Computational Analysis of the Ligand-Binding Sites of the Molecular Chaperone OppA from Yersinia pseudotuberculosis
Abstract
1. Introduction
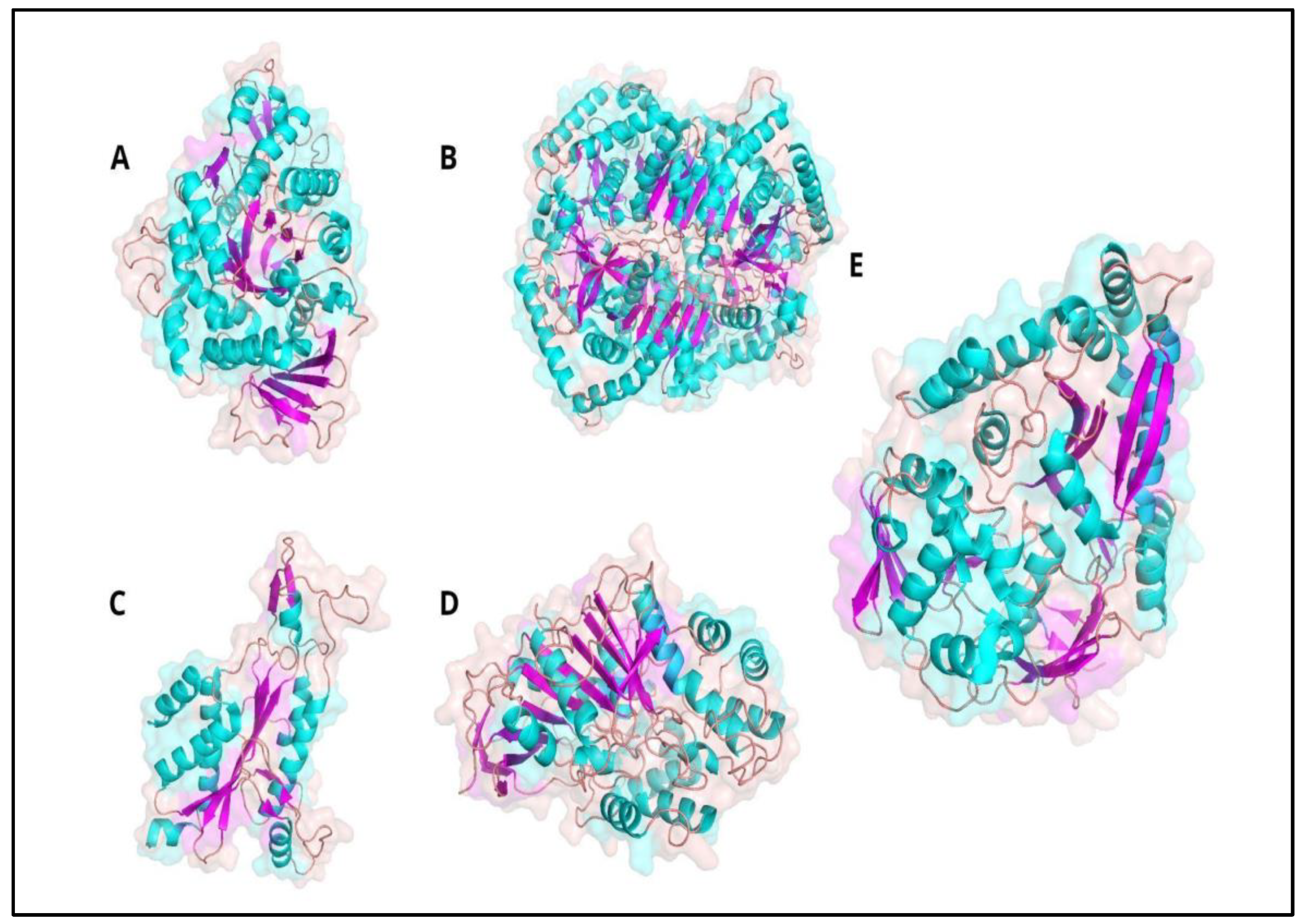
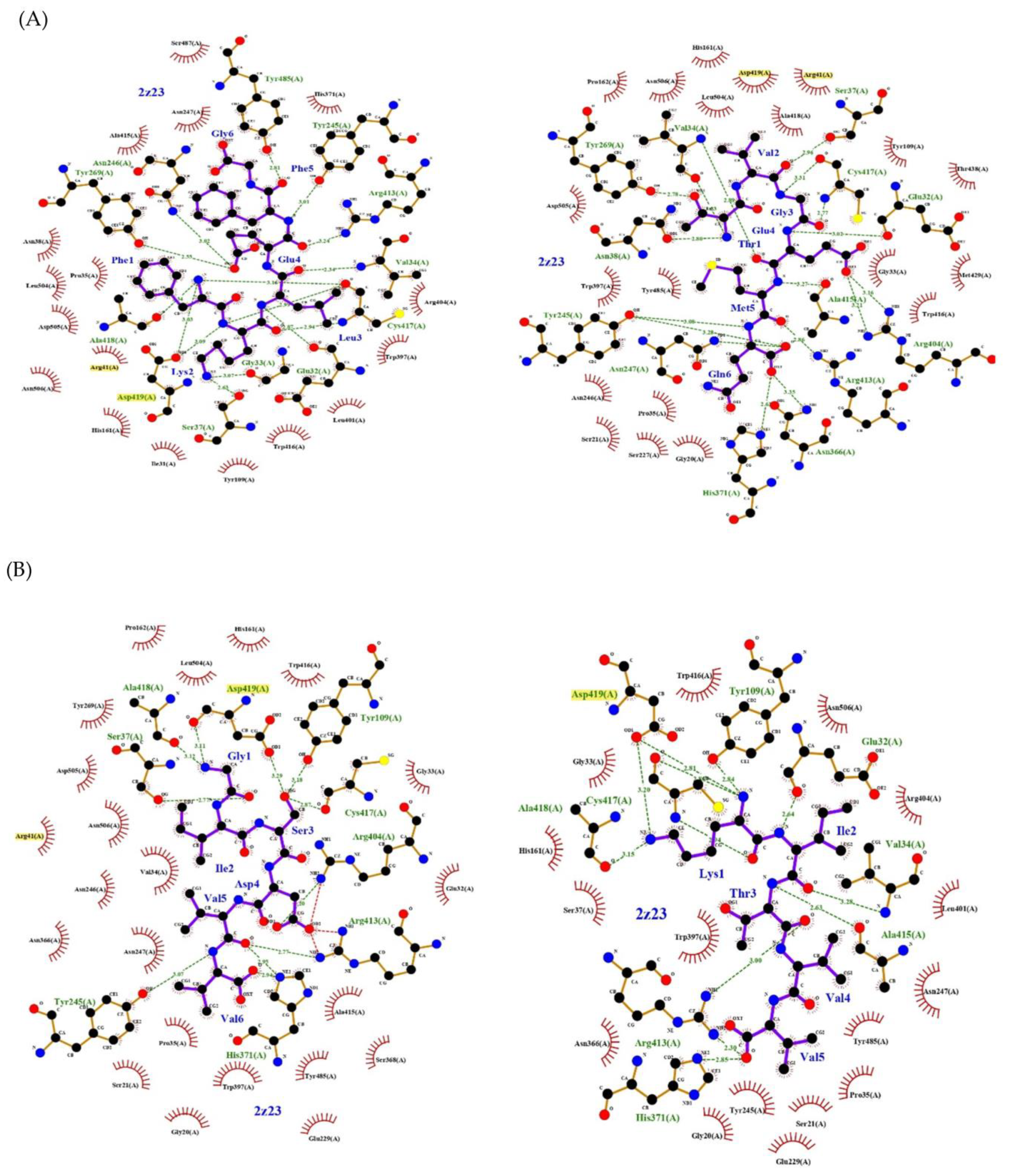
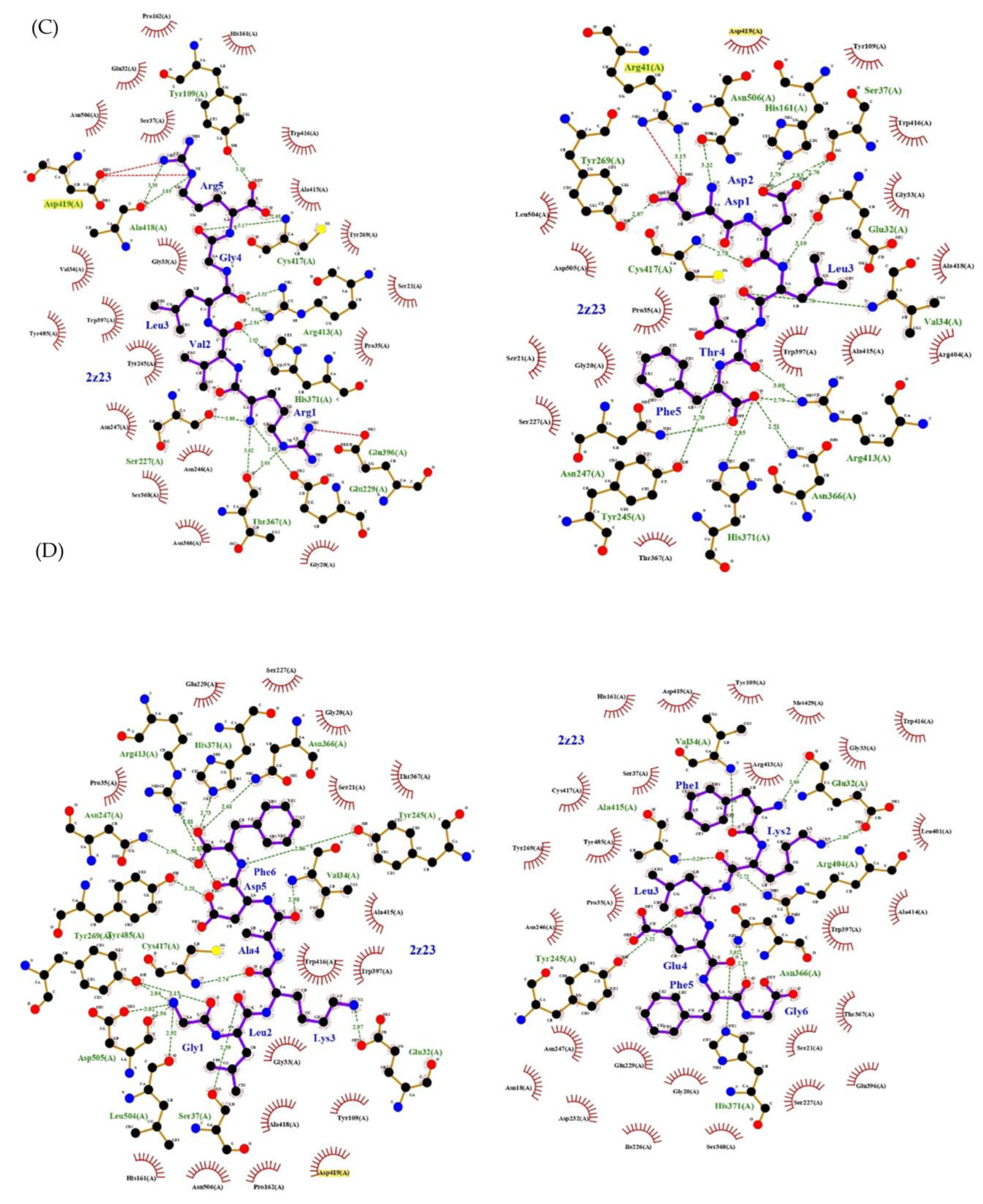
2. Results
3. Discussion
4. Materials and Methods
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Wani, A.K.; Akhtar, N.; Sher, F.; Navarrete, A.A.; Américo-Pinheiro, J. Microbial adaptation to different environmental conditions: Molecular perspective of evolved genetic and cellular systems. Arch. Microbiol. 2022, 204, 144. [Google Scholar] [CrossRef]
- Domnauer, M.; Zheng, F.; Li, L.; Zhang, Y.; Chang, C.E.; Unruh, J.R.; Conkright-Fincham, J.; McCroskey, S.; Florens, L.; Zhang, Y.; et al. Proteome plasticity in response to persistent environmental change. Mol. Cell 2021, 81, 3294–3309.e12. [Google Scholar] [CrossRef]
- Hartl, F.U.; Hayer-Hartl, M. Converging concepts of protein folding in vitro and in vivo. Nat. Struct. Mol. Biol. 2009, 16, 574–581. [Google Scholar] [CrossRef]
- Balchin, D.; Hayer-Hartl, M.; Hartl, F.U. Recent advances in understanding catalysis of protein folding by molecular chaperones. FEBS Lett. 2020, 594, 2770–2781. [Google Scholar] [CrossRef]
- Sharma, S.; Chakraborty, K.; Müller, B.K.; Astola, N.; Tang, Y.C.; Lamb, D.C.; Hayer-Hartl, M.; Hartl, F.U. Monitoring protein conformation along the pathway of chaperonin-assisted folding. Cell 2008, 133, 142–153. [Google Scholar] [CrossRef]
- Hartl, F.U.; Bracher, A.; Hayer-Hartl, M. Molecular chaperones in protein folding and proteostasis. Nature 2011, 475, 324–332. [Google Scholar] [CrossRef]
