Calling and Phasing of Single-Nucleotide and Structural Variants of the LDLR Gene Using Oxford Nanopore MinION
Abstract
1. Introduction
2. Results
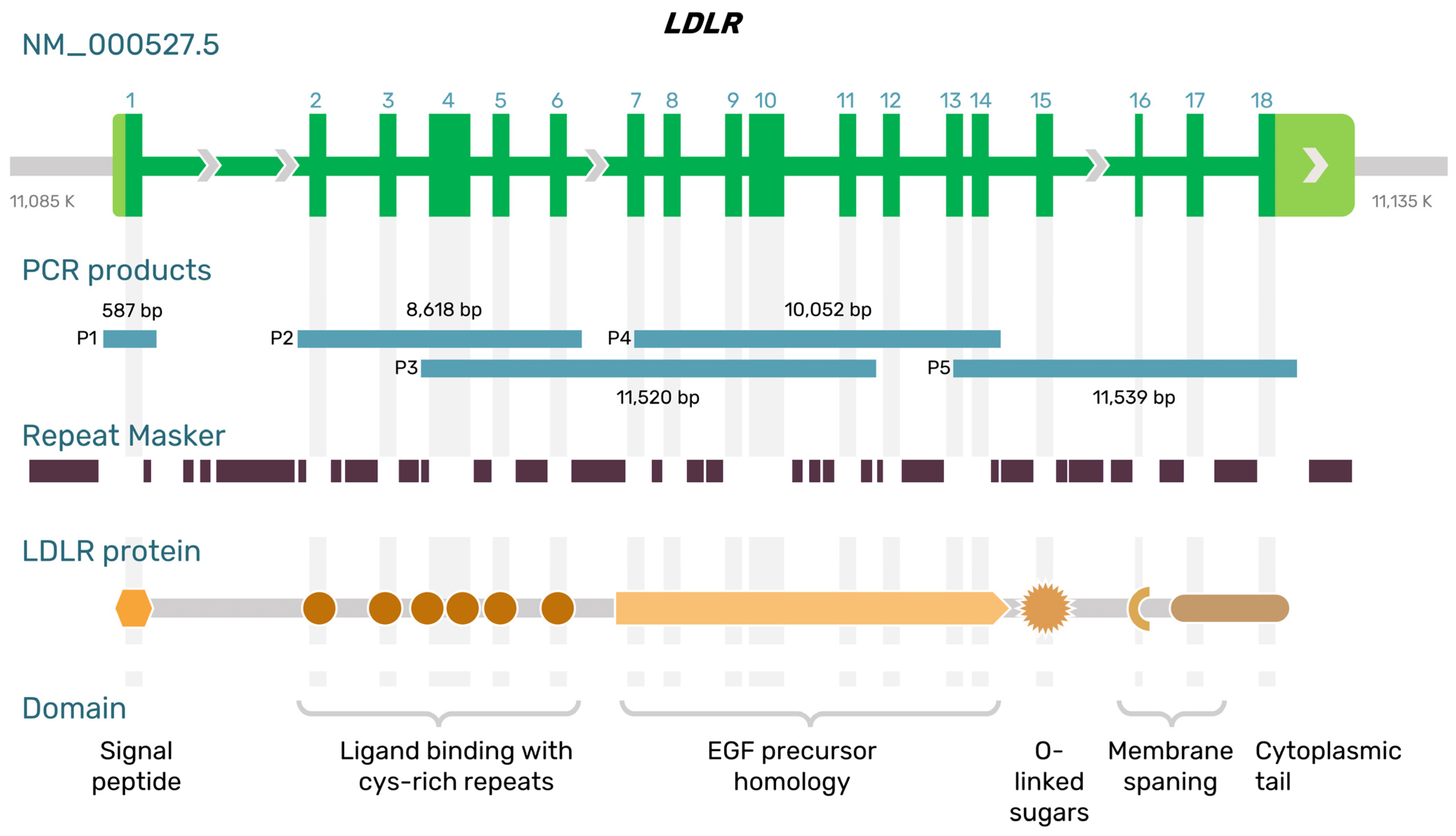
2.1. Long-Range PCR Primers for Amplifying the LDLR Gene from Exon 2 to Exon 18 with Introns
2.2. Nanopore Sequencing of SNVs and Small and Large Deletions in the LDLR Gene
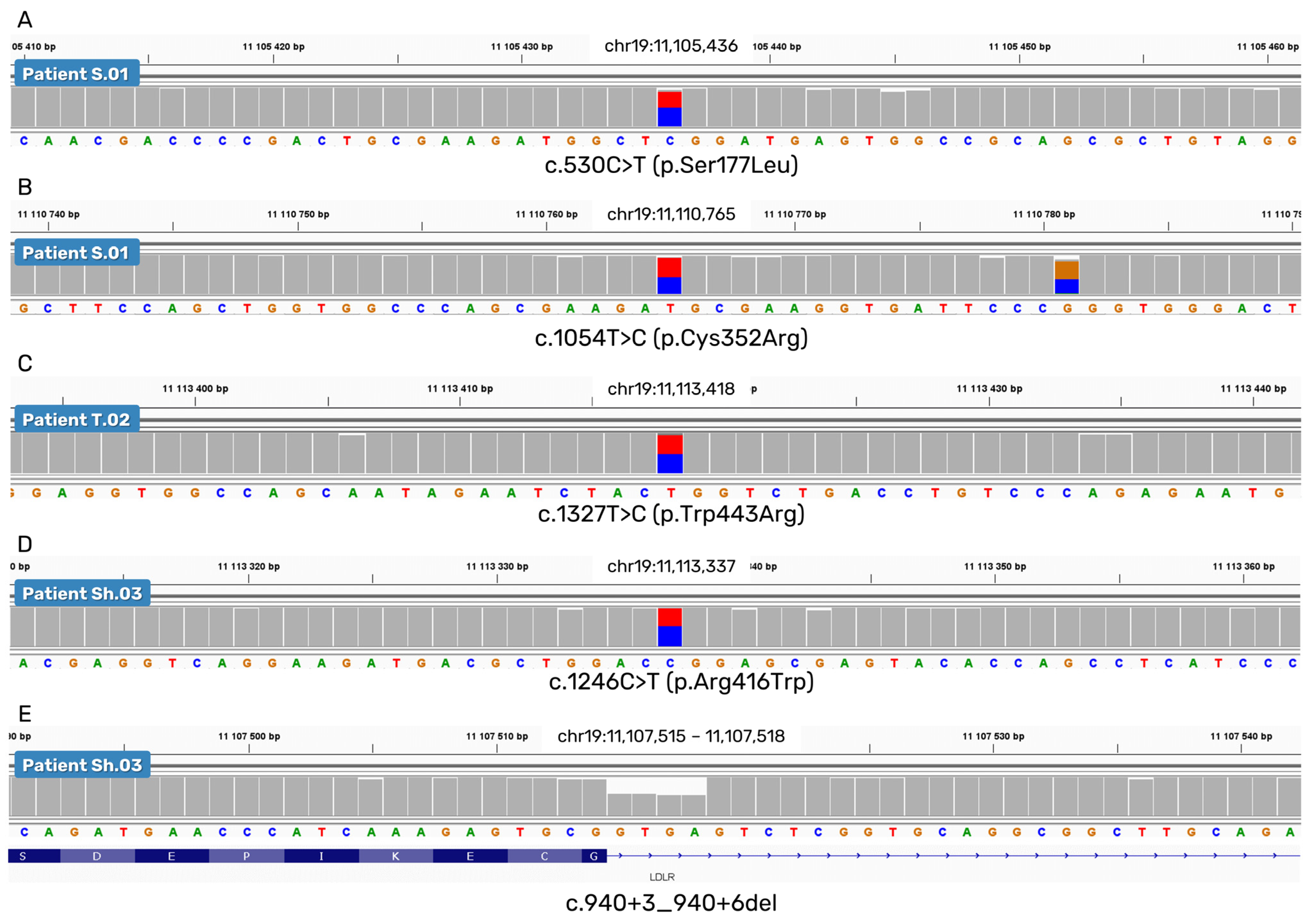
2.2.1. SNVs and a Small Deletion
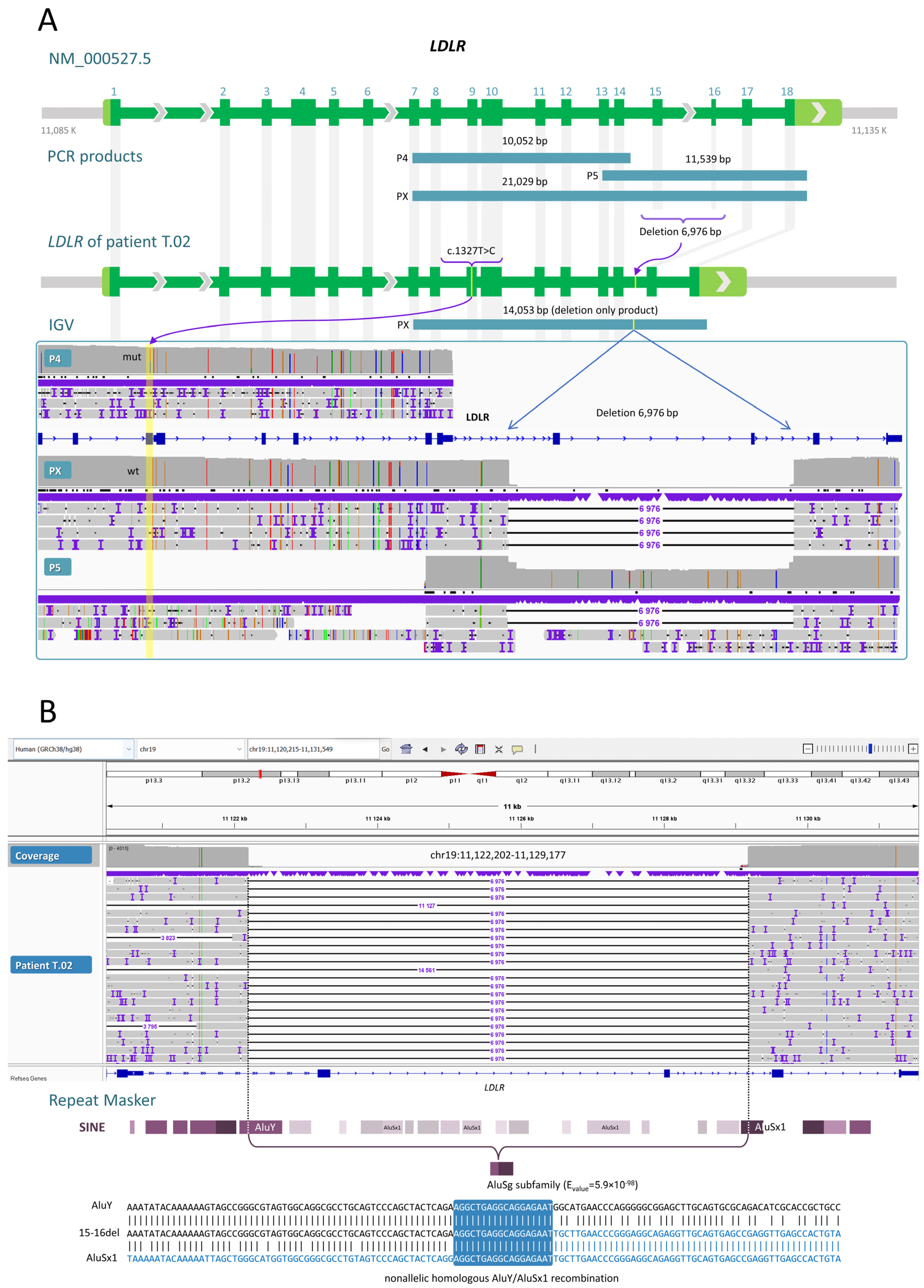
2.2.2. A Deletion of Exons 15 and 16
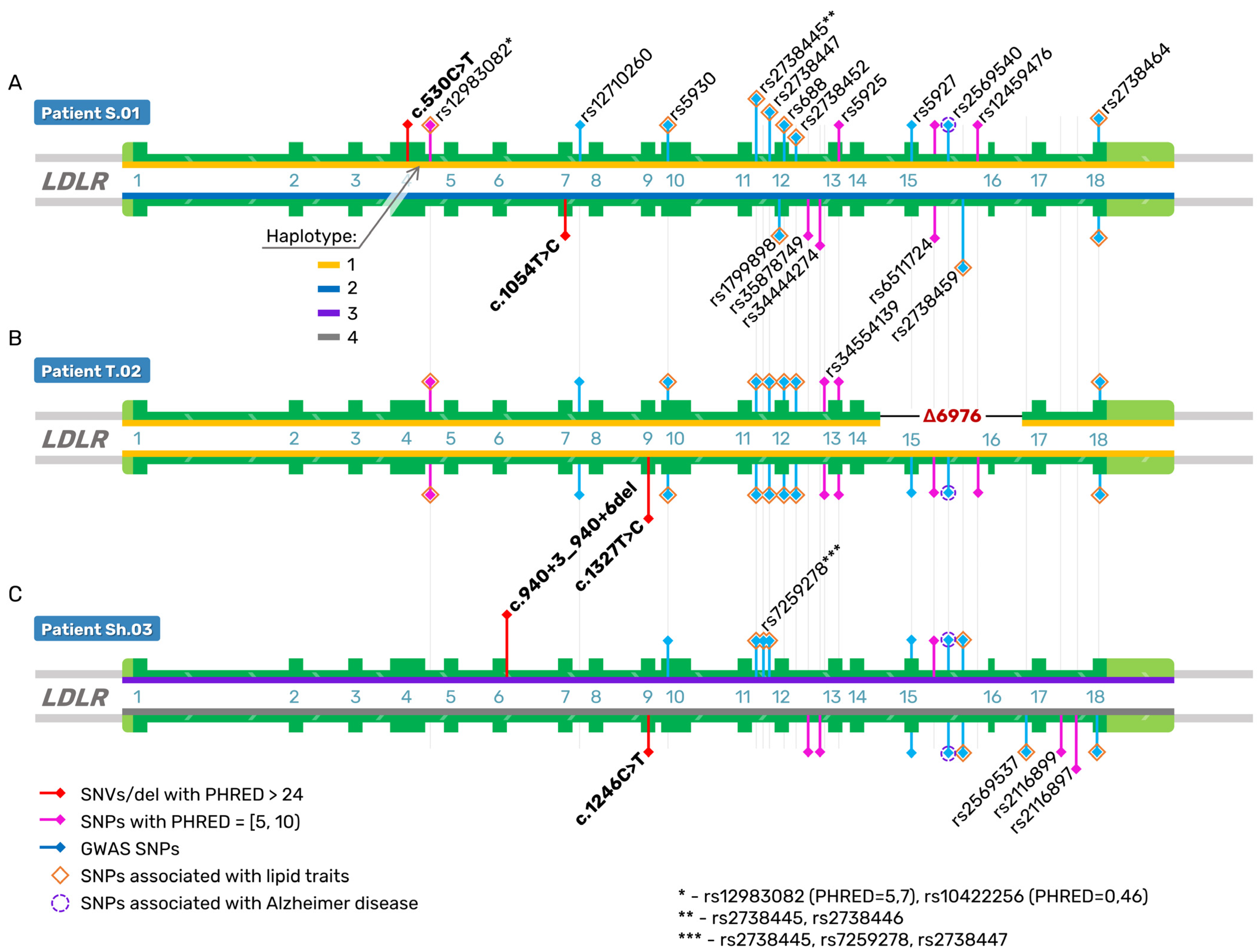
