Rapid Whole Genome Sequencing Diagnoses and Guides Treatment in Critically Ill Children in Belgium in Less than 40 Hours
Abstract
1. Introduction
2. Results
2.1. Demographic and Enrollment Criteria
2.2. Molecular Diagnostic Yield
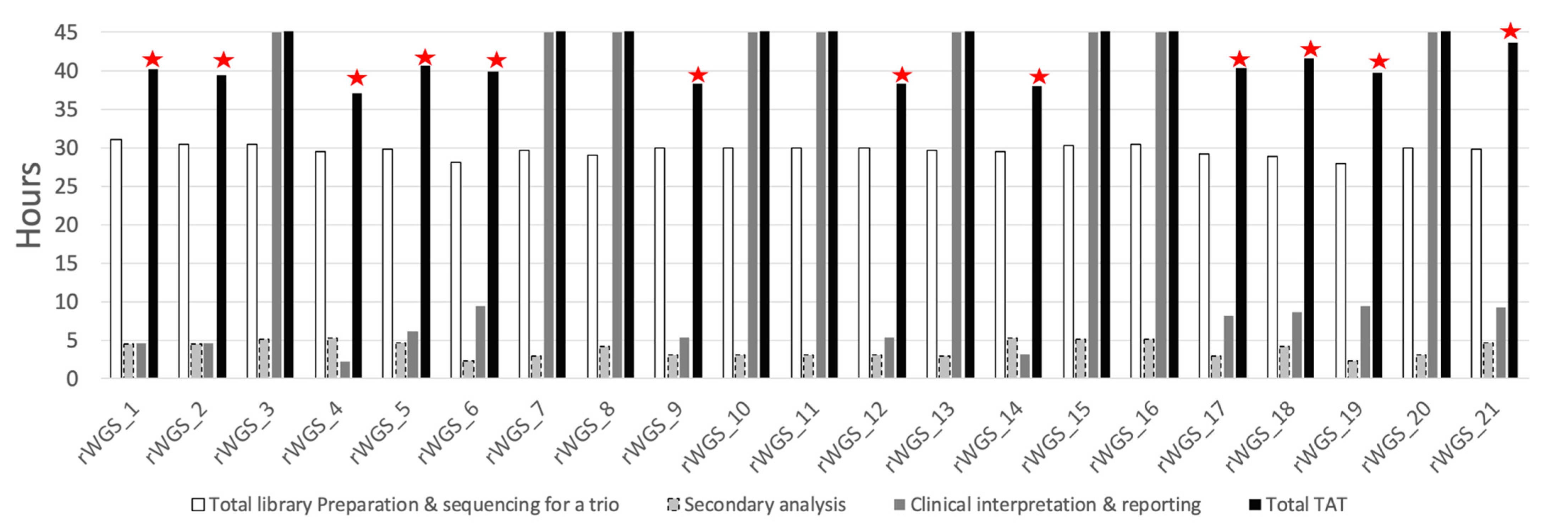
2.3. Turnaround Time
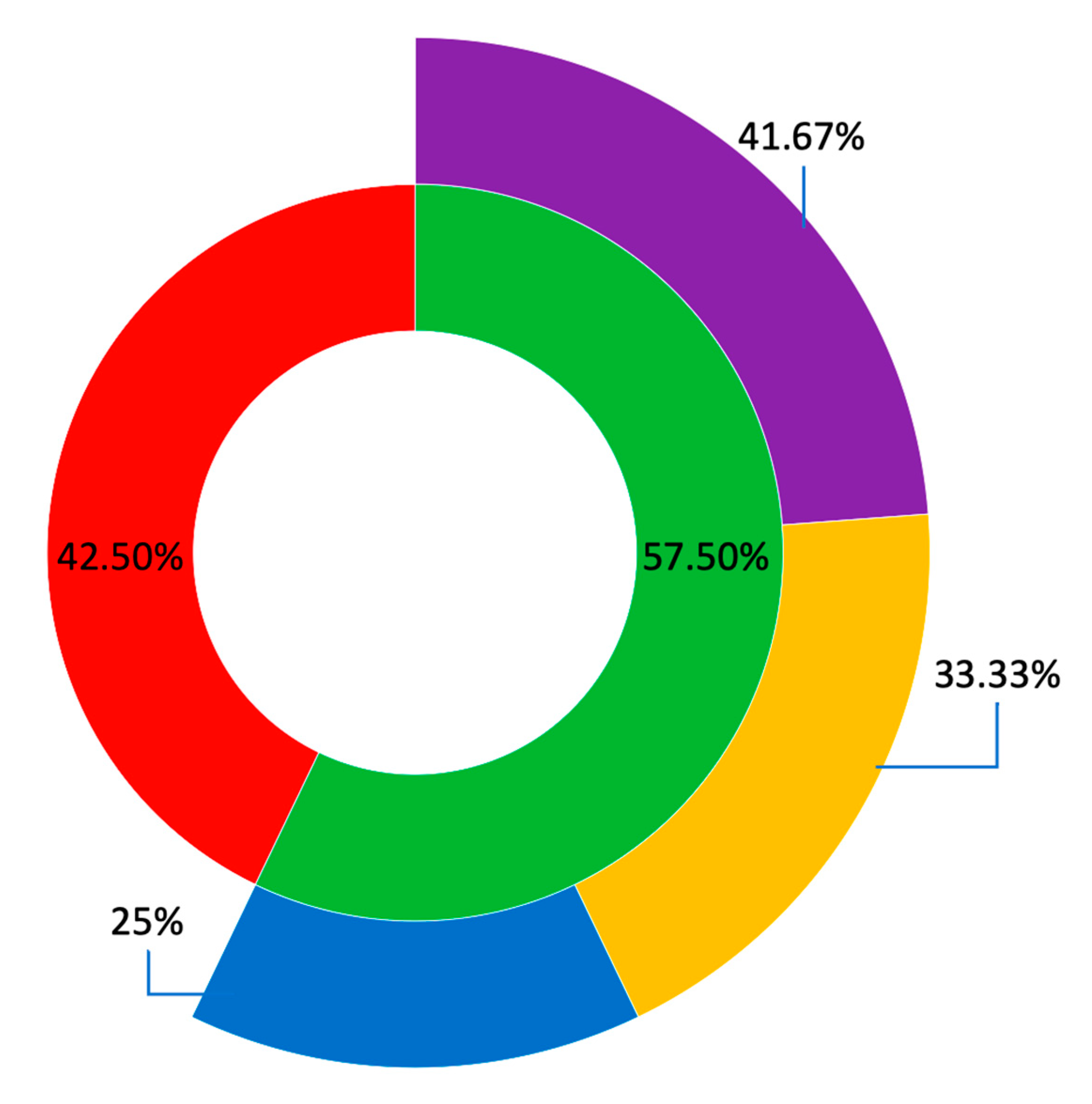
2.4. Clinical Utility
3. Discussion
3.1. Feasibility
3.2. Yield
3.3. TAT
3.4. Clinical Utility
3.5. Challenges/Limitations
4. Materials and Methods
4.1. Patients
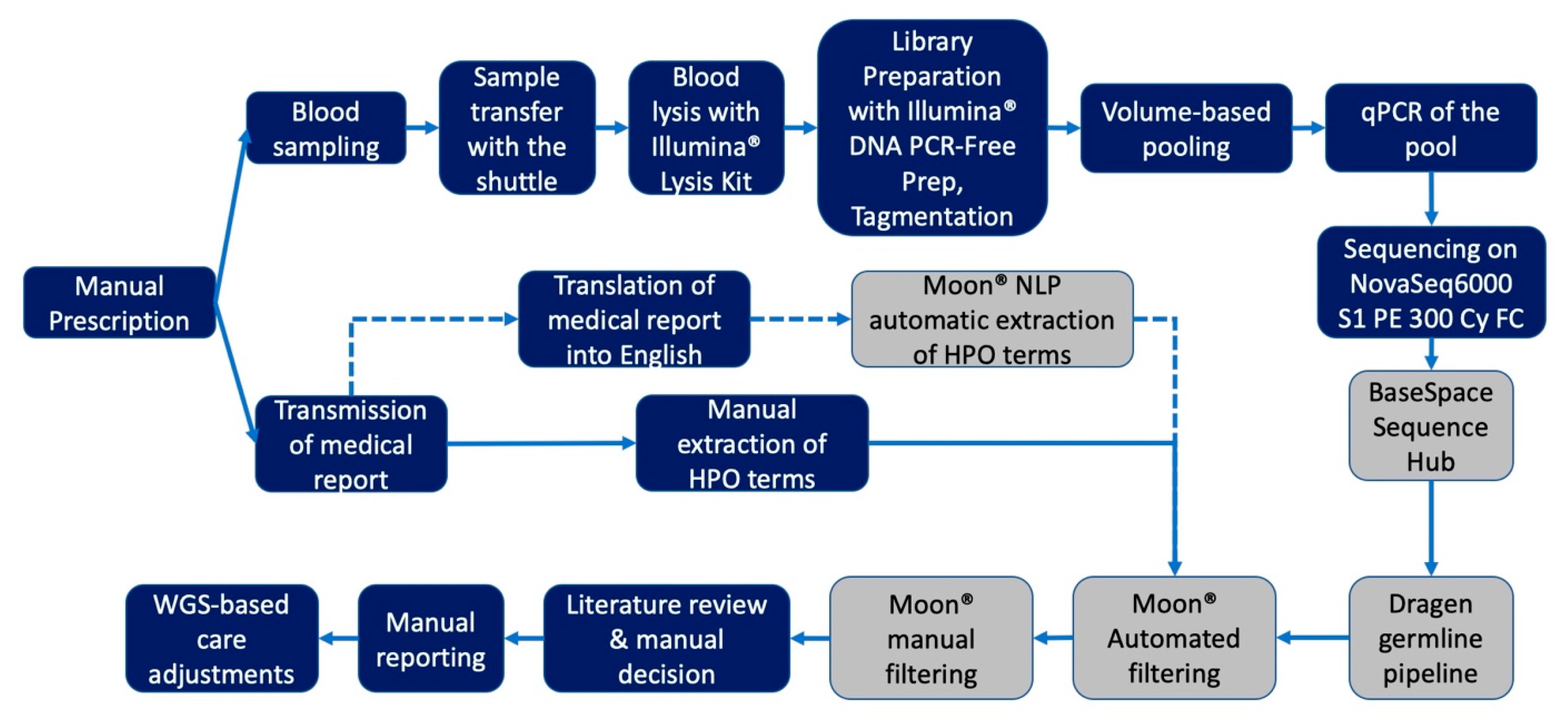
4.2. Study Workflow
4.3. Library Preparation and Sequencing
4.4. Analysis of Genomic Data
4.5. Phenotype Based Clinical Interpretation
4.6. Outcome Measures for the Clinical Utility
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Appendix A
References
- Wojcik, M.H.; Schwartz, T.S.; Thiele, K.E.; Paterson, H.; Stadelmaier, R.; Mullen, T.E.; VanNoy, G.E.; Genetti, C.A.; Madden, J.A.; Gubbels, C.S.; et al. Infant mortality: The contribution of genetic disorders. J. Perinatol. 2019, 39, 1611–1619. [Google Scholar] [CrossRef] [PubMed]
- Murphy, S.L.; Kochanek, K.D.; Xu, J. Deaths: Final Data for 2015. Natl. Vital. Stat. Rep. 2017, 66, 1–75. [Google Scholar] [PubMed]
- Meng, L.; Pammi, M.; Saronwala, A.; Magoulas, P.; Ghazi, A.R.; Vetrini, F.; Zhang, J.; He, W.; Dharmadhikari, A.V.; Qu, C.; et al. Use of exome sequencing for infants in intensive care units. JAMA Pediatr. 2017, 171, e173438. [Google Scholar] [CrossRef] [PubMed]