- Buchner, J. Molecular chaperones and protein quality control: An introduction to the JBC Reviews thematic series. J. Biol. Chem. 2019, 294, 2074–2075. [Google Scholar] [CrossRef]
- Pavlov, M.Y.; Ehrenberg, M. Optimal control of gene expression for fast proteome adaptation to environmental change. Proc. Natl. Acad. Sci. USA 2013, 110, 20527–20532. [Google Scholar] [CrossRef]
- Galindo, C.L.; Rosenzweig, J.A.; Kirtley, M.L.; Chopra, A.K. Pathogenesis of Y. enterocolitica and Y. pseudotuberculosis in Human Yersiniosis. J. Pathog. 2011, 2011, 182051. [Google Scholar] [CrossRef]
- Perry, R.D.; Fetherston, J.D. Yersinia pestis—Etiologic agent of plague. Clin. Microbiol. Rev. 1997, 10, 35–66. [Google Scholar] [CrossRef]
- De Groot, N.S.; Ventura, S. Protein aggregation profile of the bacterial cytosol. PLoS ONE 2010, 5, e9383. [Google Scholar] [CrossRef] [PubMed]
- Brady, M.F.; Yarrarapu, S.N.S.; Anjum, F. Yersinia Pseudotuberculosis; StatPearls Publishing: Tampa, FL, USA, 2022. [Google Scholar]
- Arhar, T.; Shkedi, A.; Nadel, C.M.; Gestwicki, J.E. The interactions of molecular chaperones with client proteins: Why are they so weak? J. Biol. Chem. 2021, 297, 101282. [Google Scholar] [CrossRef] [PubMed]
- Kim, H.; Wu, K.; Lee, C. Stress-Responsive periplasmic chaperones in bacteria. Front. Mol. Biosci. 2021, 8, 678697. [Google Scholar] [CrossRef] [PubMed]
- Sučec, I.; Bersch, B.; Schanda, P. How do chaperones bind (Partly) unfolded client proteins? Front. Mol. Biosci. 2021, 8, 762005. [Google Scholar] [CrossRef] [PubMed]
- Wu, C.; Chakrabarty, S.; Jin, M.; Liu, K.; Xiao, Y. Insect ATP-binding cassette (ABC) transporters: Roles in xenobiotic detoxification and Bt insecticidal activity. Int. J. Mol. Sci. 2019, 20, 2829. [Google Scholar] [CrossRef] [PubMed]
