Characterization and Genomic Analysis of a Novel Lytic Phage DCp1 against Clostridium perfringens Biofilms
Abstract
1. Introduction
2. Results
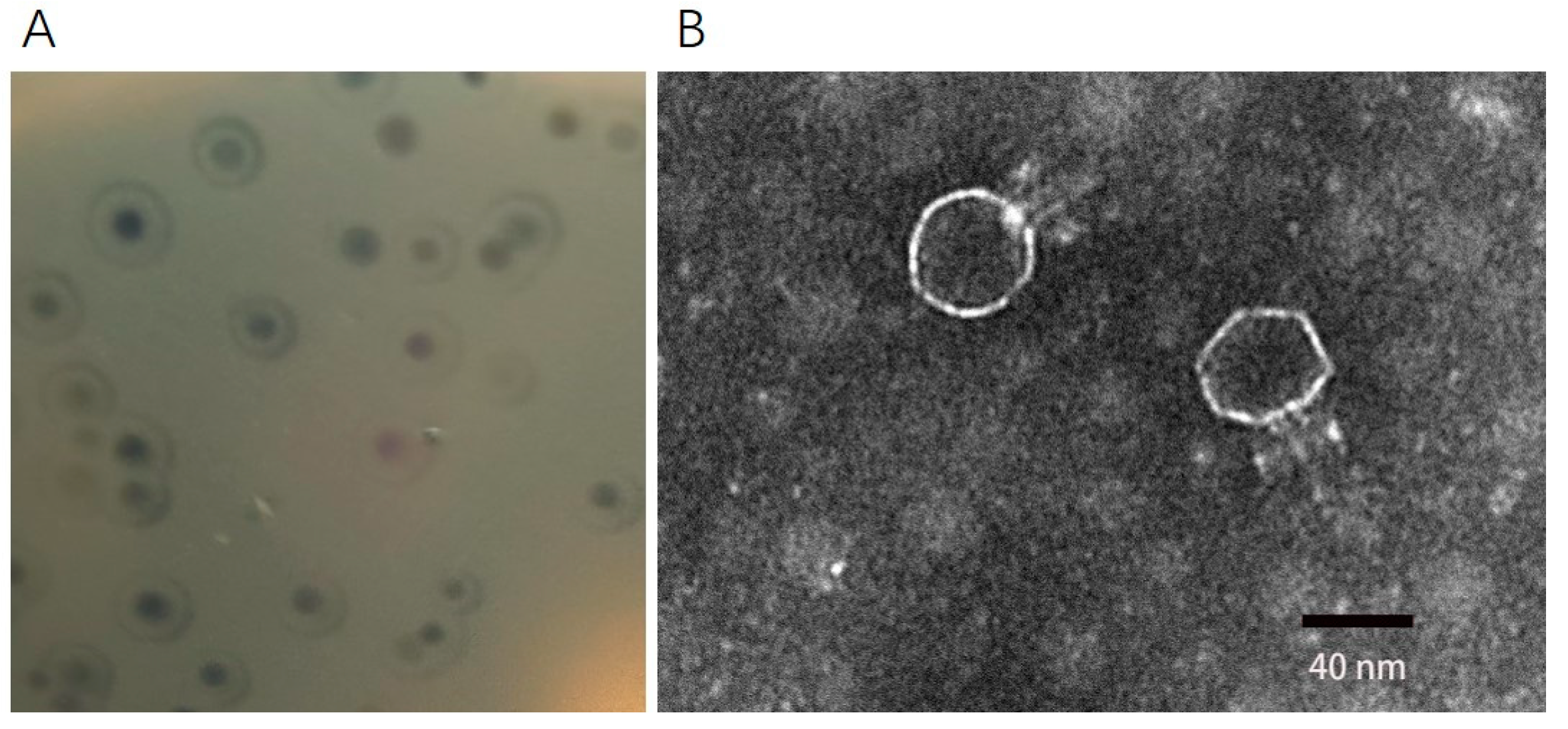
2.1. Morphology of Phage DCp1
2.2. Host Range and EOP
2.3. The Optimal MOI and One-Step Growth Curve of Phage DCp1
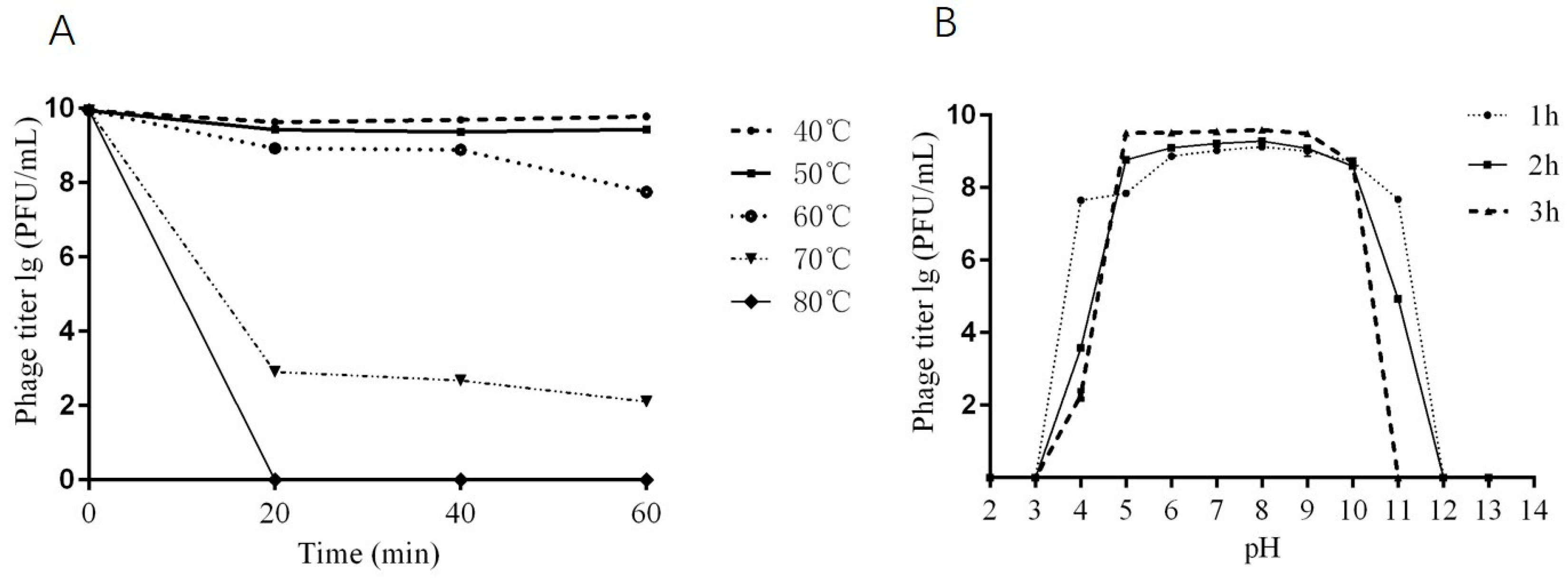
2.4. Thermal and pH Stability of Phage DCp1
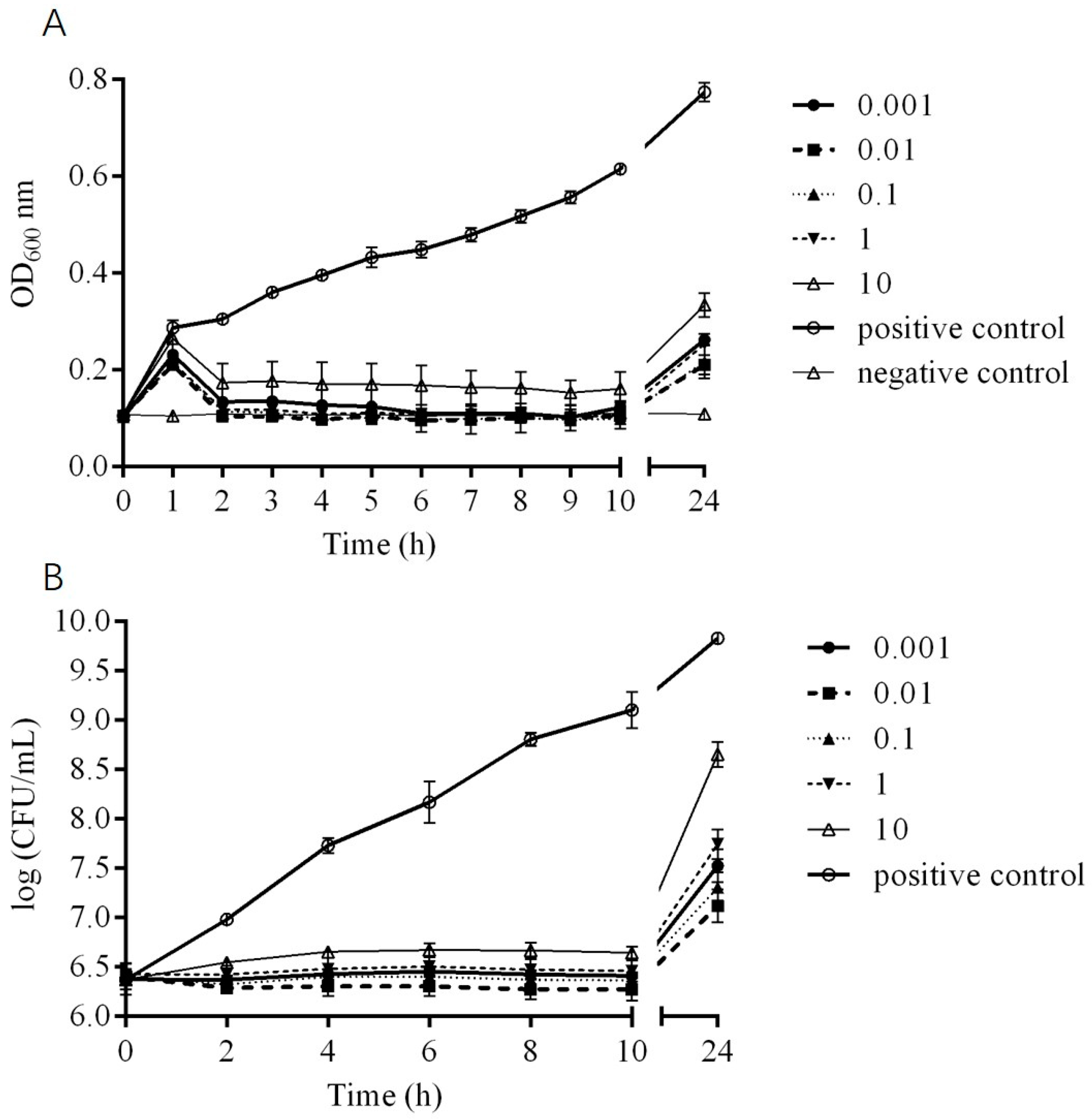
2.5. In Vitro Bactericidal Activity of Phage DCp1
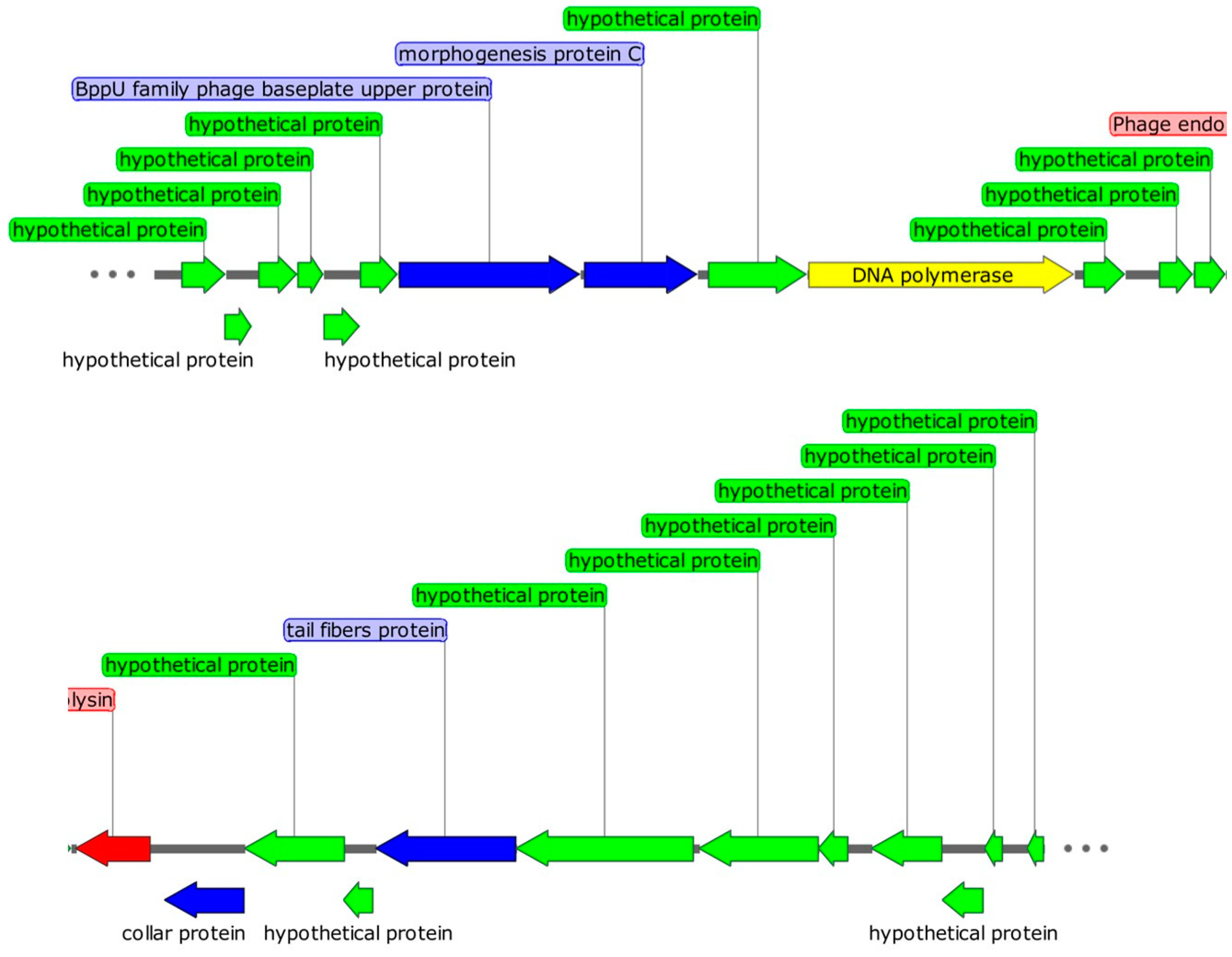
2.6. Genomic Characteristics of Phage DCp1
2.7. Comparative Analysis
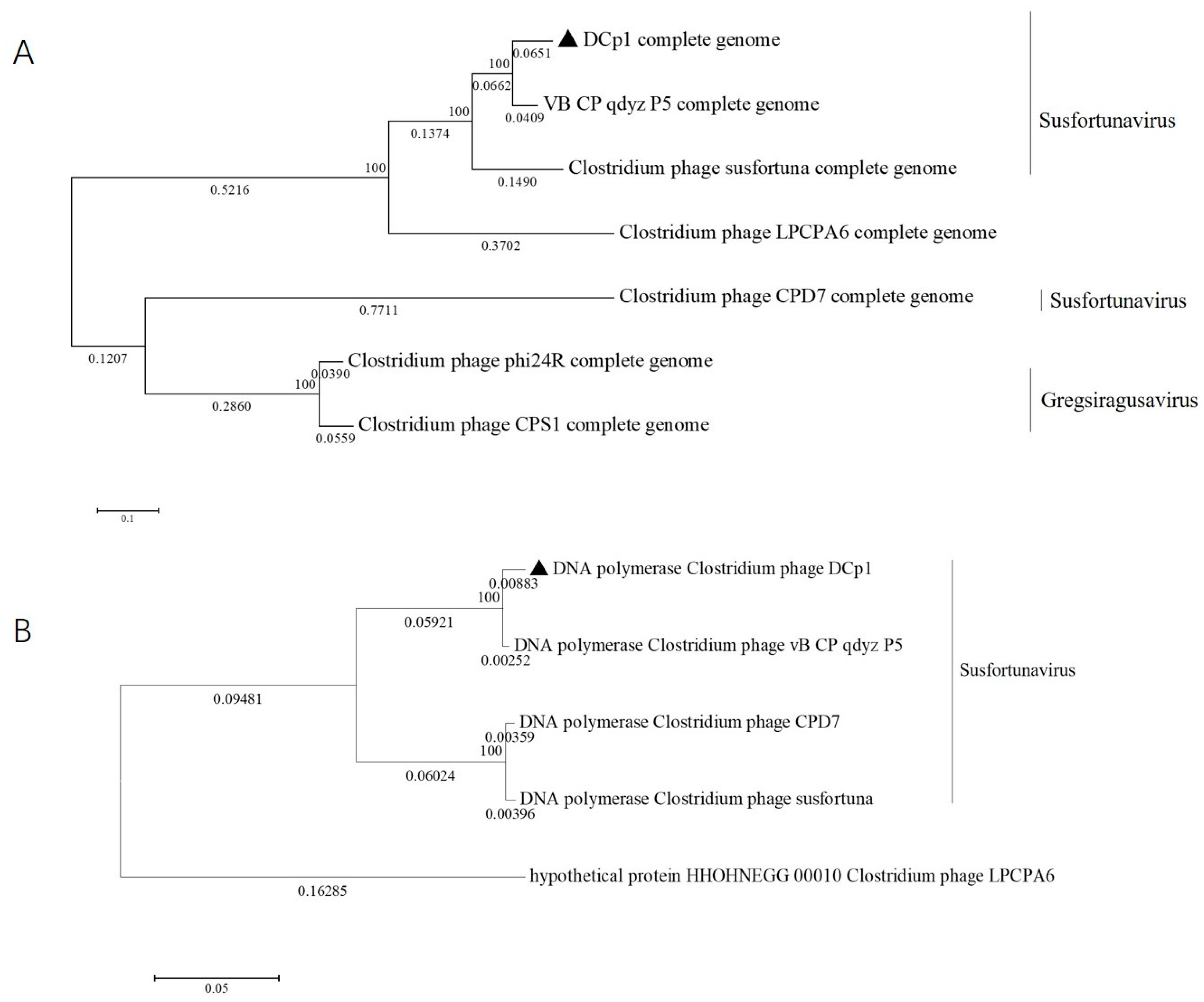
2.8. Phylogenetic Analysis
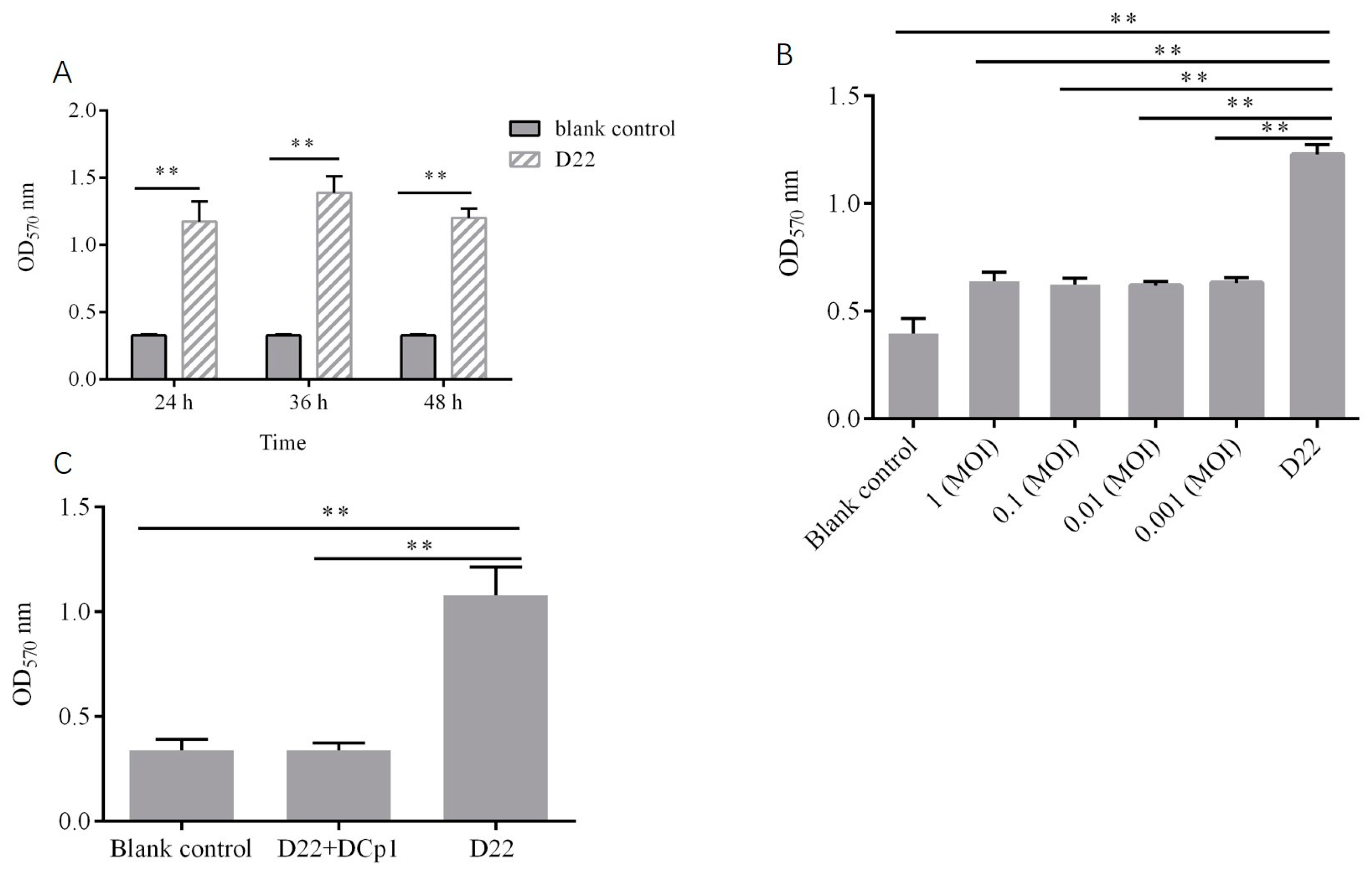
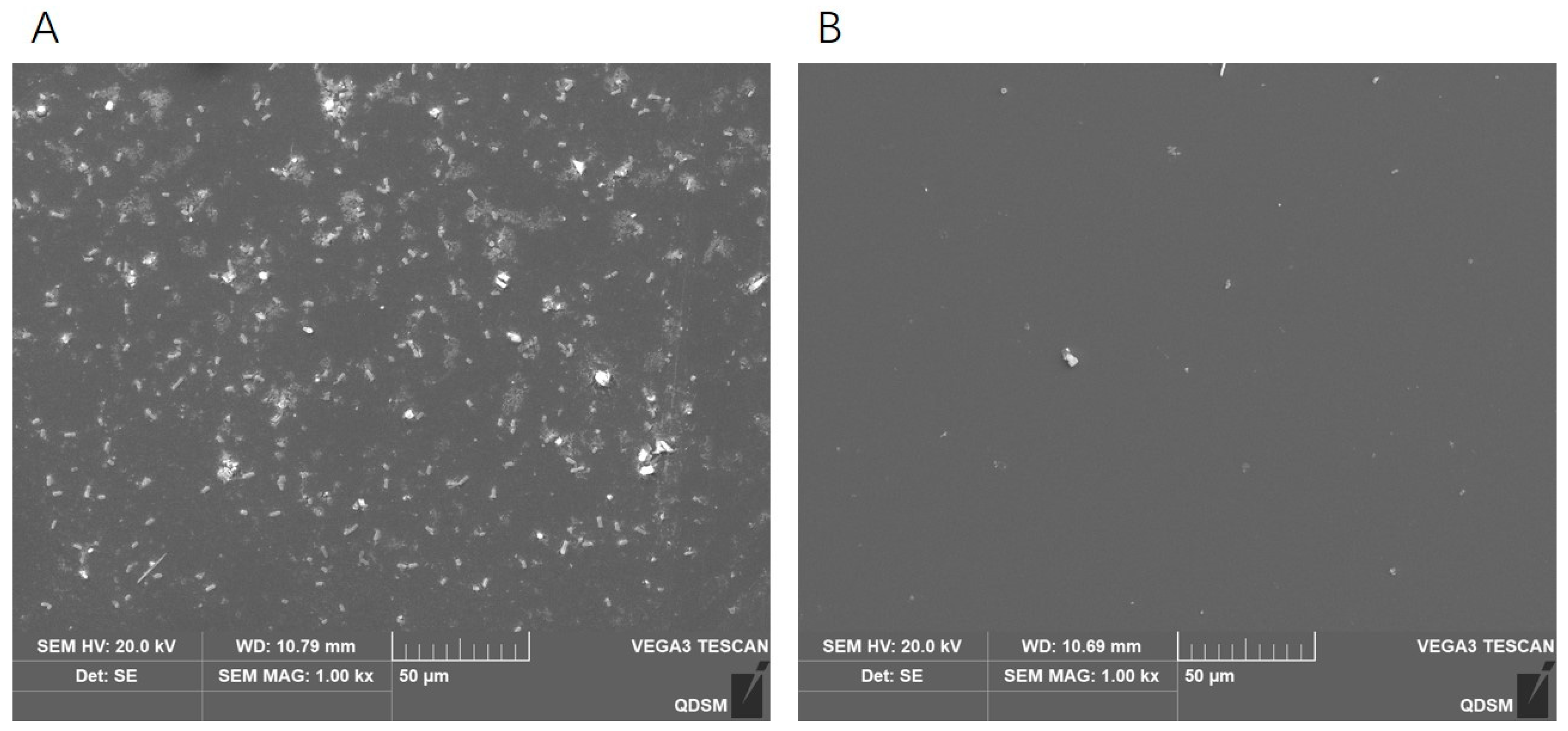
2.9. Effects of Phage DCp1 on the Biofilm of C. perfringens
3. Discussion
4. Materials and Methods
4.1. Strains and Conditions
4.2. Sample Enrichment and Bacteriophage Isolation
4.3. Electron Microscopy
4.4. Host Range and Efficiency of Plating (EOP) Measurement
4.5. Optimal Multiplicity of Infection (MOI)
4.6. One-Step Growth Curve
4.7. Thermal and pH Stability
4.8. In Vitro Bactericidal Activity
4.9. Genome Extraction, Sequencing, and Bioinformatics Analysis
4.10. Effects of Phage DCp1 on C. perfringens Biofilm
4.10.1. Biofilm Assay
4.10.2. Inhibitory Effect of Phage DCp1 on Biofilm Formation
4.10.3. Eradication of C. perfringens Biofilms by Phage DCp1
4.11. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Stanley, D.; Keyburn, A.L.; Denman, S.E.; Moore, R.J. Changes in the caecal microflora of chickens following Clostridium perfringens challenge to induce necrotic enteritis. Veter. Microbiol. 2012, 159, 155–162. [Google Scholar] [CrossRef] [PubMed]
- Feng, Y.; Gong, J.; Yu, H.; Jin, Y.; Zhu, J.; Han, Y. Identification of changes in the composition of ileal bacterial microbiota of broiler chickens infected with Clostridium perfringens. Veter. Microbiol. 2010, 140, 116–121. [Google Scholar] [CrossRef]
- Gohari, I.M.; Unterer, S.; Whitehead, A.E.; Prescott, J.F. NetF-producing Clostridium perfringens and its associated diseases in dogs and foals. J. Veter. Diagn. Investig. 2020, 32, 230–238. [Google Scholar] [CrossRef] [PubMed]
- Petit, L.; Gibert, M.; Popoff, M.R. Clostridium perfringens: Toxinotype and genotype. Trends Microbiol. 1999, 7, 104–110. [Google Scholar] [CrossRef] [PubMed]