- Halabi, N.; Ramaswamy, S.; El Naofal, M.; Taylor, A.; Yaslam, S.; Jain, R.; Alfalasi, R.; Shenbagam, S.; Bitzan, M.; Yavuz, L.; et al. Rapid whole genome sequencing of critically ill pediatric patients from genetically underrepresented populations. Genome Med. 2022, 14, 56. [Google Scholar] [CrossRef]
- French, C.E.; Delon, I.; Dolling, H.; Sanchis-Juan, A.; Shamardina, O.; Mégy, K.; Abbs, S.; Austin, T.; Bowdin, S.; Branco, R.G.; et al. Whole genome sequencing reveals that genetic conditions are frequent in intensively ill children. Intensive Care Med. 2019, 45, 627–636. [Google Scholar] [CrossRef]
- Mc Laughlin, R.; Hardiman, O. Detection of long repeat expansions from PCR-free whole-genome sequence data. Genome Res. 2017, 27, 1895–1903. [Google Scholar] [CrossRef]
- Nicholas, T.J.; Al-Sweel, N.; Farrell, A.; Mao, R.; Bayrak-Toydemir, P.; Miller, C.E.; Bentley, D.; Palmquist, R.; Moore, B.; Hernandez, E.J.; et al. Comprehensive variant calling from whole-genome sequencing identifies a complex inversion that disrupts ZFPM2 in familial congenital diaphragmatic hernia. Mol. Genet. Genom. Med. 2022, 10, e1888. [Google Scholar] [CrossRef]
