Cloning of an Albino Mutation of Arabidopsis thaliana Using Mapping-by-Sequencing
Abstract
1. Introduction
2. Results and Discussion
2.1. Characterization of an EMS-Induced Albino Mutant
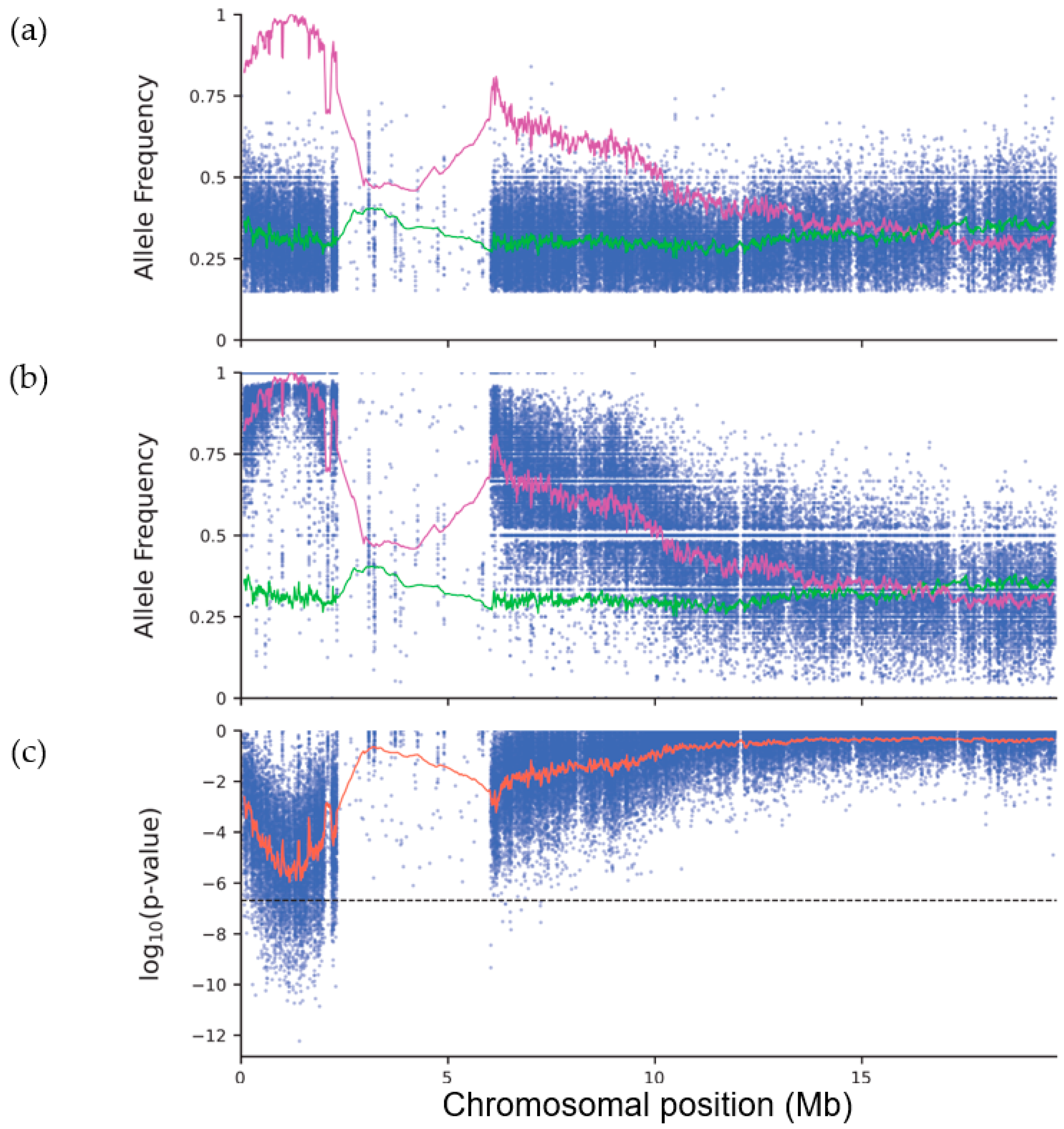
2.2. Mapping-By-Sequencing of the Albino Mutant
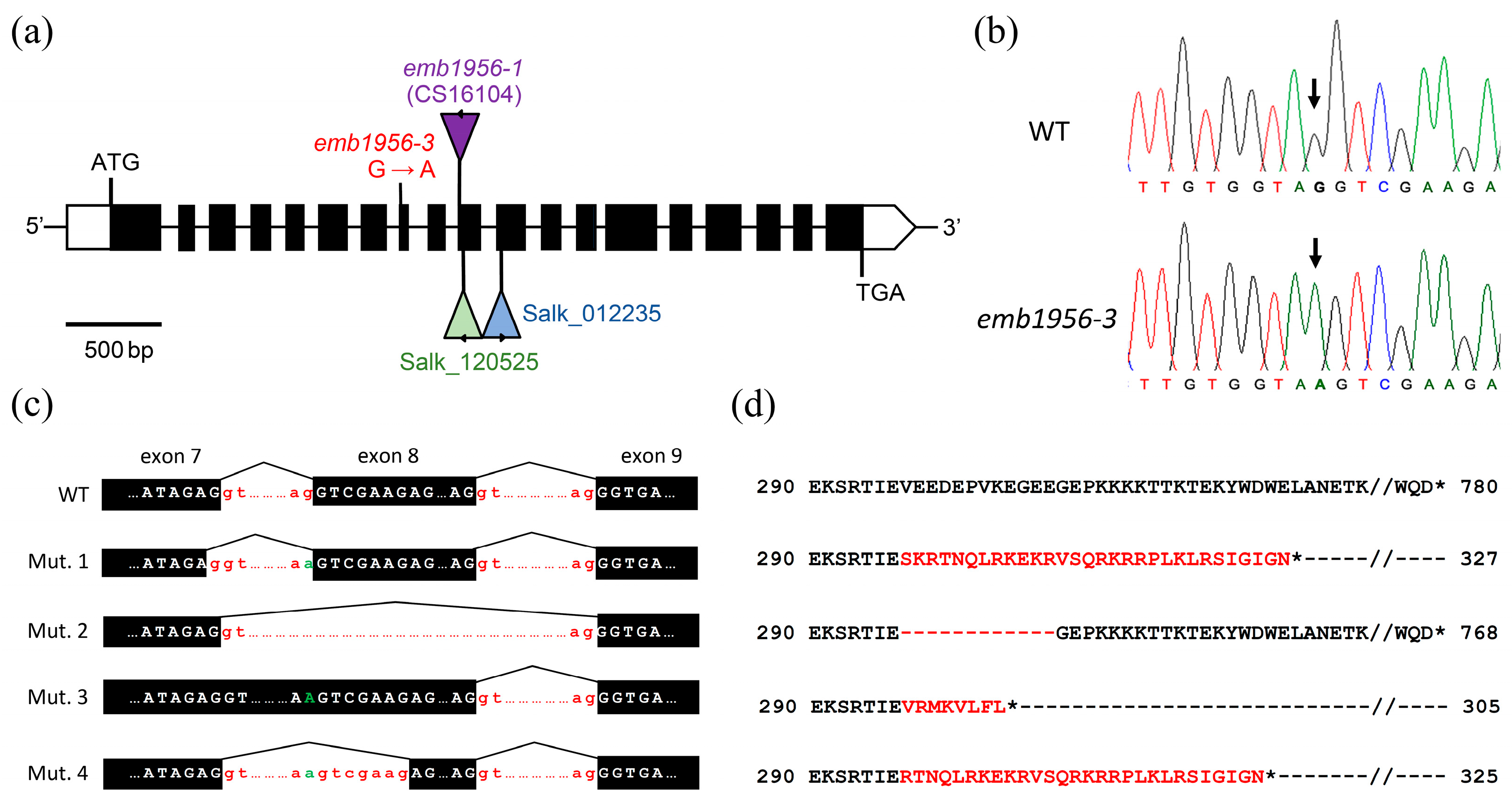
2.3. The Albino Mutant Carries a G-To-A Transition Mutation in At2g04030
2.4. Complementation Tests Show That the Albino Mutant Is Affected in At2g04030
2.5. Complementation Using a Transgene
2.6. The Albino Mutation Causes Massive Downregulation of Chloroplast-Related Functions
2.7. The Transition Mutation Causes Improper Splicing of At2g04030 Transcripts
2.8. Identification of New Protein–Protein Interactions Involving the AtHsp90.5 Protein
3. Materials and Methods
3.1. Plant Material and Growth of A. thaliana Plants
3.2. Genome Sequencing and Bioinformatic Analysis
3.3. Peak Detection
3.4. Constructs for Plant Transformation and Yeast Two-Hybrid Assays
3.5. RNA Sequencing
3.6. Yeast Two-Hybrid Screen
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Schneeberger, K.; Ossowski, S.; Lanz, C.; Juul, T.; Petersen, A.H.; Nielsen, K.L.; Jørgensen, J.-E.; Weigel, D.; Andersen, S.U. SHOREmap: Simultaneous Mapping and Mutation Identification by Deep Sequencing. Nat. Methods 2009, 6, 550–551. [Google Scholar] [CrossRef]
- Candela, H.; Casanova-Sáez, R.; Micol, J.L. Getting Started in Mapping-by-Sequencing. J. Integr. Plant Biol. 2015, 57, 606–612. [Google Scholar] [CrossRef]
- Li, Z.; Guo, Y.; Ou, L.; Hong, H.; Wang, J.; Liu, Z.; Guo, B.; Zhang, L.; Qiu, L. Identification of the Dwarf Gene GmDW1 in Soybean (Glycine max L.) by Combining Mapping-by-Sequencing and Linkage Analysis. Theor. Appl. Genet. 2018, 131, 1001–1016. [Google Scholar] [CrossRef]
- Takei, H.; Shinozaki, Y.; Yano, R.; Kashojiya, S.; Hernould, M.; Chevalier, C.; Ezura, H.; Ariizumi, T. Loss-of-Function of a Tomato Receptor-Like Kinase Impairs Male Fertility and Induces Parthenocarpic Fruit Set. Front. Plant Sci. 2019, 10, 403. [Google Scholar] [CrossRef]
