Abstract
Eosinophilic esophagitis (EoE) is a chronic, Th2-inflammatory disease of the esophagus that can severely affect food intake. Currently, diagnosis and assessing response to treatment of EoE is highly invasive and requires endoscopy with esophageal biopsies. Finding non-invasive and accurate biomarkers is important for improving patient well-being. Unfortunately, EoE is usually accompanied by other atopies, which make it difficult to identify specific biomarkers. Providing an update of circulating EoE biomarkers and concomitant atopies is therefore timely. This review summarizes the current knowledge in EoE blood biomarkers and two of its most common comorbidities, bronchial asthma (BA) and atopic dermatitis (AD), focusing on dysregulated proteins, metabolites, and RNAs. It also revises the current knowledge on extracellular vesicles (EVs) as non-invasive biomarkers for BA and AD, and concludes with the potential use of EVs as biomarkers in EoE.
1. Introduction
Eosinophilic esophagitis (EoE) is a chronic disease characterized clinically by symptoms referred to as esophageal dysfunction, and histologically by eosinophil-predominant inflammation of this organ [1]. EoE is mainly driven by food-antigens that trigger T-helper 2 (Th2) local immune response [2,3], and is considered as a particular form of food allergy that frequently appears in patients who suffer from other Th2-associated atopies [4]. EoE is recognized as the leading cause of chronic dysphagia in children and young adults, and the second cause of chronic esophagitis after gastroesophageal reflux disease (GERD) [5]; and there has been a constant rise in its incidence, with currently 34.4 new cases/100,000 subjects per year [6]. Although progress in management of the disease has significantly reduced diagnostic delay over recent years [7], endoscopy with esophageal biopsies still remains the only reliable method for EoE diagnosis, monitoring disease activity and assessing the response to treatment. Thus, finding of a non-invasive, accurate and specific biomarker is a key goal, and an area of great potential in the research of this disease. Despite the efforts being made however, minimally invasive biomarkers are not yet being applied to routine clinical practice [8].
One of the main obstacles in the search for non-invasive biomarkers is the lack of specificity. This is complicated greatly by the frequent concomitance of EoE with several atopic conditions, including atopic dermatitis (AD), bronchial asthma (BA), IgE-mediated food allergies, and allergic rhinitis, all significantly more common in EoE patients than in the general population [4,9,10]. In fact, EoE has been proposed as a late manifestation of the “atopic march”, a natural concatenation of atopic disorders over time [11], with BA and AD being among the most common comorbidities of EoE [10]. Despite this strong association, only a small proportion of studies have included atopic controls [12] as shown in Table 1, thus hampering the proper identification of suitable biomarkers.

Table 1.
Research studies including atopic control groups. AEC: absolute eosinophil count, AR: allergic rhinitis, BA: bronchial asthma, AD: atopic dermatitis, AX: food anaphylaxis, CU: contact urticaria, ND: non-determined, EGID: eosinophilic gastrointestinal disorders, EDN: eosinophil-derived neurotoxin, EoP: eosinophil progenitor.
Extracellular vesicles (EVs) are a tremendously heterogeneous group of membrane-limited entities, of nanometric size, released into the extracellular space by virtually all cell types and cellular organisms [21]. Although initially conceived as cell ‘debris’ or cellular waste, evidence of their relevance in both physiological and pathological scenarios, mainly defined by their role in intercellular communication, has driven forward research since the early 2000s [22]. In allergy, EVs intervene actively in different aspects of its pathophysiology, from induction of inflammation by activation of allergen-specific T cells, to the contribution of sustained chronic inflammation and development of fibrosis [23]. This is possible due to an orchestrated mechanism in which EVs are released and uptake by cells of the inflammatory microenvironment, whose effects are greatly determined by their content (i.e., inflammatory cytokines, enzymes, miRNAs, etc.) [23,24,25]. Moreover, EVs have arisen as the most promising source of biomarkers for several reasons. First, EVs are abundant in many bodily fluids such as blood, urine, saliva, ascites fluid, pleural effusion, breast milk, and cerebrospinal and bronchoalveolar lavage fluid [26]. Moreover, EVs’ intraluminal and extraluminal cargo (proteins, nucleic acids, lipids, and metabolites) are functional [27], and correlate well with their parental cell. They inform the cell identity and its biological status, thus acting as a peripheral representation of a pathological process. Finally, EVs have a significant advantage over other serum-biomarkers now in that their conformation extends the stability of their cargo such as RNAs and proteins, protecting them from catabolic enzymes in circulation. Despite this, the nanometric size of EVs, and the complex composition of biofluids as blood are technically challenging and need to be overcome in order to accelerate their use in diagnostics [28].