- Jiao, Q.; Sun, H.; Zhang, H.; Wang, R.; Li, S.; Sun, D.; Yang, X.A.; Jin, Y. The combination of whole-exome sequencing and copy number variation sequencing enables the diagnosis of rare neurological disorders. Clin. Genet. 2019, 96, 140–150. [Google Scholar] [CrossRef]
- Choudhry, N.; Golding, J.; Manry, M.W.; Rao, R.C. Whole genome sequencing increases molecular diagnostic yield compared with current diagnostic testing for inherited retinal disease. Ophthalmology 2016, 123, 1143–1150. [Google Scholar] [CrossRef]
- Gorzynski, J.E.; Goenka, S.D.; Shafin, K.; Jensen, T.D.; Fisk, D.G.; Grove, M.E.; Spiteri, E.; Pesout, T.; Monlong, J.; Baid, G.; et al. Ultrarapid nanopore genome sequencing in a critical care setting. N. Engl. J. Med. 2022, 386, 700–702. [Google Scholar] [CrossRef]
- Farnaes, L.; Hildreth, A.; Sweeney, N.M.; Clark, M.M.; Chowdhury, S.; Nahas, S.; Cakici, J.A.; Benson, W.; Kaplan, R.H.; Kronick, R.; et al. Rapid whole-genome sequencing decreases infant morbidity and cost of hospitalization. NPJ Genom. Med. 2018, 3, 10. [Google Scholar] [CrossRef] [PubMed]
- Farnaes, L.; Hildreth, A.; Sweeney, N.M.; Clark, M.M.; Chowdhury, S.; Nahas, S.; Cakici, J.A.; Benson, W.; Kaplan, R.H.; Kronick, R.; et al. Does genomic sequencing early in the diagnostic trajectory make a difference? A follow-up study of clinical outcomes and cost-effectiveness. Genet. Med. 2019, 21, 173–180. [Google Scholar] [CrossRef]