- Ter Beek, J.; Guskov, A.; Slotboom, D.J. Structural diversity of ABC transporters. J. Gen. Physiol. 2014, 143, 419–435. [Google Scholar] [CrossRef]
- Goemans, C.; Denoncin, K.; Collet, J.-F. Folding mechanisms of periplasmic proteins. Biochim. Biophys. Acta 2014, 1843, 1517–1528. [Google Scholar] [CrossRef]
- Poole, R.K.; Cozens, A.G.; Shepherd, M. The CydDC family of transporters. Res. Microbiol. 2019, 170, 407–416. [Google Scholar] [CrossRef]
- Jeckelmann, J.-M.; Erni, B. Transporters of glucose and other carbohydrates in bacteria. Pflug. Arch. Eur. J. Physiol. 2020, 472, 1129–1153. [Google Scholar] [CrossRef]
- Pichoff, S.; Du, S.; Lutkenhaus, J. Roles of FtsEX in cell division. Res. Microbiol. 2019, 170, 374–380. [Google Scholar] [CrossRef]
- Singh, R.; Liechti, G.; Slade, J.A.; Maurelli, A.T. Chlamydia trachomatis oligopeptide transporter performs dual functions of oligopeptide transport and peptidoglycan recycling. Infect. Immun. 2020, 88, e00086-20. [Google Scholar] [CrossRef]
- De Boer, M.; Gouridis, G.; Vietrov, R.; Begg, S.L.; Schuurman-Wolters, G.K.; Husada, F.; Eleftheriadis, N.; Poolman, B.; McDevitt, C.A.; Cordes, T. Conformational and dynamic plasticity in substrate-binding proteins underlies selective transport in ABC importers. eLife 2019, 8, e44652. [Google Scholar] [CrossRef] [PubMed]
- Matsuzaki, M.; Kiso, Y.; Yamamoto, I.; Satoh, T. Isolation of a periplasmic molecular chaperone-like protein of Rhodobacter sphaeroides f. sp. denitrificans that is homologous to the dipeptide transport protein DppA of Escherichia coli. J. Bacteriol. 1998, 180, 2718–2722. [Google Scholar] [CrossRef] [PubMed]