- Gohari, I.M.; Arroyo, L.; Macinnes, J.I.; Timoney, J.F.; Parreira, V.R.; Prescott, J.F. Characterization of Clostridium perfringens in the feces of adult horses and foals with acute enterocolitis. Can. J. Vet. Res. 2014, 78, 1–7. [Google Scholar] [PubMed]
- Choi, Y.-K.; Kang, M.-S.; Yoo, H.-S.; Lee, D.-Y.; Lee, H.-C.; Kim, D.-Y. Clostridium perfringens type A Myonecrosis in a Horse in Korea. J. Veter. Med. Sci. 2003, 65, 1245–1247. [Google Scholar] [CrossRef] [PubMed]
- Sacco, S.C.; Ortega, J.; Navarro, M.A.; Fresneda, K.C.; Anderson, M.; Woods, L.W.; Moore, J.; Uzal, F.A. Clostridium sordellii-associated gas gangrene in 8 horses, 1998–2019. J. Veter. Diagn. Investig. 2020, 32, 246–251. [Google Scholar] [CrossRef]
- Kiu, R.; Hall, L.J. An update on the human and animal enteric pathogen Clostridium perfringens. Emerg. Microbes Infect. 2018, 7, 1–15. [Google Scholar] [CrossRef]
- Millet, S.; Maertens, L. The European ban on antibiotic growth promoters in animal feed: From challenges to opportunities. Vet. J. 2011, 187, 143–144. [Google Scholar] [CrossRef]
- Altamirano, F.L.G.; Barr, J.J. Phage Therapy in the Postantibiotic Era. Clin. Microbiol. Rev. 2019, 32, e00066-18. [Google Scholar] [CrossRef]
- Kortright, K.E.; Chan, B.K.; Koff, J.L.; Turner, P.E. Phage Therapy: A Renewed Approach to Combat Antibiotic-Resistant Bacteria. Cell Host Microbe 2019, 25, 219–232. [Google Scholar] [CrossRef]
- Reardon, S. Phage therapy gets revitalized. Nature 2014, 510, 15–16. [Google Scholar] [CrossRef]
- Rea, M.C.; Dobson, A.; O’Sullivan, O.; Crispie, F.; Fouhy, F.; Cotter, P.D.; Shanahan, F.; Kiely, B.; Hill, C.; Ross, R.P. Effect of broad- and narrow-spectrum antimicrobials on Clostridium difficile and microbial diversity in a model of the distal colon. Proc. Natl. Acad. Sci. USA 2011, 108, 4639–4644. [Google Scholar] [CrossRef]
- Yuan, X.; Zhang, S.; Wang, J.; Li, C.; Li, N.; Yu, S.; Kong, L.; Zeng, H.; Yang, G.; Huang, Y.; et al. Isolation and characterization of a novel Escherichia coli Kayfunavirus phage DY1. Virus Res. 2021, 293, 198274. [Google Scholar] [CrossRef] [PubMed]
- Doolittle, M.M.; Cooney, J.J.; Caldwell, D.E. Lytic infection of Escherichia coli biofilms by bacteriophage T4. Can. J. Microbiol. 1995, 41, 12–18. [Google Scholar] [CrossRef]