- Fishman, V.; Battulin, N.; Nuriddinov, M.; Maslova, A.; Zlotina, A.; Strunov, A.; Chervyakova, D.; Korablev, A.; Serov, O.; Krasikova, A. Diagnosis of genetic diseases in seriously ill children by rapid whole-genome sequencing and automated phenotyping and interpretation. Sci. Transl. Med. 2019, 11, eaat6177. [Google Scholar] [CrossRef]
- Van Diemen, C.C.; Kerstjens-Frederikse, W.S.; Bergman, K.A.; De Koning, T.J.; Sikkema-Raddatz, B.; Van Der Velde, J.K.; Abbott, K.M.; Herkert, J.C.; Löhner, K.; Rump, P.; et al. Rapid targeted genomics in critically ill newborns. Pediatrics 2017, 140, e20162854. [Google Scholar] [CrossRef] [PubMed]
- Mestek-Boukhibar, L.; Clement, E.; Jones, W.D.; Drury, S.; Ocaka, L.; Gagunashvili, A.; Stabej, P.L.Q.; Bacchelli, C.; Jani, N.; Rahman, S.; et al. Rapid paediatric sequencing (RaPS): Comprehensive real-life workflow for rapid diagnosis of critically ill children. J. Med. Genet. 2018, 55, 721–728. [Google Scholar] [CrossRef]
- Śmigiel, R.; Biela, M.; Szmyd, K.; Błoch, M.; Szmida, E.; Skiba, P.; Walczak, A.; Gasperowicz, P.; Kosińska, J.; Rydzanicz, M.; et al. Rapid whole-exome sequencing as a diagnostic tool in a neonatal/pediatric intensive care unit. J. Clin. Med. 2020, 9, 2220. [Google Scholar] [CrossRef]
- Statbel. Births and Fertility. Belgium in Figure. Available online: https://statbel.fgov.be/en (accessed on 11 February 2022).