2.3. Direct Reconstruction of the LDLR Haplotype
3. Discussion
4. Materials and Methods
4.1. Patient Characteristics
4.2. DNA Extraction and Long-Range PCR
4.3. Library Preparation and ONT Sequencing
4.4. Bioinformatic Analyses of Nanopore Sequencing Data
4.5. Sanger Sequencing of the Promoter and Exon 1 of the LDLR Gene
4.6. In Silico Assessment of Pathogenicity and Regulatory Potential of the Variants
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Leigh, S.; Futema, M.; Whittall, R.; Taylor-Beadling, A.; Williams, M.; den Dunnen, J.T.; Humphries, S.E. The UCL low-density lipoprotein receptor gene variant database: Pathogenicity update. J. Med Genet. 2017, 54, 217–223. [Google Scholar] [CrossRef] [PubMed]
- gnomAD v2.1. Available online: https://gnomad.broadinstitute.org/gene/ENSG00000130164?dataset=gnomad_r2_1 (accessed on 1 November 2022).
- Ensembl. Available online: https://www.ensembl.org/homo_sapiens/Gene/Summary?g=ENSG00000130164&db=core (accessed on 1 November 2022).
- GWAS Catalog. Available online: https://www.ebi.ac.uk/gwas/ (accessed on 1 November 2022).