- Tian, Y.; Wu, L.; Lu, R.; Bao, H.; Zhou, Y.; Pang, M.; Brown, J.; Wang, J.; Wang, R.; Zhang, H. Virulent phage vB_CpeP_HN02 inhibits Clostridium perfringens on the surface of the chicken meat. Int. J. Food Microbiol. 2022, 363, 109514. [Google Scholar] [CrossRef] [PubMed]
- Huang, S.; Tian, Y.; Wang, Y.; García, P.; Liu, B.; Lu, R.; Wu, L.; Bao, H.; Pang, M.; Zhou, Y.; et al. The Broad Host Range Phage vB_CpeS_BG3P Is Able to Inhibit Clostridium perfringens Growth. Viruses 2022, 14, 676. [Google Scholar] [CrossRef]
- Lu, H.; Yan, P.; Xiong, W.; Wang, J.; Liu, X. Genomic characterization of a novel virulent phage infecting Shigella fiexneri and isolated from sewage. Virus Res. 2020, 283, 197983. [Google Scholar] [CrossRef] [PubMed]
- Chen, Z. Information theory based T7-like promoter models: Classification of bacteriophages and differential evolution of promoters and their polymerases. Nucleic. Acids Res. 2005, 33, 6172–6187. [Google Scholar] [CrossRef]
- Diab, S.S.; Kinde, H.; Moore, J.; Shahriar, M.F.; Odani, J.; Anthenill, L.; Songer, G.; Uzal, F.A. Pathology of Clostridium perfringens Type C Enterotoxemia in Horses. Veter. Pathol. 2012, 49, 255–263. [Google Scholar] [CrossRef]
- Liu, W.; Han, L.; Song, P.; Sun, H.; Zhang, C.; Zou, L.; Cui, J.; Pan, Q.; Ren, H. Complete genome sequencing of a Tequintavirus bacteriophage with a broad host range against Salmonella Abortus equi isolates from donkeys. Front. Microbiol. 2022, 13, 938616. [Google Scholar] [CrossRef]
- Montso, P.K.; Mlambo, V.; Ateba, C.N. Efficacy of novel phages for control of multi-drug resistant Escherichia coli O177 on artificially contaminated beef and their potential to disrupt biofilm formation. Food Microbiol. 2021, 94, 103647. [Google Scholar] [CrossRef]
- Park, D.-W.; Park, J.-H. Characterization and Food Application of the Novel Lytic Phage BECP10: Specifically Recognizes the O-polysaccharide of Escherichia coli O157:H7. Viruses 2021, 13, 1469. [Google Scholar] [CrossRef] [PubMed]
- Sui, B.; Qi, X.; Wang, X.; Ren, H.; Liu, W.; Zhang, C. Characterization of a Novel Bacteriophage swi2 Harboring Two Lysins Can Naturally Lyse Escherichia coli. Front. Microbiol. 2021, 12, 670799. [Google Scholar] [CrossRef]