This review updates current knowledge relating to potential non-invasive blood-based biomarkers described for EoE and two of its most common comorbidities: BA and AD, with the aim of identifying the common unspecific biomarkers. We also summarize state of the art use of EVs as circulating biomarkers for asthma and atopic dermatitis and speculate about the advantage of using EVs as non-invasive biomarkers for diagnosis and the management of EoE.
2. Methods
We searched the PubMed library, using the following individual and combined key words: eosinophilic esophagitis; atopic dermatitis; eczema; bronchial asthma; extracellular vesicles; exosomes; blood; circulating; serum; plasma; biomarker. Reference lists in the articles obtained were also searched in order to identify other potential sources of information. We included studies describing circulating biomarkers if performed in human samples, study subjects suffering from EoE, BA, or AD, together with other comorbidities. The results were limited to studies published and written in English and carried out on humans or human samples.
3. Current Knowledge of Circulating Biomarkers
EoE, BA, and AD are all chronic inflammatory diseases characterized by Th2 immune responses. Upon exposure to an allergen, sensitization occurs in the epithelial barrier of the esophagus (EoE), airways (BA), and skin (AD), the epithelial integrity of which is disrupted as a result of defects in cell–cell contacts [26,27]. Sensitized epithelial cells then orchestrate the immunological response by releasing alarmins (IL-25, IL-33, and TSLP) responsible for the polarization of CD4+ T cells towards Th2 phenotype [29,30,31]. Cytokines released by Th2 lymphocytes (IL-4, IL-5, and IL-13) stimulate, among others things, the proliferation of eosinophils that are subsequently recruited to the inflammatory foci from circulation, causing the eosinophilia characteristic of EoE, BA, and AD [32,33,34,35]. The cytotoxic nature of eosinophil-granule proteins released locally then promotes skin lesions and pruritus in AD [36]. As a result, remodeling and hyperresponsiveness of the airways in BA [37], and esophageal dysmotility and fibrosis in EoE occurs [38]. Unsurprisingly, there is a significant overlap in the molecular mechanisms of these three diseases that share up to 18 dysregulated genes [39]. However, EoE, BA, and AD are good examples of diverse eosinophilic disorders with different tissues being affected, mediation of serum IgE, and systemic/local inflammation. AD and BA have increased serum total IgE and allergen-specific IgE levels in common [32,34], therefore skin prick testing (SPT) is a suitable tool for diagnosis its use in EoE is limited however as IgE is not required for its pathogenesis [40]. In addition, AD is considered a systemic disease [41], while EoE and BA, with the exception of an endotype of BA [42], are characterized by local inflammation in the esophagus and the airways, respectively.
Since the pathophysiology of EoE, BA, and AD is very similar, it is likely that unspecific biomarkers could be found when using samples such as blood the most common tissue used when looking for non-invasive biomarkers and therefore, will be the focus of this review. It should be noted that overall biomarkers for EoE [43], BA [44], and AD [45] have been reviewed recently. As counts of circulating eosinophil and serum IgE levels have been widely employed in EoE, BA, and AD, with inconclusive results, they will not be reviewed here.
3.1. Bronchial Asthma (BA)
3.1.1. Proteins
The serum levels of periostin, a matricellular protein involved in eosinophilic inflammation and airway remodeling [46], are thought to be a promising biomarker for Th2-eosinophilic asthma for two main reasons: firstly, it correlates well with its expression in the airways, and secondly, it remains relatively stable in blood [47]. In fact, serum periostin can predict airway’s eosinophilia alone [48] and thus serves well as a diagnostic and predictive biomarker. Furthermore, its expression is markedly increased in children from 2 to 11 years old, compared to adults [49], despite exhibiting a limited diagnostic ability in children with severe asthma [50]. The diagnostic potential of osteopontin (OPN), another matricellular protein, was first suggested by Samitas et al., who observed increased levels of serum OPN in asthmatic patients compared to healthy controls [51]. Although such overexpression was later validated [52], a meta-analysis including 9 studies, in which 7 of them used serum or plasma OPN, concluded that there was no association between circulating OPN and asthma and was not useful for diagnostics or to reflect severity [53].
Upon activation, eosinophils release their granule proteins (ECP, EDN, EPX, MBP-1, and CLC/Gal-10), which are cytotoxic proteins involved in eosinophil-inflammation, tissue remodeling, and serve as indirect markers of inflammation [54]. In BA, serum eosinophil cationic protein (ECP), which appears overexpressed in the serum of both adult and pediatric patients, has been the most widely studied granule protein [55,56]. Although its utility as a diagnostic biomarker is limited, its correlation with disease severity and response to treatment exhibited greater potential [57,58]. The lesser studied eosinophil-derived neurotoxin (EDN) has arisen as a promising clinical biomarker, informative of diagnosis, severity, and treatment monitoring [59]; more importantly, EDN is very stable in blood samples, thus increasing its analytical reliability [60]. Blood C-reactive protein (CRP) has also been proposed as a marker of airway inflammation, but in a very limited cohort of non-smoker asthmatic patients without such additional complications as cardiovascular-related diseases, hyperlipidemia, chronic obstructive pulmonary disease (COPD), or infection [61].