- Leroy, C.; Van Leeuw, V.; Rigo, V. Grossesses Gémellaires (Santé Périnatale en Wallonie—Année 2018); Centre D’Épidémiologie Périnatale (CePIP): Bruxelles, Belgium, 2020. [Google Scholar]
- Franck, L.S.; Kriz, R.M.; Rego, S.; Garman, K.; Hobbs, C.; Dimmock, D. Implementing rapid whole-genome sequencing in critical care: A qualitative study of facilitators and barriers to new technology adoption. J. Pediatr. 2021, 237, 237–243. [Google Scholar] [CrossRef]
- Ference, B.A.; Kastelein, J.J.; Ray, K.K.; Ginsberg, H.N.; Chapman, M.J.; Packard, C.J.; Laufs, U.; Oliver-Williams, C.; Wood, A.M.; Butterworth, A.S.; et al. Feasibility of ultra-rapid exome sequencing in critically ill infants and children with suspected monogenic conditions in the Australian public health care system. JAMA 2020, 323, 2503. [Google Scholar] [CrossRef]
- Stark, Z.; Ellard, S. Rapid genomic testing for critically ill children: Time to become standard of care? Eur. J. Hum. Genet. 2022, 30, 142–149. [Google Scholar] [CrossRef]
- Minoche, A.E.; Lundie, B.; Peters, G.B.; Ohnesorg, T.; Pinese, M.; Thomas, D.M.; Zankl, A.; Roscioli, T.; Schonrock, N.; Kummerfeld, S.; et al. ClinSV: Clinical grade structural and copy number variant detection from whole genome sequencing data. Genome Med. 2021, 13, 32. [Google Scholar] [CrossRef]
- Dong, X.; Liu, B.; Yang, L.; Wang, H.; Wu, B.; Liu, R.; Chen, H.; Chen, X.; Yu, S.; Chen, B.; et al. Clinical exome sequencing as the first-tier test for diagnosing developmental disorders covering both CNV and SNV: A Chinese cohort. J. Med. Genet. 2020, 57, 558–566. [Google Scholar] [CrossRef] [PubMed]
- Wright, C.F.; Fitzgerald, T.W.; Jones, W.D.; Clayton, S.; McRae, J.F.; Van Kogelenberg, M.; King, D.A.; Ambridge, K.; Barrett, D.M.; Bayzetinova, T.; et al. Genetic diagnosis of developmental disorders in the DDD study: A scalable analysis of genome-wide research data. Lancet 2015, 385, 1305–1314. [Google Scholar] [CrossRef] [PubMed]
- Dimmock, D.; Caylor, S.; Waldman, B.; Benson, W.; Ashburner, C.; Carmichael, J.L.; Carroll, J.; Cham, E.; Chowdhury, S.; Cleary, J.; et al. Project baby bear: Rapid precision care incorporating rWGS in 5 California children’s hospitals demonstrates improved clinical outcomes and reduced costs of care. Am. J. Hum. Genet. 2021, 108, 1231–1238. [Google Scholar] [CrossRef] [PubMed]
- Van Den Bogaert, K.; Lannoo, L.; Brison, N.; Gatinois, V.; Baetens, M.; Blaumeiser, B.; Boemer, F.; Bourlard, L.; Bours, V.; De Leener, A.; et al. Outcome of publicly funded nationwide first-tier noninvasive prenatal screening. Genet. Med. 2021, 23, 1137–1142. [Google Scholar] [CrossRef]
- Illumina. Balancing Sample Coverage for Whole-Genome Sequencing. 2021. Available online: https://www.illumina.com/content/dam/illumina/gcs/assembled-assets/marketing-literature/illumina-dna-prep-pcr-free-index-correction-tech-note-m-gl-00005/illumina-dna-pcr-free-index-correction-tech-note-m-gl-00005.pdf (accessed on 28 December 2022).
- Miller, N.A.; Farrow, E.G.; Gibson, M.; Willig, L.K.; Twist, G.; Yoo, B.; Marrs, T.; Corder, S.; Krivohlavek, L.; Walter, A.; et al. A 26-hour system of highly sensitive whole genome sequencing for emergency management of genetic diseases. Genome Med. 2015, 7, 100. [Google Scholar] [CrossRef]
- Illumina. DRAGEN v3.2.8 Software Release Notes. 2019. Available online: https://support.illumina.com/content/dam/illumina-support/documents/downloads/software/dragen/dragen-v3-2-8-release-notes-1000000076767-00.pdf (accessed on 28 December 2022).