- Horiuchi, K.; Adelberg, E.A. Growth of Male-Specific Bacteriophage in Proteus mirabilis Harboring F-Genotes Derived from Escherichia coli. J. Bacteriol. 1965, 89, 1231–1236. [Google Scholar] [CrossRef] [PubMed]
- Hamza, A.; Perveen, S.; Abbas, Z.; Rehman, S.U. The Lytic SA Phage Demonstrate Bactericidal Activity against Mastitis Causing Staphylococcus aureus. Open Life Sci. 2016, 11, 39–45. [Google Scholar] [CrossRef]
- Liu, H.; Geagea, H.; Rousseau, G.M.; Labrie, S.J.; Tremblay, D.M.; Liu, X.; Moineau, S. Characterization of the Escherichia coli Virulent Myophage ST32. Viruses 2018, 10, 616. [Google Scholar] [CrossRef]
- Suarez, C.A.; Franceschelli, J.J.; Tasselli, S.E.; Morbidoni, H.R. Weirdo19ES is a novel singleton mycobacteriophage that selects for glycolipid deficient phage-resistant M. smegmatis mutants. PLoS ONE 2020, 15, e0231881. [Google Scholar] [CrossRef] [PubMed]
- Goh, S.; Hussain, H.; Chang, B.J.; Emmett, W.; Riley, T.V.; Mullany, P. Phage ϕC2 Mediates Transduction of Tn 6215, Encoding Erythromycin Resistance, between Clostridium difficile Strains. Mbio 2013, 4, e00840-13. [Google Scholar] [CrossRef]
- Wu, Y.; Wang, R.; Xu, M.; Liu, Y.; Zhu, X.; Qiu, J.; Liu, Q.; He, P.; Li, Q. A Novel Polysaccharide Depolymerase Encoded by the Phage SH-KP152226 Confers Specific Activity Against Multidrug-Resistant Klebsiella pneumoniae via Biofilm Degradation. Front. Microbiol. 2019, 10, 2768. [Google Scholar] [CrossRef]
- Rossmann, M.G.; Mesyanzhinov, V.V.; Arisaka, F.; Leiman, P.G. The bacteriophage T4 DNA injection machine. Curr. Opin. Struct. Biol. 2004, 14, 171–180. [Google Scholar] [CrossRef] [PubMed]
- Dowah, A.S.A.; Clokie, M.R.J. Review of the nature, diversity and structure of bacteriophage receptor binding proteins that target Gram-positive bacteria. Biophys. Rev. 2018, 10, 535–542. [Google Scholar] [CrossRef] [PubMed]
- Yap, M.L.; Rossmann, M.G. Structure and function of bacteriophage T4. Futur. Microbiol. 2014, 9, 1319–1327. [Google Scholar] [CrossRef] [PubMed]
- Veesler, D.; Spinelli, S.; Mahony, J.; Lichière, J.; Blangy, S.; Bricogne, G.; Legrand, P.; Ortiz-Lombardia, M.; Campanacci, V.; van Sinderen, D.; et al. Structure of the phage TP901-1 1.8 MDa baseplate suggests an alternative host adhesion mechanism. Proc. Natl. Acad. Sci. USA 2012, 109, 8954–8958. [Google Scholar] [CrossRef]