Several proteins involved in tissue damage and remodeling appear dysregulated in the blood of asthmatic patients. In a meta-analysis of 17 studies, the extracellular matrix glycoprotein YKL-40, was found upregulated in asthmatic patients compared to healthy controls, and correlated with disease severity and acute exacerbation of the disease, regardless of COPD and related syndromes [62]. Moreover, metalloproteinase-9 (MMP-9) and dipeptidyl peptidase-4 (DPP-4), involved in extracellular matrix remodeling, are both elevated in the serum of BA patients, and their expression, related to disease severity, is reduced after treatment with corticosteroids [58,63]. In fact, corticosteroid refractoriness, an important issue in the management of BA, could be identified by the downregulation of mucin-1 (MUC-1) in circulating neutrophils, a characteristic of patients with uncontrolled severe asthma [64]. Hur et al. explored the ability of several serum cytokines, periostin, EDN, calprotectin (S100A9), and folliculin to distinguish BA phenotypes, and concluded that increased serum concentrations of folliculin and calprotectin discriminated paucigranulocytic and neutrophilic phenotypes, respectively [65].
Serum levels of several cytokines, including Th2-cytokines such as interleukin (IL)-25, IL-4, IL-5, IL-13, and IL-33, are elevated in asthmatic adults and children compared to controls [66]. Increased serum levels of the chemokine CCL26/eotaxin-3 could differentiate moderate-to-severe asthma from healthy controls [67], and recent findings point out the ability of CCL17/TARC to predict type-2 eosinophilic asthma [68]. Upregulated IL-8 and TNFα were detected in acute attacks of asthma [69], and IL-1β is an indicator for the risk of pediatric allergic asthma [70].
3.1.2. Metabolites
A number of studies have identified metabolite dysregulation as useful for diagnosis, phenotyping, assessing of severity, and response to treatment [71]. However, the lack of analytical standardization sabotages replicability [72]. Therefore, only those metabolites that appear dysregulated in at least two different studies using plasma or serum samples from asthmatic patients were considered. Only linoleic acid levels resulted consistently significant in active asthma [73,74], although correlation with severity was not found [74]. In contrast, levels of glycerophosphocoline and L-valine were downregulated [75,76,77]. However, the majority of selected metabolites exhibited opposing results, appearing down- and upregulated in asthma indistinctly, which could be explained by the variability in the origin (whole blood, plasma, or serum) or population type (adult or children) of where/from whom samples were collected. The metabolites include arachidonic acid [75,77], succinate [75,78], palmitic acid [75,79], xanthine [74,75], taurine [74,76], bilirubin [74,75], arginine [75,80], histidine [80,81], and glucose [73,80].
3.1.3. RNA
Changes in gene expression overall suggest the potential of circulating RNAs as BA biomarkers [82,83,84,85], with miRNAs being the most commonly studied. In 2016, Panganiban et al. described that the differential expression of 30 miRNAs in plasma distinguished a cohort of patients with allergic rhinitis and 2 subtypes of asthma with high or low peripheral eosinophil counts from healthy controls [85]. In total, 11 miRNAs were upregulated (miR-125b, miR-126, miR-21, miR-16, miR-223, miR-148a, miR-146a, and let-7b/c/e) and 5 downregulated (miR-1, miR-299-5p, miR-570, miR-106a and miR-155) exclusively in asthma samples [85]. A further analysis compiling different studies concluded that the combination of miR-185-5p, miR-155, miR-21, miR-320, miR-1246, miR-144-5p, miR-1165-3p, and let-7a potentially served as a diagnostic biomarker for asthma [86]. In line with previous reports, Kyyaly et al. proposed a panel of circulating miRNAs with diagnostic (upregulated miR-126, miR-155, miR-21, miR-125b, miR146a, and miR-98, and downregulation of let-7 family, miR-192, miR-15a, and miR-30a) and assessment of severity potential (upregulated miR-126, miR-155, miR-125b, and miR-1165-3p, and downregulated miR-1 and miR-19b) [84].
In some cases, a correlation between certain miRNAs and lncRNAs served as biomarkers of disease exacerbation. This is the case for ANRIL/miR125a or NEAT1/miR124 [87]. Other lncRNAs are informative for BA diagnosis in adults (RP11-401.2 and LNC-000127) [88], and children (CASC2, PTTG3P, lncRNA-H19) [89,90,91]. Among mRNAs, the upregulation of PTGDR2, a prostaglandin receptor involved in the chemotaxis of leukocytes, was found to be significantly upregulated in the blood of asthmatic patients [92].