- Oh, S.E.; Yeung, C.; Babaei-Rad, R.; Zhao, R. Cosuppression of the Chloroplast Localized Molecular Chaperone HSP90.5 Impairs Plant Development and Chloroplast Biogenesis in Arabidopsis. BMC Res. Notes 2014, 7, 643. [Google Scholar] [CrossRef]
- Cao, D.; Froehlich, J.E.; Zhang, H.; Cheng, C.-L. The Chlorate-Resistant and Photomorphogenesis-Defective Mutant Cr88 Encodes a Chloroplast-Targeted HSP90. Plant J. 2003, 33, 107–118. [Google Scholar] [CrossRef]
- Feng, J.; Fan, P.; Jiang, P.; Lv, S.; Chen, X.; Li, Y. Chloroplast-Targeted Hsp90 Plays Essential Roles in Plastid Development and Embryogenesis in Arabidopsis Possibly Linking with VIPP1. Physiol. Plant. 2014, 150, 292–307. [Google Scholar] [CrossRef]
- Inoue, H.; Li, M.; Schnell, D.J. An Essential Role for Chloroplast Heat Shock Protein 90 (Hsp90C) in Protein Import into Chloroplasts. Proc. Natl. Acad. Sci. USA 2013, 110, 3173–3178. [Google Scholar] [CrossRef]
- Yang, L.; Wang, J.; Han, Z.; Lei, L.; Liu, H.L.; Zheng, H.; Xin, W.; Zou, D. Combining QTL-Seq and Linkage Mapping to Fine Map a Candidate Gene in QCTS6 for Cold Tolerance at the Seedling Stage in Rice. BMC Plant Biol. 2021, 21, 278. [Google Scholar] [CrossRef]
- Fekih, R.; Takagi, H.; Tamiru, M.; Abe, A.; Natsume, S.; Yaegashi, H.; Sharma, S.; Sharma, S.; Kanzaki, H.; Matsumura, H.; et al. MutMap+: Genetic Mapping and Mutant Identification without Crossing in Rice. PLoS ONE 2013, 8, e68529. [Google Scholar] [CrossRef]
- Mateo-Bonmatí, E.; Casanova-Sáez, R.; Candela, H.; Micol, J.L. Rapid Identification of Angulata Leaf Mutations Using Next-Generation Sequencing. Planta 2014, 240, 1113–1122. [Google Scholar] [CrossRef]
- Greene, E.A.; Codomo, C.A.; Taylor, N.E.; Henikoff, J.G.; Till, B.J.; Reynolds, S.H.; Enns, L.C.; Burtner, C.; Johnson, J.E.; Odden, A.R.; et al. Spectrum of Chemically Induced Mutations From a Large-Scale Reverse-Genetic Screen in Arabidopsis. Genetics 2003, 164, 731–740. [Google Scholar] [CrossRef]