- Loeffler, J.M.; Nelson, D.; Fischetti, V.A. Rapid Killing of Streptococcus pneumoniae with a Bacteriophage Cell Wall Hydrolase. Science 2001, 294, 2170–2172. [Google Scholar] [CrossRef]
- Simmons, M.; Donovan, D.M.; Siragusa, G.R.; Seal, B.S. Recombinant Expression of Two Bacteriophage Proteins That Lyse Clostridium perfringens and Share Identical Sequences in the C-Terminal Cell Wall Binding Domain of the Molecules but Are Dissimilar in Their N-Terminal Active Domains. J. Agric. Food Chem. 2010, 58, 10330–10337. [Google Scholar] [CrossRef]
- Bernhardt, T.G.; Wang, I.-N.; Struck, D.K.; Young, R. Breaking free: “Protein antibiotics” and phage lysis. Res. Microbiol. 2002, 153, 493–501. [Google Scholar] [CrossRef]
- Fischetti, V.A. Lysin Therapy for Staphylococcus aureus and Other Bacterial Pathogens. Poxviruses 2017, 409, 529–540. [Google Scholar] [CrossRef]
- Ha, E.; Son, B.; Ryu, S. Clostridium perfringens Virulent Bacteriophage CPS2 and Its Thermostable Endolysin LysCPS2. Viruses 2018, 10, 251. [Google Scholar] [CrossRef]
- Young, R.; Bläsi, U. Holins: Form and function in bacteriophage lysis. FEMS Microbiol. Rev. 1995, 17, 191–205. [Google Scholar] [CrossRef]
- Filée, J.; Forterre, P.; Sen-Lin, T.; Laurent, J. Evolution of DNA Polymerase Families: Evidences for Multiple Gene Exchange Between Cellular and Viral Proteins. J. Mol. Evol. 2002, 54, 763–773. [Google Scholar] [CrossRef] [PubMed]
- Morcinek-Orłowska, J.; Zdrojewska, K.; Węgrzyn, A. Bacteriophage-Encoded DNA Polymerases—Beyond the Traditional View of Polymerase Activities. Int. J. Mol. Sci. 2022, 23, 635. [Google Scholar] [CrossRef]
- Cardarelli, L.; Lam, R.; Tuite, A.; Baker, L.A.; Sadowski, P.D.; Radford, D.R.; Rubinstein, J.L.; Battaile, K.P.; Chirgadze, N.; Maxwell, K.L.; et al. The Crystal Structure of Bacteriophage HK97 gp6: Defining a Large Family of Head–Tail Connector Proteins. J. Mol. Biol. 2010, 395, 754–768. [Google Scholar] [CrossRef] [PubMed]
- Volozhantsev, N.V.; Oakley, B.B.; Morales, C.A.; Verevkin, V.V.; Bannov, V.A.; Krasilnikova, V.M.; Popova, A.V.; Zhilenkov, E.L.; Garrish, J.K.; Schegg, K.M.; et al. Molecular Characterization of Podoviral Bacteriophages Virulent for Clostridium perfringens and Their Comparison with Members of the Picovirinae. PLoS ONE 2012, 7, e38283. [Google Scholar] [CrossRef]