3.2. Atopic Dermatitis (AD)
3.2.1. Proteins
As eosinophils are active participants in the pathogenesis of AD, eosinophil granule-derived proteins are found in circulation, although their utility as biomarkers is under debate. ECP was postulated in the early 1990s as a severity biomarker [93,94], and EDN subsequently [95]. However, later studies in pediatric patients showed no significant association [96,97].
Several adhesion molecules appeared upregulated in skin biopsies of AD patients [98], and some have also been detected in circulation. Increased serum periostin has potential as a diagnostic and severity biomarker [99], and its levels even vary in response to therapy [99,100]. E-selectin and tissue remodeling matrix metalloproteinases (MMP-3/9/10/12) are also potentially useful for diagnosis [101,102].
In AD, Th2-chemokines such as CCL26/eotaxin-3 [103,104], CCL22/MDC [105], CCL18/PARC [100,106], CCL27/CTACK [107], and CCL17/TARC, have been more commonly found as blood biomarkers compared to Th2-cytokines [45], the latter of which is considered the most reliable biomarker for AD [66]. In fact, elevated serum TARC/CCL17 discriminated AD from healthy controls [102,108,109] and showed the best odds ratio (OR) when compared to eosinophil count, total IgE, serum IL-18, and lactate dehydrogenase (LDH) [110]. However, more extensive evidence promotes TARC’s utility as a biomarker of severity [111,112,113], and response to treatment [100]. Its specificity has been questioned however as it also appears upregulated in several skin diseases [45], with the exception of psoriasis [102]. A few cytokines with biomarker potential in AD include IL-13, IL-22 [100,109], and IL-18 [110,114]. Contradictory results relating to DPP-4 showed its upregulation in plasma of AD patients [115], while in circulating T-cells in AD, it exhibited a significant decrease in surface expression [116,117].
Other inflammatory molecules are: the soluble receptor IL-2 (sIL-2R), associated with severity [93,94] but with inconclusive results [102]; soluble CD23 (sCD23) [94]; the receptors of soluble Tumor Necrosis Factor (sTNFRI/II) [118]; C-reactive protein (CRP), which is upregulated in AD subjects compared to healthy controls and correlated with disease severity scores [119]; and lactate dehydrogenase (LDH), which is also elevated in AD [99,119] as a potential indicator of disease severity both in children [120], and adults [110]. Adipokines, involved in the integration of metabolism and immune function [121], have also been postulated as potential biomarkers. A significant decrease of serum resistin and adiponectin differentiated adults with AD from controls, and correlated with the SCORing Atopic Dermatitis (SCORAD) index [122]. Similarly, serum levels of YKL-40 appeared significantly upregulated in AD versus healthy controls, and correlated positively with the SCORAD index, thus, indicating its potential as a biomarker for severity [123]. Likewise, elevated serum levels of squamous cell carcinoma antigens 1 and 2 (SCCA1/2) have been described in AD and psoriatic patients [124].
3.2.2. Metabolites
A distinct metabolic signature of AD has been most commonly studied using skin biopsies rather than in circulation [125], reflecting the stronger diagnostic power of skin compared to blood [126,127]. Existing studies in serum samples identified a distinct metabolic profile between AD and healthy controls involving dysregulation of phosphatidylcholine and acylcarnitine [128], and other metabolites and lipid mediators that are linked to the pro-inflammatory state of AD [129]. In plasma, Chiu et al. showed that a metabolic signature related to the metabolism of nitrogen and amino acids discriminated AD endotypes based on filaggrin mutations and IgE levels [130]. This was further evinced in pediatric patients, regarding elevated IgE endotypes [131]. In addition, metabolic signatures have identified therapeutic responders as shown for omalizumab and dupilumab treatments [132,133]. Only recently, the increased levels of vitamin D and A5 ligands in the plasma of AD patients has been postulated as a biomarker of AD severity [134].
3.2.3. RNA
Cumulative evidence connects microRNAs (miRNAs) with the pathophysiology of AD and other skin disorders [135,136]. This has motivated the search for differential circulating miRNA-expression patterns in blood in order to seek a suitable biomarker. In an initial study involving pediatric subjects, serum levels of miR-203 and miR-484-5p were found to be significantly increased in children with AD compared to controls. However, miR-203 was upregulated exclusively in those patients who showed high serum IgE levels [118]. In addition, miR-155, known to be upregulated 4–6-fold in AD skin [137], also appeared significantly increased in the circulation of atopic children [138] and circulating CD4+ monocytes of children with AD [139].