- Wang, J.; Dron, J.S.; Ban, M.R.; Robinson, J.F.; McIntyre, A.D.; Alazzam, M.; Zhao, P.J.; Dilliott, A.A.; Cao, H.; Huff, M.W.; et al. Polygenic Versus Monogenic Causes of Hypercholesterolemia Ascertained Clinically. Arter. Thromb. Vasc. Biol. 2016, 36, 2439–2445. [Google Scholar] [CrossRef] [PubMed]
- Galicia-Garcia, U.; Benito-Vicente, A.; Uribe, K.B.; Jebari, S.; Larrea-Sebal, A.; Alonso-Estrada, R.; Aguilo-Arce, J.; Ostolaza, H.; Palacios, L.; Martin, C. Mutation type classification and pathogenicity assignment of sixteen missense variants located in the EGF-precursor homology domain of the LDLR. Sci. Rep. 2020, 10, 1727. [Google Scholar] [CrossRef]
- Ison, H.E.; Clarke, S.L.; Knowles, J.W. Familial Hypercholesterolemia. Available online: https://www.ncbi.nlm.nih.gov/books/NBK174884/ (accessed on 22 November 2022).
- Tejedor, M.T.; Cenarro, A.; Tejedor, D.; Stef, M.; Palacios, L.; de Castro, I.; García-Otín, A.L.; Monteagudo, L.V.; Civeira, F.; Pocovi, M. New contributions to the study of common double mutants in the human LDL receptor gene. Naturwissenschaften 2011, 98, 943–949. [Google Scholar] [CrossRef]
- Amsellem, S.; Briffaut, D.; Carrié, A.; Rabès, J.P.; Girardet, J.P.; Fredenrich, A.; Moulin, P.; Krempf, M.; Reznik, Y.; Vialettes, B.; et al. Intronic mutations outside of Alu-repeat-rich domains of the LDL receptor gene are a cause of familial hypercholesterolemia. Hum. Genet. 2002, 111, 501–510. [Google Scholar] [CrossRef]
- Reeskamp, L.F.; Hartgers, M.L.; Peter, J.; Dallinga-Thie, G.M.; Zuurbier, L.; Defesche, J.C.; Grefhorst, A.; Hovingh, G.K. A Deep Intronic Variant in LDLR in Familial Hypercholesterolemia. Circ. Genom. Precis. Med. 2018, 11, e002385. [Google Scholar] [CrossRef] [PubMed]
- Goldmann, R.; Tichý, L.; Freiberger, T.; Zapletalová, P.; Letocha, O.; Soska, V.; Fajkus, J.; Fajkusová, L. Genomic characterization of large rearrangements of the LDLR gene in Czech patients with familial hypercholesterolemia. BMC Med Genet. 2010, 11, 115. [Google Scholar] [CrossRef]
- Iacocca, M.A.; Wang, J.; Dron, J.S.; Robinson, J.F.; McIntyre, A.D.; Cao, H.; Hegele, R.A. Use of next-generation sequencing to detect LDLR gene copy number variation in familial hypercholesterolemia. J. Lipid Res. 2017, 58, 2202–2209. [Google Scholar] [CrossRef]
- Leija-Salazar, M.; Pittman, A.; Mokretar, K.; Morris, H.; Schapira, A.H.; Proukakis, C. Investigation of Somatic Mutations in Human Brains Targeting Genes Associated With Parkinson’s Disease. Front. Neurol. 2020, 11, 570424. [Google Scholar] [CrossRef]
- Nowak, A.; Murik, O.; Mann, T.; Zeevi, D.A.; Altarescu, G. Detection of single nucleotide and copy number variants in the Fabry disease-associated GLA gene using nanopore sequencing. Sci. Rep. 2021, 11, 22372. [Google Scholar] [CrossRef]
- Soufi, M.; Bedenbender, S.; Ruppert, V.; Kurt, B.; Schieffer, B.; Schaefer, J.R. Fast and Easy Nanopore Sequencing Workflow for Rapid Genetic Testing of Familial Hypercholesterolemia. Front. Genet. 2022, 13, 836231. [Google Scholar] [CrossRef] [PubMed]
- Leung, A.W.; Leung, H.C.; Wong, C.L.; Zheng, Z.X.; Lui, W.W.; Luk, H.M.; Lo, I.F.; Luo, R.; Lam, T.W. ECNano: A cost-effective workflow for target enrichment sequencing and accurate variant calling on 4800 clinically significant genes using a single MinION flowcell. BMC Med. Genom. 2022, 15, 43. [Google Scholar] [CrossRef] [PubMed]
- Low Density Lipoprotein Receptor. Available online: https://www.ncbi.nlm.nih.gov/gene/3949 (accessed on 1 November 2022).
- Hobbs, H.H.; Russell, D.W.; Brown, M.S.; Goldstein, J.L. The LDL receptor locus in familial hypercholesterolemia: Mutational analysis of a membrane protein. Annu. Rev. Genet. 1990, 24, 133–170. [Google Scholar] [CrossRef] [PubMed]
- Iacocca, M.A.; Hegele, R.A. Role of DNA copy number variation in dyslipidemias. Curr. Opin. Lipidol. 2018, 29, 125–132. [Google Scholar] [CrossRef] [PubMed]
- Rutkowska, L.; Pinkier, I.; Sałacińska, K.; Kępczyński, Ł.; Salachna, D.; Lewek, J.; Banach, M.; Matusik, P.; Starostecka, E.; Lewiński, A.; et al. Identification of New Copy Number Variation and the Evaluation of a CNV Detection Tool for NGS Panel Data in Polish Familial Hypercholesterolemia Patients. Genes 2022, 13, 1424. [Google Scholar] [CrossRef] [PubMed]
- eQTL Catalog. Available online: https://eqtl.onderzoek.io/index.php?page=info (accessed on 1 November 2022).
- HaploReg. Available online: https://pubs.broadinstitute.org/mammals/haploreg/haploreg.php (accessed on 1 November 2022).