- Davies, D. Understanding biofilm resistance to antibacterial agents. Nat. Rev. Drug Discov. 2003, 2, 114–122. [Google Scholar] [CrossRef] [PubMed]
- Hall-Stoodley, L.; Stoodley, P. Evolving concepts in biofilm infections. Cell. Microbiol. 2009, 11, 1034–1043. [Google Scholar] [CrossRef] [PubMed]
- Kaźmierczak, N.; Grygorcewicz, B.; Roszak, M.; Bochentyn, B.; Piechowicz, L. Comparative Assessment of Bacteriophage and Antibiotic Activity against Multidrug-Resistant Staphylococcus aureus Biofilms. Int. J. Mol. Sci. 2022, 23, 1274. [Google Scholar] [CrossRef]
- Jamal, M.; Andleeb, S.; Jalil, F.; Imran, M.; Nawaz, M.A.; Hussain, T.; Ali, M.; Rahman, S.U.; Das, C.R. Isolation, characterization and efficacy of phage MJ2 against biofilm forming multi-drug resistant Enterobacter cloacae. Folia Microbiol. 2019, 64, 101–111. [Google Scholar] [CrossRef]
- Yele, A.B.; Thawal, N.D.; Sahu, P.; Chopade, B.A. Novel lytic bacteriophage AB7-IBB1 of Acinetobacter baumannii: Isolation, characterization and its effect on biofilm. Arch. Virol. 2012, 157, 1441–1450. [Google Scholar] [CrossRef]
- Kahlmeter, G.; Giske, C.G.; Kirn, T.J.; Sharp, S.E. Point-Counterpoint: Differences between the European Committee on Antimicrobial Susceptibility Testing and Clinical and Laboratory Standards Institute Recommendations for Reporting Antimicrobial Susceptibility Results. J. Clin. Microbiol. 2019, 57, e01129-19. [Google Scholar] [CrossRef]
- Ackermann, H.W. Basic phage electron microscopy. Methods Mol. Biol. 2009, 501, 113–126. [Google Scholar] [CrossRef]
- Van Twest, R.; Kropinski, A.M. Bacteriophage enrichment from water and soil. Methods Mol. Biol. 2009, 501, 15–21. [Google Scholar] [CrossRef] [PubMed]
- Zhou, W.; Feng, Y.; Zong, Z. Two New Lytic Bacteriophages of the Myoviridae Family Against Carbapenem-Resistant Acinetobacter baumannii. Front. Microbiol. 2018, 9, 850. [Google Scholar] [CrossRef]
- Lu, Z.; Breidt, F.; Fleming, H.; Altermann, E.; Klaenhammer, T. Isolation and characterization of a Lactobacillus plantarum bacteriophage, ΦJL-1, from a cucumber fermentation. Int. J. Food Microbiol. 2003, 84, 225–235. [Google Scholar] [CrossRef]