- Escobar Garduño, E.; Scior, T.; Soto Urzúa, L.; Martínez Morales, L.J. Identification of residues for chaperone-like activity of OppA protein in Yersinia pseudotuberculosis. AMB Express 2020, 10, 153. [Google Scholar] [CrossRef] [PubMed]
- Richarme, G.; Caldas, T.D. Chaperone properties of the bacterial periplasmic substrate-binding proteins. J. Biol. Chem. 1997, 272, 15607–15612. [Google Scholar] [CrossRef] [PubMed]
- Doeven, M.K.; Abele, R.; Tampé, R.; Poolman, B. The binding specificity of OppA determines the selectivity of the oligopeptide ATP-binding cassette transporter. J. Biol. Chem. 2004, 279, 32301–32307. [Google Scholar] [CrossRef]
- Wang, G.; Li, D.; Ma, X.; An, H.; Zhai, Z.; Ren, F.; Hao, Y. Functional role of oppA encoding an oligopeptide-binding protein from Lactobacillus salivarius Ren in bile tolerance. J. Ind. Microbiol. Biotechnol. 2015, 42, 1167–1174. [Google Scholar] [CrossRef]
- Bradshaw, J.L.; Pipkins, H.R.; Keller, L.E.; Pendarvis, J.K.; McDaniel, L.S. Mucosal infections and invasive potential of nonencapsulated Streptococcus pneumoniae are enhanced by oligopeptide binding proteins AliC and AliD. MBio 2018, 9, e02097-17. [Google Scholar] [CrossRef]
- LeDeaux, J.R.; Solomon, J.M.; Grossman, A.D. Analysis of non-polar deletion mutations in the genes of the spo0K (opp) operon of Bacillus subtilis. FEMS Microbiol. Lett. 2006, 153, 63–69. [Google Scholar] [CrossRef]
- Garault, P.; Le Bars, D.; Besset, C.; Monnet, V. Three oligopeptide-binding proteins are involved in the oligopeptide transport of Streptococcus thermophilus. J. Biol. Chem. 2002, 277, 32–39. [Google Scholar] [CrossRef]
- Cumberworth, A.; Lamour, G.; Babu, M.M.; Gsponer, J. Promiscuity as a functional trait: Intrinsically disordered regions as central players of interactomes. Biochem. J. 2013, 454, 361–369. [Google Scholar] [CrossRef] [PubMed]