- Setia, N.; Saxena, R.; Arora, A.; Verma, I.C. Spectrum of mutations in homozygous familial hypercholesterolemia in India, with four novel mutations. Atherosclerosis 2016, 255, 31–36. [Google Scholar] [CrossRef]
- Bourbon, M.; Alves, A.C.; Medeiros, A.M.; Silva, S.; Soutar, A.K. Familial hypercholesterolaemia in Portugal. Atherosclerosis 2008, 196, 633–642. [Google Scholar] [CrossRef]
- Palacios, L.; Grandoso, L.; Cuevas, N.; Olano-Martín, E.; Martinez, A.; Tejedor, D.; Stef, M. Molecular characterization of familial hypercholesterolemia in Spain. Atherosclerosis 2012, 221, 137–142. [Google Scholar] [CrossRef]
- Sharifi, M.; Walus-Miarka, M.; Idzior-Waluś, B.; Malecki, M.T.; Sanak, M.; Whittall, R.; Li, K.W.; Futema, M.; Humphries, S.E. The genetic spectrum of familial hypercholesterolemia in south-eastern Poland. Metabolism 2016, 65, 48–53. [Google Scholar] [CrossRef]
- Tichý, L.; Freiberger, T.; Zapletalová, P.; Soška, V.; Ravčuková, B.; Fajkusová, L. The molecular basis of familial hypercholesterolemia in the Czech Republic: Spectrum of LDLR mutations and genotype-phenotype correlations. Atherosclerosis 2012, 223, 401–408. [Google Scholar] [CrossRef]
- Jannes, C.E.; Santos, R.D.; de Souza Silva, P.R.; Turolla, L.; Gagliardi, A.C.; Marsiglia, J.D.; Chacra, A.P.; Miname, M.H.; Rocha, V.Z.; Filho, W.S.; et al. Familial hypercholesterolemia in Brazil: Cascade screening program, clinical and genetic aspects. Atherosclerosis 2015, 238, 101–107. [Google Scholar] [CrossRef] [PubMed]
- Mohd, N.N.S.; Al-Khateeb, A.M.; Chua, Y.A.; Mohd, K.N.A.; Mohd, N.H. Heterozygous familial hypercholesterolaemia in a pair of identical twins: A case report and updated review. BMC Pediatr. 2019, 19, 106. [Google Scholar] [CrossRef] [PubMed]
- Huang, C.-C.; Niu, D.-M.; Charng, M.-J. Genetic Analysis in a Taiwanese Cohort of 750 Index Patients with Clinically Diagnosed Familial Hypercholesterolemia. J. Atheroscler. Thromb. 2022, 29, 639–653. [Google Scholar] [CrossRef] [PubMed]
- Meshkov, A.; Ershova, A.; Kiseleva, A.; Zotova, E.; Sotnikova, E.; Petukhova, A.; Zharikova, A.; Malyshev, P.; Rozhkova, T.; Blokhina, A.; et al. The LDLR, APOB, and PCSK9 Variants of Index Patients with Familial Hypercholesterolemia in Russia. Genes 2021, 12, 66. [Google Scholar] [CrossRef]
- Shakhtshneider, E.; Ivanoshchuk, D.; Timoshchenko, O.; Orlov, P.; Semaev, S.; Valeev, E.; Goonko, A.; Ladygina, N.; Voevoda, M. Analysis of Rare Variants in Genes Related to Lipid Metabolism in Patients with Familial Hypercholesterolemia in Western Siberia (Russia). J. Pers. Med. 2021, 11, 1232. [Google Scholar] [CrossRef]
- Korneva, V.A.; Kuznetsova, T.Y.; Murtazina, R.Z.; Didio, A.V.; Bogoslovskaya, T.Y.; Mandelshtam, M.Y.; Vasilyev, V.B. The Familial Hypercholesterolemia Caused by a Novel Human Low Density Lipoprotein Receptor Gene Mutation c.1327 T>C (p.W433R). Kardiologiia 2017, 57, 12–16. [Google Scholar]
- Kim, H.; Lee, C.J.; Kim, S.H.; Kim, J.Y.; Choi, S.H.; Kang, H.J.; Park, K.S.; Cho, B.R.; Kim, B.J.; Sung, K.C.; et al. Phenotypic and Genetic Analyses of Korean Patients with Familial Hypercholesterolemia: Results from the KFH Registry 2020. J. Atheroscler. Thromb. 2022, 29, 1176–1187. [Google Scholar] [CrossRef]
- Semenova, A.E.; Sergienko, I.V.; García-Giustiniani, D.; Monserrat, L.; Popova, A.B.; Nozadze, D.N.; Ezhov, M.V. Verification of Underlying Genetic Cause in a Cohort of Russian Patients with Familial Hypercholesterolemia Using Targeted Next Generation Sequencing. J. Cardiovasc. Dev. Dis. 2020, 7, 16. [Google Scholar] [CrossRef]
- Sharifi, M.; Futema, M.; Nair, D.; Humphries, S.E. Genetic Architecture of Familial Hypercholesterolaemia. Curr. Cardiol. Rep. 2017, 19, 44. [Google Scholar] [CrossRef]
- Thormaehlen, A.S.; Schuberth, C.; Won, H.H.; Blattmann, P.; Joggerst-Thomalla, B.; Theiss, S.; Asselta, R.; Duga, S.; Merlini, P.A.; Ardissino, D.; et al. Systematic cell-based phenotyping of missense alleles empowers rare variant association studies: A case for LDLR and myocardial infarction. PLoS Genet. 2015, 11, e1004855. [Google Scholar] [CrossRef]
- Banerjee, P.; Chan, K.C.; Tarabocchia, M.; Benito-Vicente, A.; Alves, A.C.; Uribe, K.B.; Bourbon, M.; Skiba, P.J.; Pordy, R.; Gipe, D.A.; et al. Functional Analysis of LDLR (Low-Density Lipoprotein Receptor) Variants in Patient Lymphocytes to Assess the Effect of Evinacumab in Homozygous Familial Hypercholesterolemia Patients With a Spectrum of LDLR Activity. Arter. Thromb. Vasc. Biol. 2019, 39, 2248–2260. [Google Scholar] [CrossRef] [PubMed]