- Scocchia, A.; Wigby, K.M.; Masser-Frye, D.; Del Campo, M.; Galarreta, C.I.; Thorpe, E.; McEachern, J.; Robinson, K.; Gross, A.; Ajay, S.S. Clinical whole genome sequencing as a first-tier test at a resource-limited dysmorphology clinic in Mexico. NPJ Genom. Med. 2019, 4, 5. [Google Scholar] [CrossRef]
| rWGS_1 | rWGS_2 | rWGS_3 | rWGS_4 | rWGS_5 | rWGS_6 | rWGS_7 | rWGS_8 | rWGS_9 | rWGS_10 | rWGS_11 | rWGS_12 | rWGS_13 | rWGS_14 | rWGS_15 | rWGS_16 | rWGS_17 | rWGS_18 | rWGS_19 | rWGS_20 | rWGS_21 | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Age in year (days) | 0.24 (86) | 1.25 (457) | 9.33 (3408) | 0.03 (11) | 0.05 (20) | 18.31 (6688) | 0.08 (28) | 0.03 (11) | 1.67 (609) | 0.02 (9) | 0.01 (2) | 1.28 (467) | 0.39 (142) | 0.07 (25) | 0.47 (170) | 1.25 (457) | 0.07 (24) | 0.27 (100) | 0.05 (18) | 1.32 (481) | 5.37 (1963) |
| Gender | F | M | F | M | M | M | F | F | F | F | M | F | M | F | M | F | M | F | F | M | M |
| Recruitment unit | PICU | NEURO | NEURO | NICU | NICU | PICU | NICU | NICU | NEURO | NICU | NICU | NEURO | NEURO | NICU | PICU | NEURO | NICU | PICU | NICU | PICU | PICU |
| Patients | Pedigree | Phenotype | OMIM Diseases | Gene | Variant Coordinate (GRCh37) | HGVS c. | Protein Position | Zygocity | Inheritance | ACMG-AMP Classification |
|---|---|---|---|---|---|---|---|---|---|---|
| rWGS_1 | Trio | Muscle hypotonia; Abnormality of brainstem morphology; Frontotemporal cerebral atrophy; Encephalopathy; Progressive encephalopathy; Apneic episodes in infancy; Strabismus; Nystagmus; Abnormal corpus callosum morphology; Enlarged sylvian cistern; Laryngomalacia; Abnormal eye movements | KLF7-related Developmental delay/intellectual disability with neuromuscular and psychiatric symptoms | KLF7 | 2:207,953,249 | NM_003709.4: c.790G > A | p.Asp264As | Heterozygous | De novo | Pathogenic |
| rWGS_2 | Trio | Seizures; Hypotonia, early; Hypotonia, generalized; Large fontanelles; Neurodevelopmental delay; Respiratory infections in early life; Recurrent respiratory infections; Dystonia; Increased serum lactate; Tented upper lips vermilions; Anteverted nares; High palate | Aromatic L-amino acid decarboxylase deficiency (AADCD) | DDC | 7:50,566,899 | NM_001082971.2: c.823G > A | p.Ala275Thr | Heterozygous | Paternaly inherited | Pathogenic |
| 7:5,054,432 | NM_005859.5: c.1037A > G | p.Tyr346Cys | Heterozygous | Maternaly inherited | Likely Pathogenic | |||||
| rWGS_3 | Trio | Mixed demyelinating and axonal polyneuropathy; Progressive polyneuropathy; Progressive degeneration of movement; Difficulty in walking; Progressive muscle weakness; Feeding difficulties; Weight loss; Sensorimotor polyneuropathy affecting arms more than legs; Motor conduction block; Gastroesophageal reflux; Quadriparesis; Toe-walking | Unsolved | |||||||
| rWGS_4 | Trio | Generalized hypotonia; Generalized neonatal hypotonia; Neonatal hypotonia; Severe muscular hypotonia; Respiratory distress | Neurodevelopmental disorder with neonatal respiratory insufficiency, hypotonia, and feeding difficulties | PURA | 5:139,494,456–139,494,458 | NM_005859.4: c.697_699delTTC | p.Phe233del | Heterozygous | De novo | Pathogenic |
| rWGS_5 | Trio | Chylothorax; Facial dysmorphism | Lymphedema, hereditary, III | PIEZO1 | 16:88,851,308 | NM_001142864.4: c.64 + 1G > A | Heterozygous | De novo | Pathogenic | |