- Tzafrir, I.; Pena-Muralla, R.; Dickerman, A.; Berg, M.; Rogers, R.; Hutchens, S.; Sweeney, T.C.; McElver, J.; Aux, G.; Patton, D.; et al. Identification of Genes Required for Embryo Development in Arabidopsis. Plant Physiol. 2004, 135, 1206–1220. [Google Scholar] [CrossRef]
- Mi, H.; Muruganujan, A.; Casagrande, J.T.; Thomas, P.D. Large-Scale Gene Function Analysis with the PANTHER Classification System. Nat. Protoc. 2013, 8, 1551–1566. [Google Scholar] [CrossRef]
- Muñoz-Nortes, T.; Pérez-Pérez, J.M.; Sarmiento-Mañús, R.; Candela, H.; Micol, J.L. Deficient Glutamate Biosynthesis Triggers a Concerted Upregulation of Ribosomal Protein Genes in Arabidopsis. Sci. Rep. 2017, 7, 6164. [Google Scholar] [CrossRef]
- Mustroph, A.; Lee, S.C.; Oosumi, T.; Zanetti, M.E.; Yang, H.; Ma, K.; Yaghoubi-Masihi, A.; Fukao, T.; Bailey-Serres, J. Cross-Kingdom Comparison of Transcriptomic Adjustments to Low-Oxygen Stress Highlights Conserved and Plant-Specific Responses. Plant Physiol. 2010, 152, 1484–1500. [Google Scholar] [CrossRef] [PubMed]
- Branco-Price, C.; Kawaguchi, R.; Ferreira, R.B.; Bailey-Serres, J. Genome-Wide Analysis of Transcript Abundance and Translation in Arabidopsis Seedlings Subjected to Oxygen Deprivation. Ann. Bot. 2005, 96, 647–660. [Google Scholar] [CrossRef] [PubMed]
- Islam, S.; Bhor, S.A.; Tanaka, K.; Sakamoto, H.; Yaeno, T.; Kaya, H.; Kobayashi, K. Impaired Expression of Chloroplast HSP90C Chaperone Activates Plant Defense Responses with a Possible Link to a Disease-Symptom-Like Phenotype. Int. J. Mol. Sci. 2020, 21, 4202. [Google Scholar] [CrossRef]
- Willmund, F.; Dorn, K.V.; Schulz-Raffelt, M.; Schroda, M. The Chloroplast DnaJ Homolog CDJ1 of Chlamydomonas Reinhardtii Is Part of a Multichaperone Complex Containing HSP70B, CGE1, and HSP90C. Plant Physiol. 2008, 148, 2070–2082. [Google Scholar] [CrossRef]