A subsequent study performed with a small cohort of adult patients with AD, psoriasis, and healthy controls, showed that serum levels of miR146a, miR-203, and miR-205 had no discriminative ability for AD compared to controls; however, it exclusively identified significant downregulation of serum miR-125b in AD patients [140]. These results contradict the aforementioned miR-203 upregulation in AD [118]. A recent study carried out in pediatric patients suffering from AD found a significant increment in serum miR-146a expression, and a lower ratio of Th1/Th2 compared to controls [141]. Another study that selected miR-29b based on a miRNA microarray of skin biopsies, demonstrated its upregulated expression in the serum and correlation with the disease severity score [142].
One of the two studies to date that used plasma instead of serum as a source of circulating biomarkers for AD identified 25 differentially regulated miRNAs by high-throughput sequencing of samples from 700 subjects (including adult and pediatric patients, and healthy controls without history of atopies). Of those upregulated sequences, plasma miR-151a was significantly increased in AD when confirmed by RT-qPCR [143]. The second study, using plasma samples of children with AD, showed the dysregulation of 40 miRNAs and proposed the most differentially expressed miRNA, hsa-miR-194-5p, as a potential biomarker of AD, based on area under the receiver operating characteristic (ROC) analysis [144].
In addition to microRNAs, a significant increase of mRNAs encoding for pro-inflammatory cytokine IL-17 and retinoic acid receptor (ROR)γt in circulating Th17- CD4+ cells obtained from AD patients suggest their potential as a diagnostic biomarker for the disease [139].
3.3. Eosinophilic Esophagitis (EoE)
3.3.1. Proteins
Periostin is markedly overexpressed in the esophagus of EoE patients [145], and represents a promising non-invasive biomarker. Slightly increased levels of serum periostin differentiated EoE patients from controls in correlation with high serum IL-13 [146]. However, this is, to our knowledge, the only study assessing serum periostin. Alternatively, given the disruption of the epithelial barrier and esophageal fibrosis in EoE, the use of autoantibodies against epithelial adhesion molecules has been hypothesized as serving as disease biomarkers. Indeed, antibodies against transmembrane desmoglein-3 (DSG3) and collagen XVII (NC16A) appeared to be increased in the serum of EoE patients, with a more prominent increase of NC16A [147].
ECP and EDN are the eosinophil granule proteins most commonly studied in circulating blood in the context of EoE. Although no utility of ECP or EDN as biomarkers was reported in a small number of studies [148,149], this evidence was overshadowed by others showing the upregulation of EDN [16,150,151] and ECP [150,152,153] in the blood of EoE patients compared to healthy controls. Consequently, treatment with mepolizumab led to a reduction in ECP and EDN levels [154], and topical corticosteroids [155] or diet restriction also reduced ECP levels [156] with variable results [157]. Despite the lack of differences in circulating MBP-1 initially reported [148], more recent works have shown a significant upregulation of MBP-1 as being helpful in discriminating EoE from healthy controls, and after treatment [158], and even in combination with CLC-GAL10 [153]. Strikingly, in contrast to common upregulation of granule proteins reported to date, Wright et al. observed lower levels of granule proteins in the serum of EoE patients that exhibited a marked degranulation within the tissue [149]. They postulated that circulating eosinophils in EoE might retain their granules, therefore suggesting that downregulation of serum EPX is a biomarker of EoE [149].
Cytokines, as mediators of inflammation, have been extensively studied as inflammatory disease biomarkers. The blood levels of cytokines: IL-4, IL-13, IL-5, IL-6, IL-12p70, CD40ligand, IL-1α, and IL-17 distinguished EoE from non-EoE patients [159], but such changes could not be validated in subsequent studies [66]. Among cytokines, IL-5 has raised the highest interest given its key role in the proliferation, maturation, and differentiation of eosinophils within the bone marrow [160]. However, many studies could not demonstrate differences in serum IL-5 [148,150,151,158,161]. Only Lu et al. [162] observed a significant increase of Th2 cytokines, specifically IL-10, in EoE patients, together with increased levels of absolute eosinophil counts and serum levels of TNFα and IL-12 cytokines. Similarly, despite the increased esophageal expression of eotaxin-3 [163], a chemokine important for eosinophil migration and tissue infiltration, differences in circulation have not been detected [150,153,158]. The exception is a study performed in pediatric patients, where eotaxin-3 was increased in plasma, along with increased absolute eosinophil counts and tissue eosinophilia [16].
3.3.2. Metabolites
Just two studies highlighted dysregulated metabolites in circulation as being possibly useful for EoE diagnosis. This is the case with 15(S)-hydroxyeicosatetraenoic acid (15(S)-HETE), a metabolite detected in peripheral blood derived from arachidonate 15-lipoxygenase (ALOX15), which is upregulated in the esophagus of EoE patients, and also found to be increased by 2.4-fold in the serum of EoE patients [164,165]. A further study identified several urea cycle metabolites (dimethylarginine, putrescine and N-acetylputrescine) as potential biomarkers for EoE diagnosis in children [166].