- Dugay, A.R.; Silhavy, T.J. Quality control in the bacterial periplasm. Biochim. Biophys. Acta 2004, 1694, 121–134. [Google Scholar] [CrossRef] [PubMed]
- Taylor, A.; Zakai, S.A.; Kelly, D. The periplasmic chaperone network of Campylobacter jejuni: Evidence that SalC (Cj1289) and PpiD (Cj0694) are involved in maintaining outer membrane integrity. Front. Microbiol. 2017, 8, 531. [Google Scholar] [CrossRef] [PubMed]
- Mas, G.; Thoma, J.; Hiller, S. The Periplasmic Chaperones Skp and SurA. In Bacterial Cell Walls and Membranes; Sub-Cellular Biochemistry; Springer: Berlin/Heidelberg, Germany, 2019; Volume 92, pp. 169–186. [Google Scholar] [CrossRef]
- Papadopoulos, A.; Busch, M.; Reiners, J.; Hachani, E.; Bäumer, M.; Schmitt, L.; Jaeger, K.-E.; Kovacic, F.; Smits SH, J.; Kedrov, A. The periplasmic chaperone Skp prevents misfolding of the secretory Lipase A from Pseudomonas aeruginosa. Front. Mol. Biosci. 2022, 9, 1026724. [Google Scholar] [CrossRef]
- Johansen, J.; Rasmussen, A.A.; Overgaard, M.; Valentin-Hansen, P. Conserved small non-coding RNAs that belong to the σE regulon: Role in down-regulation of outer membrane proteins. J. Mol. Biol. 2006, 364, 1–8. [Google Scholar] [CrossRef]
- Picon, A.; Kunji, E.R.; Lanfermeijer, F.C.; Konings, W.N.; Poolman, B. Specificity mutants of the binding protein of the oligopeptide transport system of Lactococcus lactis. J. Bacteriol. 2000, 182, 1600–1608. [Google Scholar] [CrossRef]
- Tame, J.R.; Murshudov, G.N.; Dodson, E.J.; Neil, T.K.; Dodson, G.G.; Higgins, C.F.; Wilkinson, A.J. The structural basis of sequence-independent peptide binding by OppA protein. Science 1994, 264, 1578–1581. [Google Scholar] [CrossRef]