- de Luna-Valdez, L.A.; Villaseñor-Salmerón, C.I.; Cordoba, E.; Vera-Estrella, R.; León-Mejía, P.; Guevara-García, A.A. Functional Analysis of the Chloroplast GrpE (CGE) Proteins from Arabidopsis thaliana. Plant Physiol. Biochem. 2019, 139, 293–306. [Google Scholar] [CrossRef]
- Kim, Y.; Schumaker, K.S.; Zhu, J.-K. EMS Mutagenesis of Arabidopsis. In Arabidopsis Protocols; Salinas, J., Sanchez-Serrano, J.J., Eds.; Humana Press: Totowa, NJ, USA, 2006; pp. 101–103. ISBN 978-1-59745-003-4. [Google Scholar]
- Murashige, T.; Skoog, F. A Revised Medium for Rapid Growth and Bio Assays with Tobacco Tissue Cultures. Physiol. Plant. 1962, 15, 473–497. [Google Scholar] [CrossRef]
- Bolger, A.M.; Lohse, M.; Usadel, B. Trimmomatic: A Flexible Trimmer for Illumina Sequence Data. Bioinformatics 2014, 30, 2114–2120. [Google Scholar] [CrossRef]
- Lamesch, P.; Berardini, T.Z.; Li, D.; Swarbreck, D.; Wilks, C.; Sasidharan, R.; Muller, R.; Dreher, K.; Alexander, D.L.; Garcia-Hernandez, M.; et al. The Arabidopsis Information Resource (TAIR): Improved Gene Annotation and New Tools. Nucleic Acids Res. 2012, 40, D1202–D1210. [Google Scholar] [CrossRef]
- Langmead, B.; Trapnell, C.; Pop, M.; Salzberg, S.L. Ultrafast and Memory-Efficient Alignment of Short DNA Sequences to the Human Genome. Genome Biol. 2009, 10, R25. [Google Scholar] [CrossRef]
- Langmead, B.; Salzberg, S.L. Fast Gapped-Read Alignment with Bowtie 2. Nat. Methods 2012, 9, 357–359. [Google Scholar] [CrossRef]
- Li, H.; Handsaker, B.; Wysoker, A.; Fennell, T.; Ruan, J.; Homer, N.; Marth, G.; Abecasis, G.; Durbin, R. 1000 Genome Project Data Processing Subgroup The Sequence Alignment/Map Format and SAMtools. Bioinformatics 2009, 25, 2078–2079. [Google Scholar] [CrossRef]
- Danecek, P.; Bonfield, J.K.; Liddle, J.; Marshall, J.; Ohan, V.; Pollard, M.O.; Whitwham, A.; Keane, T.; McCarthy, S.A.; Davies, R.M.; et al. Twelve Years of SAMtools and BCFtools. GigaScience 2021, 10, giab008. [Google Scholar] [CrossRef]
