Phage Engineering for Targeted Multidrug-Resistant Escherichia coli
Abstract
1. Introduction
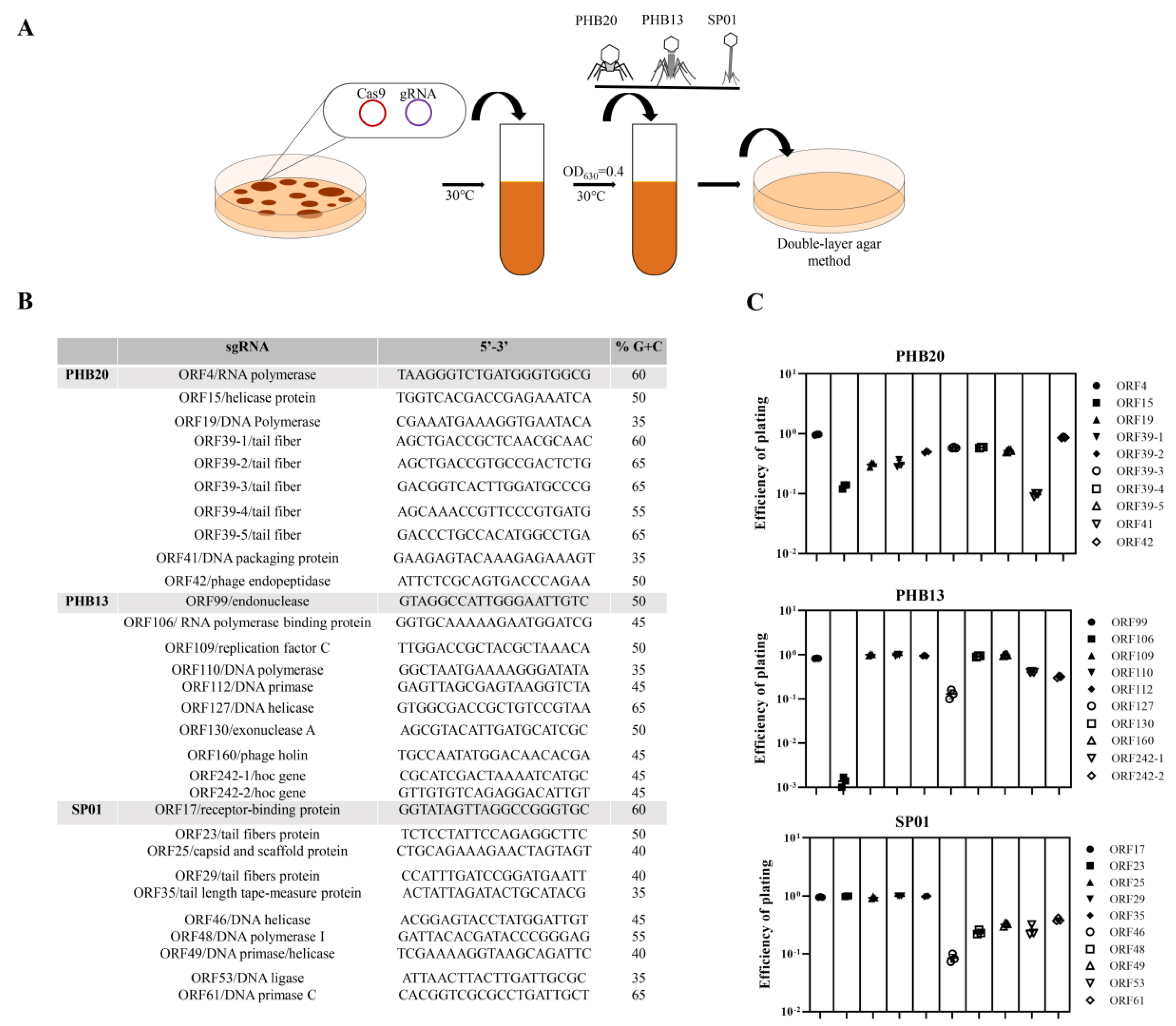
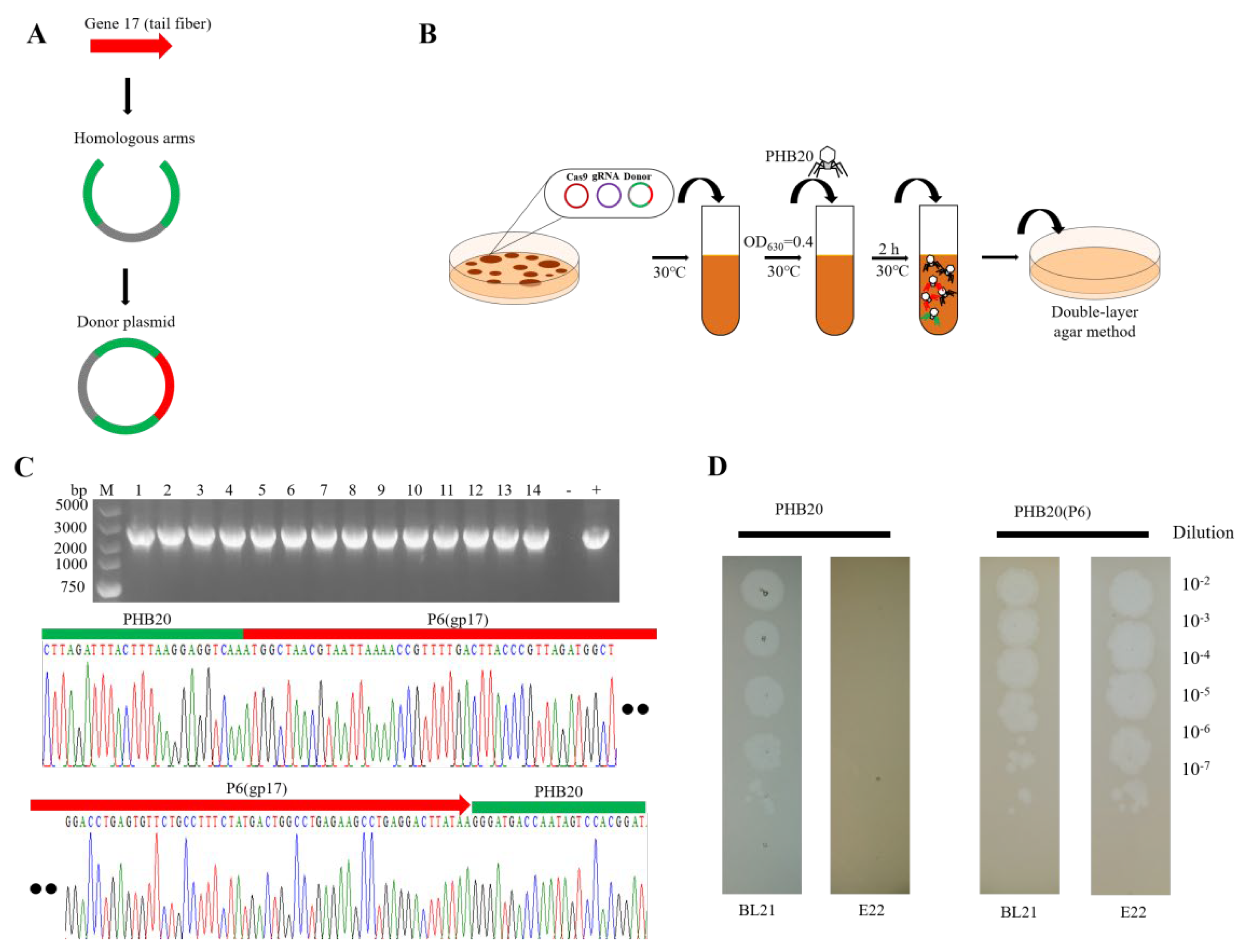
2. Results and Discussion
3. Materials and Methods
3.1. E. coli Growth Media
3.2. Transformation of E. coli BL21
3.3. Phage Infection E. coli BL21
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Araújo, S.; Tacão, M.; Baraúna, R.; Ramos, R.; Silva, A.; Henriques, I. Genome analysis of two multidrug-resistant Escherichia coli O8:H9-ST48 strains isolated from lettuce. Gene 2021, 785, 145603. [Google Scholar] [CrossRef] [PubMed]
- Díaz-Jiménez, D.; García-Meniño, I.; Fernández, J.; García, V.; Mora, A. Chicken and turkey meat: Consumer exposure to multidrug-resistant Enterobacteriaceae including mcr-carriers, uropathogenic E. coli and high-risk lineages such as ST131. Int. J. Food Microbiol. 2020, 331, 108750. [Google Scholar] [CrossRef] [PubMed]
- Tadese, N.D.; Gebremedhi, E.Z.; Moges, F.; Borana, B.M.; Marami, L.M.; Sarba, E.J.; Abebe, H.; Kelbesa, K.A.; Atalel, D.; Tessema, B. Occurrence and Antibiogram of Escherichia coli O157: H7 in Raw Beef and Hygienic Practices in Abattoir and Retailer Shops in Ambo Town, Ethiopia. Vet. Med. Int. 2021, 2021, 8846592. [Google Scholar] [CrossRef] [PubMed]
- Benameur, Q.; Tali-Maamar, H.; Assaous, F.; Guettou, B.; Tahrat, N.; Aggoune, N.; Rahal, K.; Ben-Mahdi, M.H. Isolation of Escherichia coli carrying the bla(CTX-M-1) and qnrS1 genes from reproductive organs of broiler breeders and internal contents of hatching eggs. J. Vet. Med. Sci. 2018, 80, 1540–1543. [Google Scholar] [CrossRef] [PubMed]
- Bhardwaj, D.K.; Taneja, N.K.; Dp, S.; Chakotiya, A.; Patel, P.; Taneja, P.; Sachdev, D.; Gupta, S.; Sanal, M.G. Phenotypic and genotypic characterization of biofilm forming, antimicrobial resistant, pathogenic Escherichia coli isolated from Indian dairy and meat products. Int. J. Food Microbiol. 2021, 336, 108899. [Google Scholar] [CrossRef]