- Tame, J.R.; Dodson, E.J.; Murshudov, G.; Higgins, C.F.; Wilkinson, A.J. The crystal structures of the oligopeptide-binding protein OppA complexed with tripeptide and tetrapeptide ligands. Structure 1995, 3, 1395–1406. [Google Scholar] [CrossRef]
- Davies, T.G.; Hubbard, R.E.; Tame, J.R. Relating structure to thermodynamics: The crystal structures and binding affinity of eight OppA-peptide complexes. Protein Sci. 1999, 8, 1432–1444. [Google Scholar] [CrossRef]
- Klepsch, M.M.; Kovermann, M.; Löw, C.; Balbach, J.; Permentier, H.P.; Fusetti, F.; de Gier, J.W.; Slotboom, D.J.; Berntsson, R.A. Escherichia coli peptide binding protein OppA has a preference for positively charged peptides. J. Mol. Biol. 2011, 414, 75–85. [Google Scholar] [CrossRef]
- Lazar, S.W.; Kolter, R. SurA assists the folding of Escherichia coli outer membrane proteins. J. Bacteriol. 1996, 178, 1770–1773. [Google Scholar] [CrossRef] [PubMed]
- Lanfermeijer, F.C.; Detmers, F.J.; Konings, W.N.; Poolman, B. On the binding mechanism of the peptide receptor of the oligopeptide transport system of Lactococcus lactis. EMBO J. 2000, 19, 3649–3656. [Google Scholar] [CrossRef] [PubMed]
- Tanabe, M.; Mirza, O.; Bertrand, T.; Atkins, H.S.; Titball, R.W.; Iwata, S.; Brown, K.A.; Byrne, B. Structures of OppA and PstS from Yersinia pestis indicate variability of interactions with transmembrane domains. Acta Crystallogr. Sect. D Biol. Crystallogr. 2007, 63, 1185–1193. [Google Scholar] [CrossRef] [PubMed]
- Yamamoto, K.; Miyake, H.; Kusunoki, M.; Osaki, S. Crystal structure of isomaltase from Saccharomyces cerevisiae. FEBS J. 2010, 277, 4205–4214. [Google Scholar] [CrossRef]