- Danecek, P.; Auton, A.; Abecasis, G.; Albers, C.A.; Banks, E.; DePristo, M.A.; Handsaker, R.E.; Lunter, G.; Marth, G.T.; Sherry, S.T.; et al. The Variant Call Format and VCFtools. Bioinformatics 2011, 27, 2156–2158. [Google Scholar] [CrossRef]
- Virtanen, P.; Gommers, R.; Oliphant, T.E.; Haberland, M.; Reddy, T.; Cournapeau, D.; Burovski, E.; Peterson, P.; Weckesser, W.; Bright, J.; et al. SciPy 1.0: Fundamental Algorithms for Scientific Computing in Python. Nat. Methods 2020, 17, 261–272. [Google Scholar] [CrossRef]
- Clough, S.J.; Bent, A.F. Floral Dip: A Simplified Method for Agrobacterium -Mediated Transformation of Arabidopsis thaliana. Plant J. 1998, 16, 735–743. [Google Scholar] [CrossRef]
- Cheng, C.-Y.; Krishnakumar, V.; Chan, A.P.; Thibaud-Nissen, F.; Schobel, S.; Town, C.D. Araport11: A Complete Reannotation of the Arabidopsis thaliana Reference Genome. Plant J. 2017, 89, 789–804. [Google Scholar] [CrossRef]
- Kim, D.; Paggi, J.M.; Park, C.; Bennett, C.; Salzberg, S.L. Graph-Based Genome Alignment and Genotyping with HISAT2 and HISAT-Genotype. Nat. Biotechnol. 2019, 37, 907–915. [Google Scholar] [CrossRef]
- Trapnell, C.; Hendrickson, D.G.; Sauvageau, M.; Goff, L.; Rinn, J.L.; Pachter, L. Differential Analysis of Gene Regulation at Transcript Resolution with RNA-Seq. Nat. Biotechnol. 2013, 31, 46–53. [Google Scholar] [CrossRef]
- de la Fuente Cantó, C.; Vigouroux, Y. Evaluation of Nine Statistics to Identify QTLs in Bulk Segregant Analysis Using next Generation Sequencing Approaches. BMC Genomics 2022, 23, 490. [Google Scholar] [CrossRef]

| Allele | Family | Wild-Type | Albino | χ2 | p-Value 1 |
|---|---|---|---|---|---|
| emb1956-3 (this work) | 1 | 49 | 19 | 0.314 | 0.575 |
| 2 | 61 | 30 | 3.081 | 0.079 | |
| 3 | 67 | 16 | 1.450 | 0.229 | |
| 4 | 46 | 32 | 10.684 | 0.001 * | |
| SALK_120525 | 1 | 54 | 15 | 0.391 | 0.532 |
| 2 | 96 | 35 | 0.206 | 0.650 | |
| 3 | 93 | 31 | 0.000 | 1.000 | |
| 4 | 125 | 51 | 1.485 | 0.223 | |
| CS16104 | 1 | 119 | 41 | 0.033 | 0.855 |