- Bruttin, A.; Brüssow, H. Human volunteers receiving Escherichia coli phage T4 orally: A safety test of phage therapy. Antimicrob. Agents Chemother. 2005, 49, 2874–2878. [Google Scholar] [CrossRef]
- Nale, J.Y.; Clokie, M.R. Preclinical data and safety assessment of phage therapy in humans. Curr. Opin. Biotechnol. 2021, 68, 310–317. [Google Scholar] [CrossRef]
- Mu, A.; McDonald, D.; Jarmusch, A.K.; Martino, C.; Brennan, C.; Bryant, M.; Humphrey, G.C.; Toronczak, J.; Schwartz, T.; Nguyen, D.; et al. Assessment of the microbiome during bacteriophage therapy in combination with systemic antibiotics to treat a case of staphylococcal device infection. Microbiome 2021, 9, 92. [Google Scholar] [CrossRef]
- Squires, R.A. Bacteriophage therapy for challenging bacterial infections: Achievements, limitations and prospects for future clinical use by veterinary dermatologists. Vet. Dermatol. 2021, 32, 587-e158. [Google Scholar] [CrossRef]
- Alomari, M.M.M.; Dec, M.; Nowaczek, A.; Puchalski, A.; Wernicki, A.; Kowalski, C.; Urban-Chmiel, R. Therapeutic and Prophylactic Effect of the Experimental Bacteriophage Treatment to Control Diarrhea Caused by E. coli in Newborn Calves. ACS Infect. Dis. 2021, 7, 2093–2101. [Google Scholar] [CrossRef]
- Chen, Y.; Li, X.; Wang, S.; Guan, L.; Li, X.; Hu, D.; Gao, D.; Song, J.; Chen, H.; Qian, P. A Novel Tail-Associated O91-Specific Polysaccharide Depolymerase from a Podophage Reveals Lytic Efficacy of Shiga Toxin-Producing Escherichia coli. Appl. Environ. Microbiol. 2020, 86, e00145-20. [Google Scholar] [CrossRef] [PubMed]
- Petsong, K.; Benjakul, S.; Vongkamjan, K. Optimization of wall material for phage encapsulation via freeze-drying and antimicrobial efficacy of microencapsulated phage against Salmonella. J. Food Sci. Technol. 2021, 58, 1937–1946. [Google Scholar] [CrossRef] [PubMed]
- Sabzali, S.; Bouzari, M. Isolation, identification and some characteristics of two lytic bacteriophages against Salmonella enterica serovar Paratyphi B and S. enterica serovar Typhimurium from various food sources. FEMS Microbiol. Lett. 2021, 368, fnab037. [Google Scholar] [CrossRef]