| 16:88,792,770 | NM_001142864.4: c.3890T > C | p.Leu1297Pro | Heterozygous | Paternaly inherited | VUS | |||||
| 16:88,783,093 | NM_001142864.4: c.6800C > T | p.Thr2267Met | Heterozygous | Maternaly inherited | VUS | |||||
| rWGS_6 | Duo | Brain calcification; Intellectual impairment; Thrombocytopenia; Leukopenia; Hepatic cirrhosis; Brain imaging abnormality; Neurodevelopmental delay; Retinitis pigmentosa; Speech delay | COACH syndrome 1 | TMEM67 | 8:94,811,875 | NM_153704.6: c.2130G > A | p.Met710Ile | Homozygous | Biparental | VUS |
| Urbach-Wiethe disease | ECM1 | 1:150,480,710 | NM_004425.4: c.25T > A | p.Leu9Met | Homozygous | Biparental | VUS | |||
| rWGS_7 | Trio | Intrauterine growth retardation; Hydrocephaly; Aqueduct of sylvius stenosis; Severe intrauterine growth retardation; Leukopenia; Cholestasis; Thrombocytopenia; Neutropenia; Metabolic acidosis; Atrial septum defect | Unsolved | |||||||
| rWGS_8 | Trio | Severe muscular hypotonia; Lactic acidosis; Hyperammonemia, acute; Hyperammonemia; Neonatal asphyxia | Unsolved | |||||||
| rWGS_9 | Trio | Renal insufficiency; Failure to thrive; Hyperlaxity; Hemiparesis; Unilateral multifocal epileptiform discharges; Hemiclonic seizures; Seizure; Global developmental delay | Mitochondrial DNA depletion syndrome 4A (Alpers type) | POLG | 15:89,864,238 | NM_002693.3: c.2740A > C | p.Thr914Pro | Heterozygous | Maternaly inherited | Pathogenic |
| 15:89,870,432 | NM_002693.2: c.1399G > A | p.Ala467Thr | Heterozygous | Paternaly inherited | Pathogenic | |||||
| rWGS_10 | Trio | Abnormality of the brainstem white matter; Increased urine alpha-ketoglutarate concentration; Leukodystrophy; Multifocal seizures; Generalized clonic seizure; Tonic-clonic convulsions; Generalized convulsive status epilepticus; Bilateral tonic-clonic seizure with generalised onset | Unsolved | |||||||
| rWGS_11 | Trio | Brain imaging abnormality; Increased troponin i level in blood; Abnormal metabolic brain imaging; EEG with burst suppression; Opacification of the corneal epithelium; 3-methylglutaric aciduria; Lactic acidosis; Leukoencephalopathy; Abnormality of the basal ganglia; Corneal opacity | Unsolved | |||||||
| rWGS_12 | Duo | High arched palate; Hypertelorism; Hypotonia; Facial dysmorphism; Abnormal interventricular septum morphology; Poor speech; Failure to thrive | Noonan syndrome 1 | PTPN11 | 12:112,915,524 | NM_002834.5: c.923A > G | p.Asn308Ser | Heterozygous | Absent in mother, father not available for testing | Pathogenic |
| rWGS_13 | Trio | Hypernatremia; metabolic acidosis; generalized seizures, loss of consciousness | Unsolved | |||||||
| rWGS_14 | Trio | Cholestasis; Aortic valve stenosis; Conjugated hyperbilirubinemia; Jaundice; Episodic vomiting; Feeding difficulties in infancy; Meconium ileus; Elevated gamma-glutamyltransferase level; Prolonged neonatal jaundice; Nasogastric tube feeding in infancy | Alagille syndrome 1 | JAG1 | 20:10,621,792 | NM_000214.3: c.3016_3017delCC | p.Pro1006PhefsTer5 | Heterozygous | De novo | Pathogenic |
| rWGS_15 | Trio | Retinal dystrophy; Generalized hypotonia; Tracheomalacia; Sensorineural hearing impairment; Generalized neonatal hypotonia; Ptosis; Unilateral ptosis; Feeding difficulties; Absent or decreased deep tendon reflexes; Respiratory distress | Unsolved | |||||||