3.3.3. RNA
Lu et al. tested for the first time the potential of circulating microRNAs as a reflection of the RNA expression profile in tissue. They found that increased levels of miR-146a, miR-146b, and miR-223 significantly differentiated between healthy controls and EoE patients, while downregulation of miR-146a and miR-233 indicated EoE remission induced by glucocorticoid therapy [167]. Likewise, significant upregulation of serum miR-21 30-fold in EoE correlated with an increased expression 40-fold in the esophageal tissue [19]. Unfortunately, there is still little evidence of circulating microRNAs in EoE, and other authors failed when they tried to detect them in the serum [168]. Differences in mRNAs have also been explored as potential circulating biomarkers for EoE [169,170,171]. The presence of IgE receptor I (FcεRI) mRNAs was found to be significantly reduced in the blood of EoE patients compared to GERD and healthy controls [169], a finding that aligns with the fact that EoE is a non-IgE-mediated allergy [40]. Similarly, IL-15R mRNAs were less abundant in the blood of EoE compared to GERD patients [169]. Despite IL-15 expression being induced in the esophagus of active EoE [172], the authors hypothesized that the reduction in circulating lymphocytes with IL-15 receptors was due to their preferential recruitment to the tissue. On the other hand, upregulation of mRNAs encoding for eosinophil surface molecules such as CD101, CXCR6, and CD274 (PDL1) served to discriminate EoE from GERD patients [170,171].
Common circulating biomarkers for either BA and AD, BA and EoE, AD and EoE, or BA, AD, and EoE are shown in Figure 1.
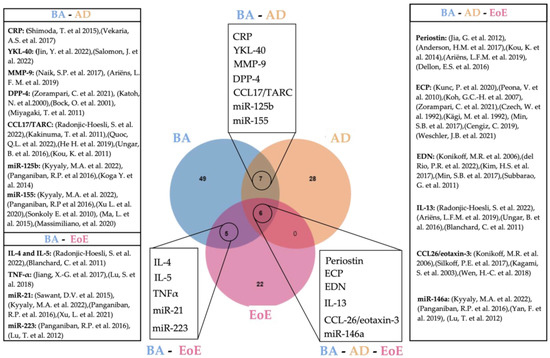
Figure 1.
Venn diagram showing overlapping circulating biomarkers for BA, AD, and EoE [16,19,47,48,49,54,55,56,57,58,59,60,61,62,65,66,67,68,83,84,85,92,93,94,99,100,101,102,103,107,108,114,118,122,123,136,137,138,139,144,148,150,151,152,153,154,157,160,165]. Created with Jvenn diagram viewer [173]. BA: bronchial asthma, AD: atopic dermatitis, EoE: eosinophilic esophagitis.
4. Extracellular Vesicles (EVs) as Circulating Biomarkers
4.1. EVs and the Immune System
The term extracellular vesicle was coined in 2011 [174] to define any non-replicating extracellular entities delimited by a lipidic membrane that are released by cells. Although the classification is complex and continues to grow as the field develops [22], EVs can be generally divided in two broad categories based on their biogenesis: endosomal origin (or exosomes), and plasma-membrane (or ectosomes) [25]. Regardless of their biogenesis, EVs perform fundamental functions in homeostasis and pathological processes that range from the removal/recycling of unnecessary molecules from the cell to the delivery of intercellular signals. The biogenesis of EVs, as well as the identity and state of the parental cell, largely contribute to defining their cargo and, consequently, their function [175].
The ability of EVs to mediate intercellular communication is of enormous relevance in immune signaling. In inflammation and innate immunity, EVs exert pro- and anti-inflammatory functions mediated by their load of bioactive lipids (i.e., arachidonic acid) [176]; cytokines [177]; damage-associated molecular patterns (DAMPs) [178]; pro-inflammatory microRNAs [179]; or soluble mediators (i.e., C-reactive protein) [180]. Activation of immune responses upon microbial or allergen intrusion also involves EVs, as they carry allergens and microbial-associated molecular patterns [25]. In addition, a fundamental function in adaptive immunity, antigen presentation, can be mediated by MHC-loaded EVs, contributing to a more sophisticated regulation of immunity [181,182]. Viewing the published data, it is logical to think that EVs can be utilized as biomarkers, especially in the context of immune mediated disorders, where EVs seem to have ubiquitous roles. We will now examine the data suggesting the utility of circulating EVs as biomarkers of AS and AD.
4.2. Circulating EVs as Biomarkers in BA
One of the first hints indicating an association between asthma and EVs was the increment of such particles in the circulation of asthmatics [183]. Duarte et al. found elevated levels of platelet microparticles in the circulation of a group of 20 asthmatics under corticosteroid treatment compared to 15 healthy volunteers, and further studies showed similar upregulation induced by pollution [183,184,185]. Nonetheless, elevated EV levels do not correspond specifically to asthma, as was observed in asthmatics and non-asthmatics from a cohort of type-1 allergic patients with rhinoconjunctivitis [186].