- Favrin, S.J.; Jassim, S.A.; Griffiths, M.W. Application of a novel immunomagnetic separation-bacteriophage assay for the detection of Salmonella enteritidis and Escherichia coli O157:H7 in food. Int. J. Food Microbiol. 2003, 85, 63–71. [Google Scholar] [CrossRef] [PubMed]
- Smartt, A.E.; Xu, T.; Jegier, P.; Carswell, J.J.; Blount, S.A.; Sayler, G.S.; Ripp, S. Pathogen detection using engineered bacteriophages. Anal. Bioanal. Chem. 2011, 402, 3127–3146. [Google Scholar] [CrossRef] [PubMed]
- Ando, H.; Lemire, S.; Pires, D.P.; Lu, T.K. Engineering Modular Viral Scaffolds for Targeted Bacterial Population Editing. Cell Syst. 2015, 1, 187–196. [Google Scholar] [CrossRef]
- Yehl, K.; Lemire, S.; Yang, A.C.; Ando, H.; Mimee, M.; Torres, M.T.; de la Fuente-Nunez, C.; Lu, T.K. Engineering Phage Host-Range and Suppressing Bacterial Resistance through Phage Tail Fiber Mutagenesis. Cell 2019, 179, 459–469.e459. [Google Scholar] [CrossRef]
- Dedrick, R.M.; Guerrero-Bustamante, C.A.; Garlena, R.A.; Russell, D.A.; Ford, K.; Harris, K.; Gilmour, K.C.; Soothill, J.; Jacobs-Sera, D.; Schooley, R.T.; et al. Engineered bacteriophages for treatment of a patient with a disseminated drug-resistant Mycobacterium abscessus. Nat. Med. 2019, 25, 730–733. [Google Scholar] [CrossRef]
- Park, J.Y.; Moon, B.Y.; Park, J.W.; Thornton, J.A.; Park, Y.H.; Seo, K.S. Genetic engineering of a temperate phage-based delivery system for CRISPR/Cas9 antimicrobials against Staphylococcus aureus. Sci. Rep. 2017, 7, 44929. [Google Scholar] [CrossRef]
- Cobb, L.H.; Park, J.; Swanson, E.A.; Beard, M.C.; McCabe, E.M.; Rourke, A.S.; Seo, K.S.; Olivier, A.K.; Priddy, L.B. CRISPR-Cas9 modified bacteriophage for treatment of Staphylococcus aureus induced osteomyelitis and soft tissue infection. PLoS One 2019, 14, e0220421. [Google Scholar] [CrossRef]