- Strøm, T.B.; Tveten, K.; Laerdahl, J.K.; Leren, T.P. Mutation G805R in the transmembrane domain of the LDL receptor gene causes familial hypercholesterolemia by inducing ectodomain cleavage of the LDL receptor in the endoplasmic reticulum. FEBS Open Bio 2014, 4, 321–327. [Google Scholar] [CrossRef] [PubMed]
- Strøm, T.B.; Laerdahl, J.K.; Leren, T.P. Mutations affecting the transmembrane domain of the LDL receptor: Impact of charged residues on the membrane insertion. Hum. Mol. Genet. 2017, 26, 1634–1642. [Google Scholar] [CrossRef] [PubMed]
- Shu, H.Y.; Zhang, W.; Zheng, C.C.; Gao, M.Y.; Li, Y.C.; Wang, Y.G. Identification and Functional Characterization of a Low-Density Lipoprotein Receptor Gene Pathogenic Variant in Familial Hypercholesterolemia. Front. Genet. 2021, 12, 650077. [Google Scholar] [CrossRef] [PubMed]
- Aalto-Setälä, K.; Helve, E.; Kovanen, P.T.; Kontula, K. Finnish type of low density lipoprotein receptor gene mutation (FH-Helsinki) deletes exons encoding the carboxy-terminal part of the receptor and creates an internalization-defective phenotype. J. Clin. Investig. 1989, 84, 499–505. [Google Scholar] [CrossRef]
- Koivisto, P.V.; Koivisto, U.M.; Kovanen, P.T.; Gylling, H.; Miettinen, T.A.; Kontula, K. Deletion of exon 15 of the LDL receptor gene is associated with a mild form of familial hypercholesterolemia. FH-Espoo. Arterioscler. Thromb. A J. Vasc. Biol. 1993, 13, 1680–1688. [Google Scholar] [CrossRef]
- Rødningen, O.K.; Røshy, O.; Tonstad, S.; Ose, L.; Berg, K.; Leren, T.P. A 9.6 kilobase deletion in the low density lipoprotein receptor gene in Norwegian familial hypercholesterolemia subjects. Clin. Genet. 2008, 42, 288–295. [Google Scholar] [CrossRef]
- Etxebarria, A.; Benito-Vicente, A.; Palacios, L.; Stef, M.; Cenarro, A.; Civeira, F.; Ostolaza, H.; Martin, C. Functional characterization and classification of frequent low-density lipoprotein receptor variants. Hum. Mutat. 2015, 36, 129–141. [Google Scholar] [CrossRef]
- Gao, F.; Ihn, H.E.; Medina, M.W.; Krauss, R.M. A common polymorphism in the LDL receptor gene has multiple effects on LDL receptor function. Hum. Mol. Genet. 2013, 22, 1424–1431. [Google Scholar] [CrossRef]
- Ling, I.F.; Gopalraj, R.K.; Simpson, J.F.; Estus, S. Expression and regulation of a low-density lipoprotein receptor exon 12 splice variant. J. Neurochem. 2010, 115, 614–624. [Google Scholar] [CrossRef]
- Lee, J.D.; Hsiao, K.M.; Wang, T.C.; Lee, T.H.; Kuo, Y.W.; Huang, Y.C.; Hsu, H.L.; Lin, Y.H.; Wu, C.Y.; Huang, Y.C.; et al. Mutual effect of rs688 and rs5925 in regulating low-density lipoprotein receptor splicing. DNA Cell Biol. 2014, 33, 869–875. [Google Scholar] [CrossRef] [PubMed]
- Khamis, A.; Palmen, J.; Lench, N.; Taylor, A.; Badmus, E.; Leigh, S.; Humphries, S.E. Functional analysis of four LDLR 5′UTR and promoter variants in patients with familial hypercholesterolaemia. Eur. J. Hum. Genet. 2015, 23, 790–795. [Google Scholar] [CrossRef] [PubMed]
- Fairoozy, R.H.; White, J.; Palmen, J.; Kalea, A.Z.; Humphries, S.E. Identification of the Functional Variant(s) that Explain the Low-Density Lipoprotein Receptor (LDLR) GWAS SNP rs6511720 Association with Lower LDL-C and Risk of CHD. PLoS ONE 2016, 11, e0167676. [Google Scholar] [CrossRef]
- Rentzsch, P.; Schubach, M.; Shendure, J.; Kircher, M. CADD-Splice-improving genome-wide variant effect prediction using deep learning-derived splice scores. Genome Med. 2021, 13, 31. [Google Scholar] [CrossRef] [PubMed]
- Caruz, A.; Neukam, K.; Rivero-Juárez, A.; Herrero, R.; Real, L.M.; Camacho, A.; Barreiro, P.; Labarga, P.; Rivero, A.; Pineda, J.A. Association of low-density lipoprotein receptor genotypes with hepatitis C viral load. Genes Immun. 2014, 15, 16–24. [Google Scholar] [CrossRef]
- Wilson, G.M.; Vasa, M.Z.; Deeley, R.G. Stabilization and cytoskeletal-association of LDL receptor mRNA are mediated by distinct domains in its 3′ untranslated region. J. Lipid Res. 1998, 39, 1025–1032. [Google Scholar] [CrossRef]