| rWGS_16 | Trio | Hypoplasia of the maxilla; Rhinitis; Central apnea; Hypotonia; Growth delay; Hypothermia; Night sweats; Dysautonomia; Failure to thrive; Cerebellar hemorrhage; Delayed gross motor development | Unsolved | |||||||
| rWGS_17 | Trio | Thick vermilion border; Plagiocephaly; Polyhydramnios; Seizure; Hypotonia; Prominent nose; Respiratory distress; Anteverted ears; Preauricular pit; Broad nose; Thick upper lip vermilion; Status epilepticus | Epileptic encephalopathy, early infantile, 14 | KCNT1 | 9:138,656,902 | NM_020822.3: c.1061T > A | p.Met354Lys | Heterozygous | De novo | Likely Pathogenic |
| rWGS_18 | Trio | Dysphagia; Respiratory distress; Stridor; Facial paralysis; Psychomotor retardation; Retrognathia; Camptodactyly; Agenesis of cerebellar vermis HP | TUBB3-related spectrum syndrome | TUBB3 | 16:90,001,644 | NM_006086.4: c.785G > A | p.Arg262His | Heterozygous | De novo | Pathogenic |
| rWGS_19 | Trio | Hypoglycemia; Metabolic acidosis; Lactic acidosis;3-methylglutaconic aciduria;Noncompaction cardiomyopathy; Neonatal hypoglycemia; Hyperammonemia | 3-hydroxy-3-methylglutaryl-Coa lyase deficiency | HMGCL | 1:24,143,241 | NM_000191.3: c.272T > A | p.Val91Asp | Homozygous | Likely Pathogenic | |
| rWGS_20 | Trio | Renal insufficiency; Increased intracranial pressure; Lactic acidosis; Acute rhabdomyolysis; Convulsive status epilepticus; Recurrent hypoglycemia; Cerebral edema | Unsolved | |||||||
| rWGS_21 | Trio | Conjunctivitis; Failure to thrive; Macrocytic anemia; Steatorrhea; Abnormality of the face; Short stature; Decreased body weight; low vitamine A and D; hypoalbuminemia | Infantile isolated exocrine pancreatic insufficiency, SPINK1-related | SPINK1 | 5:147,211,114 | NM_001379610.1: c.27delC | p.Ser10ValfsTer6 | Homozygous | Biparental | Pathogenic |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lumaka, A.; Fasquelle, C.; Debray, F.-G.; Alkan, S.; Jacquinet, A.; Harvengt, J.; Boemer, F.; Mulder, A.; Vaessen, S.; Viellevoye, R.; et al. Rapid Whole Genome Sequencing Diagnoses and Guides Treatment in Critically Ill Children in Belgium in Less than 40 Hours. Int. J. Mol. Sci. 2023, 24, 4003. https://doi.org/10.3390/ijms24044003
Lumaka A, Fasquelle C, Debray F-G, Alkan S, Jacquinet A, Harvengt J, Boemer F, Mulder A, Vaessen S, Viellevoye R, et al. Rapid Whole Genome Sequencing Diagnoses and Guides Treatment in Critically Ill Children in Belgium in Less than 40 Hours. International Journal of Molecular Sciences. 2023; 24(4):4003. https://doi.org/10.3390/ijms24044003
Chicago/Turabian StyleLumaka, Aimé, Corinne Fasquelle, Francois-Guillaume Debray, Serpil Alkan, Adeline Jacquinet, Julie Harvengt, François Boemer, André Mulder, Sandrine Vaessen, Renaud Viellevoye, and et al. 2023. "Rapid Whole Genome Sequencing Diagnoses and Guides Treatment in Critically Ill Children in Belgium in Less than 40 Hours" International Journal of Molecular Sciences 24, no. 4: 4003. https://doi.org/10.3390/ijms24044003
APA StyleLumaka, A., Fasquelle, C., Debray, F.-G., Alkan, S., Jacquinet, A., Harvengt, J., Boemer, F., Mulder, A., Vaessen, S., Viellevoye, R., Palmeira, L., Charloteaux, B., Brysse, A., Bulk, S., Rigo, V., & Bours, V. (2023). Rapid Whole Genome Sequencing Diagnoses and Guides Treatment in Critically Ill Children in Belgium in Less than 40 Hours. International Journal of Molecular Sciences, 24(4), 4003. https://doi.org/10.3390/ijms24044003