- Tao, P.; Mahalingam, M.; Kirtley, M.L.; van Lier, C.J.; Sha, J.; Yeager, L.A.; Chopra, A.K.; Rao, V.B. Mutated and bacteriophage T4 nanoparticle arrayed F1-V immunogens from Yersinia pestis as next generation plague vaccines. PLoS Pathog. 2013, 9, e1003495. [Google Scholar] [CrossRef] [PubMed]
- Tao, P.; Zhu, J.; Mahalingam, M.; Batra, H.; Rao, V.B. Bacteriophage T4 nanoparticles for vaccine delivery against infectious diseases. Adv. Drug Deliv. Rev. 2019, 145, 57–72. [Google Scholar] [CrossRef] [PubMed]
- Brüssow, H.; Hendrix, R.W. Phage genomics: Small is beautiful. Cell 2002, 108, 13–16. [Google Scholar] [CrossRef] [PubMed]
- Ackermann, H.W. 5500 Phages examined in the electron microscope. Arch. Virol. 2007, 152, 227–243. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.; Li, X.; Wang, S.; Qian, P.; Li, Y. CRISPR-Cas9-Mediated Genome Editing in Escherichia coli Bacteriophages. CRISPR-Cas Methods 2021, 2, 325–334. [Google Scholar]
- Veesler, D.; Cambillau, C. A common evolutionary origin for tailed-bacteriophage functional modules and bacterial machineries. Microbiol. Mol. Biol. Rev. 2011, 75, 423–433. [Google Scholar] [CrossRef]

Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Song, J.; Liu, Z.; Zhang, Q.; Liu, Y.; Chen, Y. Phage Engineering for Targeted Multidrug-Resistant Escherichia coli. Int. J. Mol. Sci. 2023, 24, 2459. https://doi.org/10.3390/ijms24032459
Song J, Liu Z, Zhang Q, Liu Y, Chen Y. Phage Engineering for Targeted Multidrug-Resistant Escherichia coli. International Journal of Molecular Sciences. 2023; 24(3):2459. https://doi.org/10.3390/ijms24032459
Chicago/Turabian StyleSong, Jiaoyang, Zhengjie Liu, Qing Zhang, Yuqing Liu, and Yibao Chen. 2023. "Phage Engineering for Targeted Multidrug-Resistant Escherichia coli" International Journal of Molecular Sciences 24, no. 3: 2459. https://doi.org/10.3390/ijms24032459
APA StyleSong, J., Liu, Z., Zhang, Q., Liu, Y., & Chen, Y. (2023). Phage Engineering for Targeted Multidrug-Resistant Escherichia coli. International Journal of Molecular Sciences, 24(3), 2459. https://doi.org/10.3390/ijms24032459