Identifying significant differences in the EV-cargo is one of the main hopes when looking for EV biomarkers, and EV-miRNAs are among the most commonly studied molecules in this regard. Increased levels of serum EV miR-126 in allergic asthmatic patients has suggested its potential as a diagnostic biomarker [187]. MiRNA-125b in serum EVs appeared upregulated in patients with different levels of asthma severity, and was even able to discriminate mild-bronchial asthma from healthy controls [188]. Consistently, upregulation of EV-miR-125b correlated with increased CRP and IgE serum levels, which together with downregulated EV- miR-124, miR-133, and miR-130, could differentiate subjects with severe asthma under corticosteroid treatment from healthy controls [189]. The let-7 family of microRNA is dysregulated in bronchial asthma [84]. Accordingly, Zheng et al. observed that increased let-7i-5p levels in plasma EVs correlated with patient exposure to fine particulate matter in a cohort of asthmatic children and, in combination with serum IgE, exhibited a greater diagnostic performance [185]. Plasma EVs significantly enriched in miR-223 and miR-21 also differentiated moderate asthmatic patients from healthy controls [190]. The only study, to our knowledge, that employed RNAseq in circulating human EVs for plasma did not find overall changes in the miRNA content; however, an insufficient cohort size could partly explain these results. Nonetheless, the authors identified the upregulation of miR-122-5p in all groups of eosinophilic asthma compared to controls [191].
Although less explored, proteins in EVs harbor potential biomarkers for defining asthma endotypes, as hypothesized by Suzuki et al. [192]. Without purification of EVs, the proteome of plasma samples from COPD or patients with severe asthma showed different profiles differentiating each disease and asthma endotypes, significantly enriched in extracellular vesicle markers, and thus suggesting an association between EVs and protein groups [192]. Similarly, a pattern of upregulated (TNFα, IL-4, IL-5, IL-6, IL-17F, CCL2, and CCL17/TARC) and downregulated (IL-11, IL-27, and CCL20) EV-associated cytokines discriminated healthy controls from allergic patients, including asthmatics [186].
4.3. Circulating EVs as Biomarkers in AD
The pathogenesis of AD has been associated with the colonization of the skin by different microbes, one of the best characterized being Staphylococcus aureus [193]. The fact that bacteria can release EVs [194] has sustained the research around microbe-derived EVs as potential pathogenic effectors and biomarkers. A metagenomic analysis on serum and urine EVs from AD and healthy donors identified high homogeneity between both type of biofluids, and significant downregulation of several lactic acid bacteria genera in comparison to controls in urine EVs [195]. A similar study including a greater number of healthy controls, and using only serum EVs, identified Escherichia–Shigella and Enterococcus as upregulated in AD [196], which suggests great potential for microbial EVs as biomarkers for AD.
EVs derived from immune cells can reflect their origin and the activation state of the donor cell by their content and surface marker [25]. Based on this premise, Ryutaro Oba et al. identified several EV-subsets from T cells: CD3+CD4+ EVs for CD4+ T cells, CD3+CD8+ EVs for CD8+ T cells, and CD3+HLA-DR+ EVs for Th1-type T cells [197]. Interestingly, differential distribution of such EV subsets could discriminate AD from healthy adults as CD3+CD4+ EVs were significantly upregulated in AD, while CD3+HLA-DR+ EVs were downregulated. However, when AD was compared to other inflammatory diseases such as osteoarthritis or rheumatoid arthritis, no differences were found in the aforementioned EV-subsets. EVs derived from mast cells have been identified in the serum of both AD and non-atopic controls, although a significant overexpression of the miR103a-3p characterized EVs from AD patients [198]. RNAseq of plasma derived EVs from pediatric AD patients revealed 10 differentially regulated genes, among which the transfer RNA Fragment tRF-28-QSZ34KRQ590K (tRFs are a novel class of non-coding RNAs with regulatory roles [199]) was significantly downregulated in AD [200]. To date, a single study has characterized the proteomic differences in serum EVs in the context of AD [201]. Over a thousand proteins were identified, of which 19 were unique to AD-EVs, with overrepresented functions linked to pro-inflammatory cytokine production such as platelet activation or Rap1 signaling [202].
Common biomarkers between BA, BA-EVs, AD, and AD-EVs are shown in Figure 2.
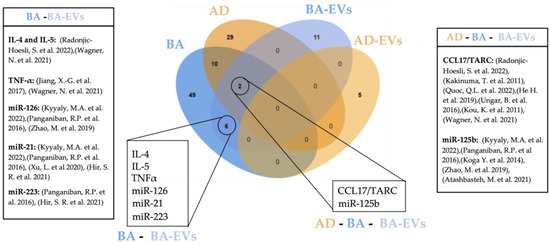
Figure 2.
