91–99. [Google Scholar] [CrossRef] [PubMed]
- Wang, F.; Liu, R.; Lee, S.W.; Sloss, C.M.; Couget, J.; Cusack, J.C. Heparin-binding EGF-like growth factor is an early response gene to chemotherapy and contributes to chemotherapy resistance. Oncogene 2007, 26, 2006–2016. [Google Scholar] [CrossRef]
- Koshikawa, N.; Mizushima, H.; Minegishi, T.; Eguchi, F.; Yotsumoto, F.; Nabeshima, K.; Miyamoto, S.; Mekada, E.; Seiki, M. Proteolytic activation of heparin-binding EGF-like growth factor by membrane-type matrix metalloproteinase-1 in ovarian carcinoma cells. Cancer Sci. 2011, 102, 111–116. [Google Scholar] [CrossRef]
- Stawowczyk, M.; Wellenstein, M.D.; Lee, S.B.; Yomtoubian, S.; Durrans, A.; Choi, H.; Narula, N.; Altorki, N.K.; Gao, D.; Mittal, V. Matrix Metalloproteinase 14 promotes lung cancer by cleavage of Heparin-Binding EGF-like Growth Factor. Neoplasia 2017, 19, 55–64. [Google Scholar] [CrossRef] [PubMed]


Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lee, H.; Ibrahimi, L.; Azar, D.T.; Han, K.-Y. The Role of Membrane-Type 1 Matrix Metalloproteinase–Substrate Interactions in Pathogenesis. Int. J. Mol. Sci. 2023, 24, 2183. https://doi.org/10.3390/ijms24032183
Lee H, Ibrahimi L, Azar DT, Han K-Y. The Role of Membrane-Type 1 Matrix Metalloproteinase–Substrate Interactions in Pathogenesis. International Journal of Molecular Sciences. 2023; 24(3):2183. https://doi.org/10.3390/ijms24032183
Chicago/Turabian StyleLee, Hyun, Lucas Ibrahimi, Dimitri T. Azar, and Kyu-Yeon Han. 2023. "The Role of Membrane-Type 1 Matrix Metalloproteinase–Substrate Interactions in Pathogenesis" International Journal of Molecular Sciences 24, no. 3: 2183. https://doi.org/10.3390/ijms24032183
APA StyleLee, H., Ibrahimi, L., Azar, D. T., & Han, K.-Y. (2023). The Role of Membrane-Type 1 Matrix Metalloproteinase–Substrate Interactions in Pathogenesis. International Journal of Molecular Sciences, 24(3), 2183. https://doi.org/10.3390/ijms24032183