- McPherson, A. Crystal structure of monoclinic rabbit muscle Lactate Dehydrogenase with four tetramers as the asymmetric unit. PDB 2020. [Google Scholar] [CrossRef]
- Kim, Y.; Grable, J.; Love, R.; Greene, P.; Rosenberg, J. X-ray structure of the DNA-Eco RI endonuclease-DNA recognition complex: The recognition network and the integration of recognition and cleavage. PDB 1995. [Google Scholar] [CrossRef]
- Grochulski, P.; Cygler, M. Two conformational states of Candida rugosa lipase. PDB 1994. [Google Scholar] [CrossRef]
- Yan, Y.; Tao, H.; He, J.Y.; Huang, S.-Y. The HDOCK server for integrated protein-protein docking. Nat. Protoc. 2020, 15, 1829–1852. [Google Scholar] [CrossRef]
- Tovchigrechko, A.; Vakser, I.A. GRAMM-X Public web server for Protein-to-Protein docking. Nucleic Acids Res. 2006, 34 (Suppl. 2), 310–314. [Google Scholar] [CrossRef]
- Pettersen, E.F.; Goddard, T.D.; Huang, C.C.; Couch, G.S.; Greenblatt, D.M.; Meng, E.C.; Ferrin, T.E. UCSF Chimera: A visualization system for exploratory research and analysis. Comput. Chem. 2004, 25, 1605–1612. [Google Scholar] [CrossRef]
- Trott, O.; Olson, A.J. AutoDock Vina: Improved docking speed and accuracy with a new scoring feature, efficient and multi-threaded optimization. J. Comput. Chem. 2010, 31, 455–461. [Google Scholar] [CrossRef] [PubMed]
- Wallace, A.C.; Laskowski, R.A.; Thornton, J.M. LIGPLOT: A program to generate schematic diagrams of protein-ligand interactions. Protein Eng. 1996, 8, 127–134. [Google Scholar] [CrossRef] [PubMed]

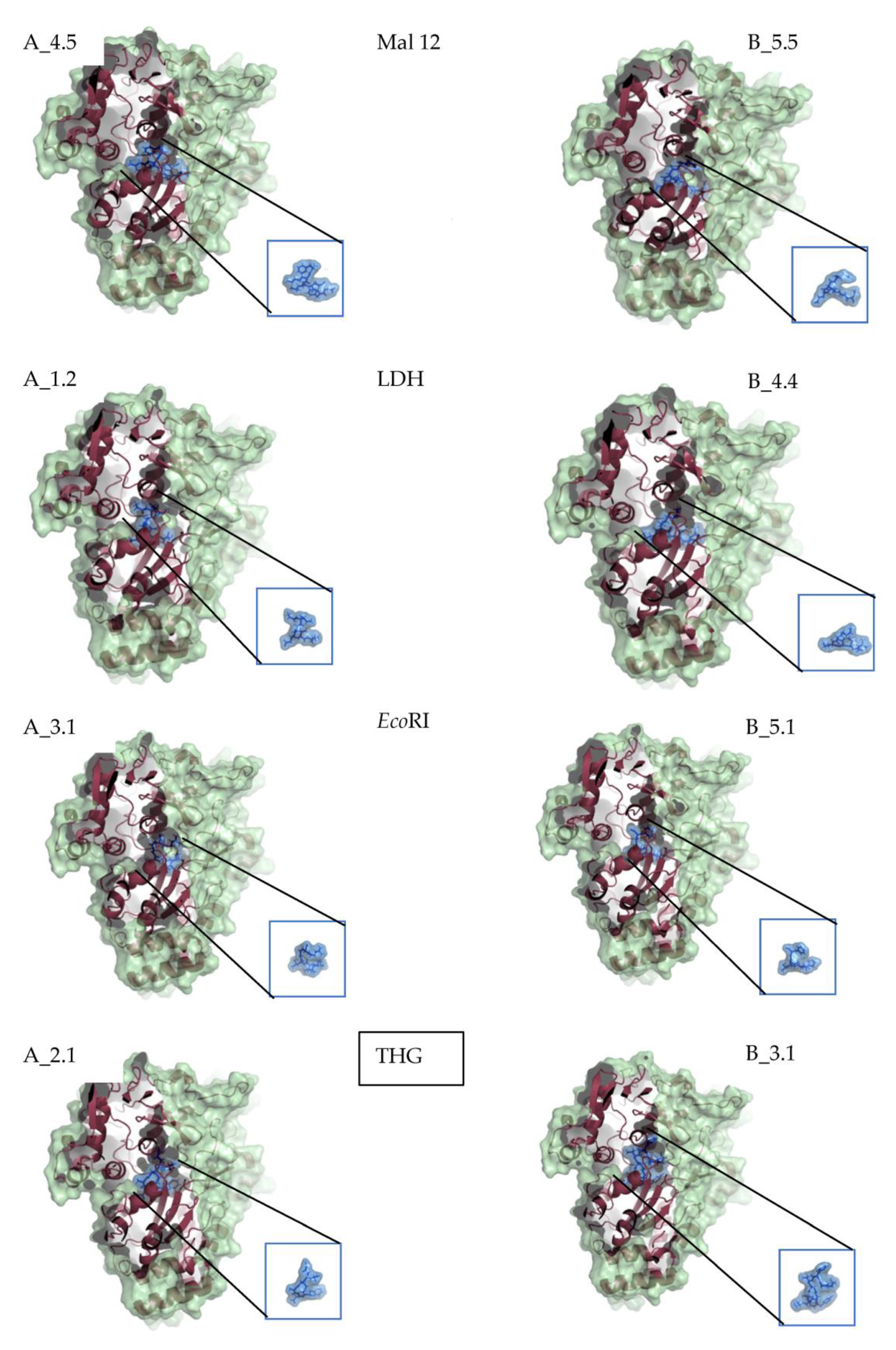
| Number of Ligands | Number of Conformations | Vina Score | Number Hydrogen Bonds | Number Salt Bridges | Average Hydrogen Bonds Distances in Å by LigProt | aa with Hydrogen Interactions | |
|---|---|---|---|---|---|---|---|