| 2 | 90 | 39 | 1.884 | 0.170 | |
| 3 | 119 | 44 | 0.346 | 0.557 | |
| 4 | 126 | 52 | 1.685 | 0.194 | |
| SALK_012235 | 1 | 86 | 32 | 0.282 | 0.595 |
| 2 | 117 | 35 | 0.316 | 0.574 | |
| 3 | 104 | 37 | 0.116 | 0.734 | |
| 4 | 130 | 53 | 1.532 | 0.216 |
| Cross | Wild-Type | Albino | χ2 | p-Value * |
|---|---|---|---|---|
| CS16104 × EMB1956/emb1956-3 | 45 | 23 | 2.824 | 0.093 |
| SALK_012235 × EMB1956/emb1956-3 | 46 | 16 | 0.022 | 0.883 |
| SALK_120525 × EMB1956/emb1956-3 | 28 | 9 | 0.009 | 0.924 |
| EMB1956/emb1956-3 × CS16104 | 22 | 10 | 0.667 | 0.414 |
| EMB1956/emb1956-3 × SALK_012235 | 49 | 9 | 2.782 | 0.095 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Rodríguez-Alcocer, E.; Ruiz-Pérez, E.; Parreño, R.; Martínez-Guardiola, C.; Berna, J.M.; Çakmak Pehlivanlı, A.; Jover-Gil, S.; Candela, H. Cloning of an Albino Mutation of Arabidopsis thaliana Using Mapping-by-Sequencing. Int. J. Mol. Sci. 2023, 24, 4196. https://doi.org/10.3390/ijms24044196
Rodríguez-Alcocer E, Ruiz-Pérez E, Parreño R, Martínez-Guardiola C, Berna JM, Çakmak Pehlivanlı A, Jover-Gil S, Candela H. Cloning of an Albino Mutation of Arabidopsis thaliana Using Mapping-by-Sequencing. International Journal of Molecular Sciences. 2023; 24(4):4196. https://doi.org/10.3390/ijms24044196
Chicago/Turabian StyleRodríguez-Alcocer, Eva, Erundina Ruiz-Pérez, Ricardo Parreño, César Martínez-Guardiola, José Marcos Berna, Ayça Çakmak Pehlivanlı, Sara Jover-Gil, and Héctor Candela. 2023. "Cloning of an Albino Mutation of Arabidopsis thaliana Using Mapping-by-Sequencing" International Journal of Molecular Sciences 24, no. 4: 4196. https://doi.org/10.3390/ijms24044196
APA StyleRodríguez-Alcocer, E., Ruiz-Pérez, E., Parreño, R., Martínez-Guardiola, C., Berna, J. M., Çakmak Pehlivanlı, A., Jover-Gil, S., & Candela, H. (2023). Cloning of an Albino Mutation of Arabidopsis thaliana Using Mapping-by-Sequencing. International Journal of Molecular Sciences, 24(4), 4196. https://doi.org/10.3390/ijms24044196