- Selvaraj, M.S.; Li, X.; Li, Z.; Pampana, A.; Zhang, D.Y.; Park, J.; Aslibekyan, S.; Bis, J.C.; Brody, J.A.; Cade, B.E.; et al. Whole genome sequence analysis of blood lipid levels in >66,000 individuals. Nat. Commun. 2022, 13, 5995. [Google Scholar] [CrossRef]
- Kulseth, M.A.; Berge, K.E.; Bogsrud, M.P.; Leren, T.P. Analysis of LDLR mRNA in patients with familial hypercholesterolemia revealed a novel mutation in intron 14, which activates a cryptic splice site. J. Hum. Genet. 2010, 55, 676–680. [Google Scholar] [CrossRef]
- Payer, L.M.; Steranka, J.P.; Ardeljan, D.; Walker, J.; Fitzgerald, K.C.; Calabresi, P.A.; Cooper, T.A.; Burns, K.H. Alu insertion variants alter mRNA splicing. Nucleic Acids Res. 2019, 47, 421–431. [Google Scholar] [CrossRef]
- Payer, L.M.; Steranka, J.P.; Kryatova, M.S.; Grillo, G.; Lupien, M.; Rocha, P.P.; Burns, K.H. Alu insertion variants alter gene transcript levels. Genome Res. 2021, 31, 2236–2248. [Google Scholar] [CrossRef]
- Van Iperen, E.P.; Sivapalaratnam, S.; Boekholdt, S.M.; Hovingh, G.K.; Maiwald, S.; Tanck, M.W.; Soranzo, N.; Stephens, J.C.; Sambrook, J.G.; Levi, M.; et al. Common genetic variants do not associate with CAD in familial hypercholesterolemia. Eur. J. Hum. Genet. 2014, 22, 809–813. [Google Scholar] [CrossRef] [PubMed]
- Mach, F.; Baigent, C.; Catapano, A.L.; Koskinas, K.C.; Casula, M.; Badimon, L.; Chapman, M.J.; Backer, G.G.D.; Delgado, V.; Ference, B.A.; et al. 2019 ESC/EAS Guidelines for the management of dyslipidaemias: Lipid modification to reduce cardiovascular risk. Eur. Heart J. 2019, 41, 111–188. [Google Scholar] [CrossRef] [PubMed]
- Zakharova, I.S.; Shevchenko, A.I.; Tmoyan, N.A.; Elisaphenko, E.A.; Zubkova, E.S.; Sleptcov, A.A.; Nazarenko, M.S.; Ezhov, M.V.; Kukharchuk, V.V.; Parfyonova, Y.V.; et al. Induced pluripotent stem cell line ICGi038-A, obtained by reprogramming peripheral blood mononuclear cells from a patient with familial hypercholesterolemia due to compound heterozygous c.1246C > T/c.940 + 3_940 + 6del mutations in LDLR. Stem Cell Res. 2022, 60, 102702. [Google Scholar] [CrossRef] [PubMed]
- Zakharova, I.S.; Shevchenko, A.I.; Tmoyan, N.A.; Elisaphenko, E.A.; Kalinin, A.P.; Sleptcov, A.A.; Nazarenko, M.S.; Ezhov, M.V.; Kukharchuk, V.V.; Parfyonova, Y.V.; et al. Induced pluripotent stem cell line ICGi037-A, obtained by reprogramming peripheral blood mononuclear cells from a patient with familial hypercholesterolemia due to heterozygous p.Trp443Arg mutations in LDLR. Stem Cell Res. 2022, 60, 102703. [Google Scholar] [CrossRef] [PubMed]
- Zakharova, I.S.; Shevchenko, A.I.; Tmoyan, N.A.; Elisaphenko, E.A.; Zubkova, E.S.; Sleptcov, A.A.; Nazarenko, M.S.; Ezhov, M.V.; Kukharchuk, V.V.; Parfyonova, Y.V.; et al. Induced pluripotent stem cell line ICGi036-A generated by reprogramming peripheral blood mononuclear cells from a patient with familial hypercholesterolemia caused due to compound heterozygous p.Ser177Leu/p.Cys352Arg mutations in LDLR. Stem Cell Res. 2022, 59, 102653. [Google Scholar] [CrossRef]
- PrimerQuest. Available online: https://www.idtdna.com/pages/tools/primerquest (accessed on 1 November 2022).
- Owczarzy, R.; Tataurov, A.V.; Wu, Y.; Manthey, J.A.; McQuisten, K.A.; Almabrazi, H.G.; Pedersen, K.F.; Lin, Y.; Garretson, J.; McEntaggart, N.O.; et al. IDT SciTools: A suite for analysis and design of nucleic acid oligomers. Nucleic Acids Res. 2008, 36, W163–W169. [Google Scholar] [CrossRef]
- Wick, R.R.; Judd, L.M.; Holt, K.E. Performance of neural network basecalling tools for Oxford Nanopore sequencing. Genome Biol. 2019, 20, 129. [Google Scholar] [CrossRef]
- Li, H. New strategies to improve minimap2 alignment accuracy. Bioinformatics 2021, 37, 4572–4574. [Google Scholar] [CrossRef]
- Samtools. Available online: https://github.com/samtools/ (accessed on 1 November 2022).
- Bedtools. Available online: https://github.com/ryanlayer/bedtools (accessed on 1 November 2022).
- Epi2me. Available online: https://github.com/epi2me-labs/wf-human-variation (accessed on 1 November 2022).
- IGV. Available online: https://software.broadinstitute.org/software/igv/home (accessed on 1 November 2022).
- Ewels, P.; Magnusson, M.; Lundin, S.; Käller, M. MultiQC: Summarize analysis results for multiple tools and samples in a single report. Bioinformatics 2016, 32, 3047–3048. [Google Scholar] [CrossRef]
- Varsome. Available online: https://varsome.com/ (accessed on 1 November 2022).