- Yang, Z.; Liu, X.; Shi, Y.; Yin, S.; Shen, W.; Chen, J.; Chen, Y.; Chen, Y.; You, B.; Gong, Y.; et al. Characterization and genome annotation of a newly detected bacteriophage infecting multidrug-resistant Acinetobacter baumannii. Arch. Virol. 2019, 164, 1527–1533. [Google Scholar] [CrossRef] [PubMed]
- Bae, D.; Lee, J.-W.; Chae, J.-P.; Kim, J.-W.; Eun, J.-S.; Lee, K.-W.; Seo, K.-H. Characterization of a novel bacteriophage φCJ22 and its prophylactic and inhibitory effects on necrotic enteritis and Clostridium perfringens in broilers. Poult. Sci. 2021, 100, 302–313. [Google Scholar] [CrossRef] [PubMed]
- Bankevich, A.; Nurk, S.; Antipov, D.; Gurevich, A.A.; Dvorkin, M.; Kulikov, A.S.; Lesin, V.M.; Nikolenko, S.I.; Pham, S.; Prjibelski, A.D.; et al. SPAdes: A new genome assembly algorithm and its applications to single-cell sequencing. J. Comput. Biol. 2012, 19, 455–477. [Google Scholar] [CrossRef]
- Aziz, R.K.; Bartels, D.; Best, A.A.; DeJongh, M.; Disz, T.; Edwards, R.A.; Formsma, K.; Gerdes, S.; Glass, E.M.; Kubal, M.; et al. The RAST server: Rapid annotations using subsystems technology. BMC Genom. 2008, 9, 75. [Google Scholar] [CrossRef]
- Besemer, J.; Borodovsky, M. GeneMark: Web software for gene finding in prokaryotes, eukaryotes and viruses. Nucleic. Acids Res. 2005, 33, W451–W454. [Google Scholar] [CrossRef]
- Schattner, P.; Brooks, A.N.; Lowe, T.M. The tRNAscan-SE, snoscan and snoGPS web servers for the detection of tRNAs and snoRNAs. Nucleic. Acids Res. 2005, 33, W686–W689. [Google Scholar] [CrossRef]
- Tamura, K.; Peterson, D.; Peterson, N.; Stecher, G.; Nei, M.; Kumar, S. MEGA5: Molecular Evolutionary Genetics Analysis Using Maximum Likelihood, Evolutionary Distance, and Maximum Parsimony Methods. Mol. Biol. Evol. 2011, 28, 2731–2739. [Google Scholar] [CrossRef]
- Darling, A.E.; Mau, B.; Perna, N.T. progressiveMauve: Multiple Genome Alignment with Gene Gain, Loss and Rearrangement. PLoS ONE 2010, 5, e11147. [Google Scholar] [CrossRef]
- Echarlebois, A.; Ejacques, M.; Earchambault, M. Biofilm formation of Clostridium perfringens and its exposure to low-dose antimicrobials. Front. Microbiol. 2014, 5, 183. [Google Scholar] [CrossRef]