| Mal 12 | 4 | 5 | −7.3 | 15 | 2.8 | TYR_485, TYR_245, ARG_413 | |
| 5 | 5 | −9.4 | 12 | 2.8 | TYR_109, CYS_417, ALA_415 | ||
| LDH | 1 | 2 | −8 | 11 | 3 | 3.0 | ASP_419, TYR_109, CYS_417 |
| 4 | 4 | −7.8 | 12 | 2.8 | TYR_109, GLU_32, VAL_34 | ||
| EcoRI | 3 | 1 | −7 | 13 | 3 | 2.9 | TYR_109, CYS_417, ARG_413 |
| 5 | 1 | −9.6 | 16 | 1 | 2.9 | TYR_269, ARG_41, ASN_506 | |
| THG | 2 | 1 | −7.7 | 14 | 2 | 2.9 | HIS_161, CYS_417, ARG_404 |
| 3 | 1 | −8.2 | 9 | 2.8 | GLU_32, ARG_404, ASN_366 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ramírez, M.B.; Urzúa, L.S.; Martínez, M.d.l.Á.M.; Morales, L.J.M. Computational Analysis of the Ligand-Binding Sites of the Molecular Chaperone OppA from Yersinia pseudotuberculosis. Int. J. Mol. Sci. 2023, 24, 4023. https://doi.org/10.3390/ijms24044023
Ramírez MB, Urzúa LS, Martínez MdlÁM, Morales LJM. Computational Analysis of the Ligand-Binding Sites of the Molecular Chaperone OppA from Yersinia pseudotuberculosis. International Journal of Molecular Sciences. 2023; 24(4):4023. https://doi.org/10.3390/ijms24044023
Chicago/Turabian StyleRamírez, Mirian Becerril, Lucía Soto Urzúa, María de los Ángeles Martínez Martínez, and Luis Javier Martínez Morales. 2023. "Computational Analysis of the Ligand-Binding Sites of the Molecular Chaperone OppA from Yersinia pseudotuberculosis" International Journal of Molecular Sciences 24, no. 4: 4023. https://doi.org/10.3390/ijms24044023
APA StyleRamírez, M. B., Urzúa, L. S., Martínez, M. d. l. Á. M., & Morales, L. J. M. (2023). Computational Analysis of the Ligand-Binding Sites of the Molecular Chaperone OppA from Yersinia pseudotuberculosis. International Journal of Molecular Sciences, 24(4), 4023. https://doi.org/10.3390/ijms24044023