- Richards, S.; Aziz, N.; Bale, S.; Bick, D.; Das, S.; Gastier-Foster, J.; Grody, W.W.; Hegde, M.; Lyon, E.; Spector, E.; et al. Standards and guidelines for the interpretation of sequence variants: A joint consensus recommendation of the American College of Medical Genetics and Genomics and the Association for Molecular Pathology. Genet. Med. 2015, 17, 405–424. [Google Scholar] [CrossRef]
- Riggs, E.R.; Andersen, E.F.; Cherry, A.M.; Kantarci, S.; Kearney, H.; Patel, A.; Raca, G.; Ritter, D.I.; South, S.T.; Thorland, E.C.; et al. Technical standards for the interpretation and reporting of constitutional copy-number variants: A joint consensus recommendation of the American College of Medical Genetics and Genomics (ACMG) and the Clinical Genome Resource (ClinGen). Genet. Med. 2020, 22, 245–257. [Google Scholar] [CrossRef] [PubMed]
- SpliceAI. Available online: https://spliceailookup.broadinstitute.org/# (accessed on 1 November 2022).
| Subjects’ IDs: | S.01 | T.02 | Sh.03 |
|---|---|---|---|
| Sex | Female | Female | Female |
| Age, years | 36 | 31 | 36 |
| Family history of FH or CAD | Yes (mother and daughter with FH; brother, MI under 50 y.o.) | Yes (father and mother both with FH; maternal uncle with FH) | Yes (father and mother both with FH) |
| Total cholesterol, mmol/L (before/after treatment) | 19/15.64–17.61 | 23/16.73–18.41 | 17.25/5.31–4.84 |
| LDL-C, mmol/L (before/after treatment) | 17/14.45–16.24 | NA/15.2–17.25 | 15.15/3.86–2.98 |
| Triglycerides, mmol/L (before/after treatment) | NA/0.62–0.86 | NA/1.67–1.1 | NA/0.57–0.75 |
| Tendon xanthomas | Yes | Yes | Yes |
| Xanthelasma | Yes | Yes | No |
| Lipoic corneal arcus | Yes | No | No |
| Clinical and instrumental manifestations of CAD | Yes (MI at 30 y.o.) | Yes (CABG at 23 y.o., CAS at 26 y.o., TAVI at 31 y.o., death from COVID-19 at 32 y.o.) | Yes (CABG at 26 y.o.) |
| Lipid-lowering therapy | Rosuvastatin 40 mg, Ezetimibe 10 mg. Inclisiran 300 mg. | Rosuvastatin 40 mg, Ezetimibe 10 mg. Inclisiran 300 mg. | Rosuvastatin 40 mg, Ezetimibe 10 mg. Inclisiran 300 mg. |
| Mutations in the LDLR gene (NM_000527.5) | c.530C>T or FH Puerto Rico (exon 4)/c.1054T>C (exon 7) | c.2141-966_2390-330del (6976 bp deletion of exons 15–16)/c.1327T>C (exon 9) | c.1246C>T (exon 9)/c.940+3_940+6del (4 bp, intron 6) |
| Variant annotation in silico (Varsome) | Pathogenic/Pathogenic | -/Pathogenic | Pathogenic/Likely Pathogenic |
| Amino acid change (NP_000518.1) | p.Ser177Leu/p.Cys352Arg | p.Glu714_Ile796del/p.Trp443Arg | p.Arg416Trp/NA |
| rsID | rs121908026/rs879254769 | NA/rs773566855 | rs570942190/NA |
| ID | PCR Products | Forward (5′-3′) | Reverse (5′-3′) | Expected Length (bp) |
|---|---|---|---|---|
| P2 | Intron 1 to intron 6 | GTTCCTTCTTTGTGTCCTCCA | GTCTTTCAGTATCCACCACAGAG | 8618 |
| P3 | Intron 3 to intron 11 | TGGTGTTGGGAGACTTCACA | CTCTCCAATGGGCAGGTAGG | 11,520 |
| P4 | Exon 7 to intron 14 | CCTTAAGATCGGCTACGAGTG | TCAGAAATCAGATCACCTCTTCAG | 10,052 |
| P5 | Exon 13 to 3′UTR | GAGGATATGGTTCTCTTCCACAA | GGCTTAGAGATTGGTGGATGAG | 11,539 |
| PX | Exon 7 to 3′UTR 1 | CCTTAAGATCGGCTACGAGTG | GGCTTAGAGATTGGTGGATGAG | 21,029 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Nazarenko, M.S.; Sleptcov, A.A.; Zarubin, A.A.; Salakhov, R.R.; Shevchenko, A.I.; Tmoyan, N.A.; Elisaphenko, E.A.; Zubkova, E.S.; Zheltysheva, N.V.; Ezhov, M.V.; et al. Calling and Phasing of Single-Nucleotide and Structural Variants of the LDLR Gene Using Oxford Nanopore MinION. Int. J. Mol. Sci. 2023, 24, 4471. https://doi.org/10.3390/ijms24054471
Nazarenko MS, Sleptcov AA, Zarubin AA, Salakhov RR, Shevchenko AI, Tmoyan NA, Elisaphenko EA, Zubkova ES, Zheltysheva NV, Ezhov MV, et al. Calling and Phasing of Single-Nucleotide and Structural Variants of the LDLR Gene Using Oxford Nanopore MinION. International Journal of Molecular Sciences. 2023; 24(5):4471. https://doi.org/10.3390/ijms24054471
Chicago/Turabian StyleNazarenko, Maria S., Aleksei A. Sleptcov, Aleksei A. Zarubin, Ramil R. Salakhov, Alexander I. Shevchenko, Narek A. Tmoyan, Eugeny A. Elisaphenko, Ekaterina S. Zubkova, Nina V. Zheltysheva, Marat V. Ezhov, and et al. 2023. "Calling and Phasing of Single-Nucleotide and Structural Variants of the LDLR Gene Using Oxford Nanopore MinION" International Journal of Molecular Sciences 24, no. 5: 4471. https://doi.org/10.3390/ijms24054471
APA StyleNazarenko, M. S., Sleptcov, A. A., Zarubin, A. A., Salakhov, R. R., Shevchenko, A. I., Tmoyan, N. A., Elisaphenko, E. A., Zubkova, E. S., Zheltysheva, N. V., Ezhov, M. V., Kukharchuk, V. V., Parfyonova, Y. V., Zakian, S. M., & Zakharova, I. S. (2023). Calling and Phasing of Single-Nucleotide and Structural Variants of the LDLR Gene Using Oxford Nanopore MinION. International Journal of Molecular Sciences, 24(5), 4471. https://doi.org/10.3390/ijms24054471