- Mahmud, S.A.; Roy, R.; Sugiokto, F.; Islam, N.; Lin, M.-D.; Lin, L.-C.; Lin, N.-T. Phage φAB6-Borne Depolymerase Combats Acinetobacter baumannii Biofilm Formation and Infection. Antibiotics 2021, 10, 279. [Google Scholar] [CrossRef] [PubMed]
- Ning, H.; Lin, H.; Wang, J.; He, X.; Lv, X.; Ju, L. Characterizations of the endolysin Lys84 and its domains from phage qdsa002 with high activities against Staphylococcus aureus and its biofilms. Enzym. Microb. Technol. 2021, 148, 109809. [Google Scholar] [CrossRef] [PubMed]


| Genome Characteristics | Phage | ||||||
|---|---|---|---|---|---|---|---|
| DCp1 | vB_CP_ qdyz_P5 | Susfortuna | CPD7 | phi24R | CPS1 | LPCPA6 | |
| Genome size (bp) | 18,555 | 18,888 | 19,046 | 18,958 | 18,919 | 19,089 | 18,554 |
| G + C (%) | 28.20 | 28.80 | 29.22 | 29.12 | 27.80 | 28.26 | 30.56 |
| Predicted ORFs | 25 | 27 | 27 | 26 | 21 | 26 | 25 |
| tRNAs | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| Similarity with DCp1 (%) | 100 | 96.95 | 86.23 | 86.04 | 77.64 | 75.13 | 73.63 |
| Coverage with DCp1 (%) | 100 | 85 | 74 | 74 | 0 | 1 | 21 |
| Accession no. | OP256049 | OP894055.1 | NC_048712 | MK017820 | JN800508 | NC_048661 | OM638104 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Tang, Z.; Li, X.; Wang, X.; Zhang, C.; Zou, L.; Ren, H.; Liu, W. Characterization and Genomic Analysis of a Novel Lytic Phage DCp1 against Clostridium perfringens Biofilms. Int. J. Mol. Sci. 2023, 24, 4191. https://doi.org/10.3390/ijms24044191
Tang Z, Li X, Wang X, Zhang C, Zou L, Ren H, Liu W. Characterization and Genomic Analysis of a Novel Lytic Phage DCp1 against Clostridium perfringens Biofilms. International Journal of Molecular Sciences. 2023; 24(4):4191. https://doi.org/10.3390/ijms24044191
Chicago/Turabian StyleTang, Zhaohui, Xiaojing Li, Xinwei Wang, Can Zhang, Ling Zou, Huiying Ren, and Wenhua Liu. 2023. "Characterization and Genomic Analysis of a Novel Lytic Phage DCp1 against Clostridium perfringens Biofilms" International Journal of Molecular Sciences 24, no. 4: 4191. https://doi.org/10.3390/ijms24044191
APA StyleTang, Z., Li, X., Wang, X., Zhang, C., Zou, L., Ren, H., & Liu, W. (2023). Characterization and Genomic Analysis of a Novel Lytic Phage DCp1 against Clostridium perfringens Biofilms. International Journal of Molecular Sciences, 24(4), 4191. https://doi.org/10.3390/ijms24044191