Venn diagram showing overlapping circulating biomarkers for BA, AD, BA-EVs, and AD-EVs [65,66,67,68,83,84,85,101,107,108,138,184,185,186,187,188]. Created with Jvenn diagram viewer [173]. BA: bronchial asthma, BA-EVs: bronchial asthma extracellular vesicles, AD: atopic dermatitis, AD-EVs: atopic dermatitis extracellular vesicles.
4.4. Potential of EVs as Biomarker of EoE
Although the role of EVs remains unexplored in EoE, a number of studies have collected evidence of the functional relationship of EVs, with two areas closely linked to EoE: allergy and inflammation [203], and esophageal disorders (mostly Barrett’s esophagus) [204]. Development of Barrett’s esophagus is often preceded by sustained GERD, which exhibits overlapping symptoms with EoE but a different pathogenesis [205]. Preliminary work by Uemura et al. showed the diagnostic potential of EV-miRNAs in a rat model of GERD, in which serum EVs were withdrawn from male rats at different stages after the development of reflux esophagitis (acute, sub-acute, and chronic phases) [206]. By microarray analysis of the EVs, miR-223-3p and miR-29-3p were identified as differentially expressed among the different phases. Interestingly, the upregulation of circulating miR-223 has previously been found upregulated in EoE patients compared to healthy controls [167].
The highly conserved mechanisms of autophagy coordinate the recycling and degradation of cellular material to maintain protein homeostasis, activated in response to certain stress stimuli such as inflammation [207]. In line with this, a study employing human biopsies and murine models of EoE demonstrated that inflammatory stimuli mediated by IL-13 and TNFα cytokines upregulated autophagy in esophageal epithelial cells, causing the accumulation of autophagic vesicles (AVs) with distinct cargo between EoE and control samples [208]. Autophagy and EVs are tightly related, due to the confluence in their mechanisms of biogenesis and functionality, since the autophagy–EV crosstalk is relevant in homeostatic and pathologic scenarios [209]. Moreover, the release of extracellular vesicles and particles with autophagic cargo controlled by the secretory autophagy pathway has been described [210,211]. Future studies might consider exploring the release of vesicles to the extracellular milieu, that potentially contain autophagy-derived material, as being useful as biomarkers in EoE.
The total of specific markers that define the tissue origin of EVs is still largely unknown. According to Li et al., about 0.2% of EVs in circulation belong to different tissues, including EVs that derive from the esophagus, as several specific genes were identified in plasma-EVs [212]. However, the vast majority of circulating EVs are generated by blood cells (i.e., monocytes, lymphocytes, platelets, etc.), suggesting that immune cell-derived EVs are the most promising as a source of circulating biomarkers. In fact, every immune cell with a role in inflammation can secrete EVs [25]. This is the case for eosinophils [213]. Studies employing EVs derived from circulating eosinophils demonstrated that, upon cytokine stimulation (IFNγ, eotaxin-1, or ΤΝFα), these cells increase the release of EVs carrying several eosinophil-granule proteins, which are more prominent in eosinophils from asthmatics than in healthy controls. Moreover, eosinophil-EVs participate in tissue remodeling and cell migration, indicating their active role in disease pathophysiology [214]. Despite the potential of eosinophil-derived EVs as biomarkers for eosinophilic disorders, the characterization of these EVs is currently limited and studies reporting the detection of eosinophil-EVs in biofluids are lacking.
Changes in the microbiota of the mucosa have been linked to the initiation and maintenance of inflammation. The flora of the healthy esophagus, commonly colonized by the genus Streptococci [215], is imbalanced in EoE esophagi towards an enrichment in Neisseria, Corynebacterium [216], and Haemophilus [217]; and downregulation of Phorphyromonas [218]. Both commensal and pathogenic bacteria can secrete bacterial extracellular vesicles (BEVs) into different biofluids, thus becoming a reflection of the microbiota composition in distant sites, and potent biomarkers of disease diagnosis and monitoring [219]. For example, the metagenome of serum-derived EVs showed correlation with bodily microbiota in a murine model of Alzheimer’s disease that differed from the wild type controls [220]; and the presence of Sphingomonadaceae in urine-EVs of children with chronic rhinitis and asthma correlated with its upregulation in the airways [221]; and serum bacterial-EVs positively correlated with bacteria in paranasal sinus of patients with rhinosinusitis [222]. In the latter study, microbiota composition varied depending on whether the patients had eosinophilic inflammation or not, thus suggesting a link between microbiota and immune response of the host [222]. Therefore, circulating BEVs as potential biomarkers of dysbiosis in the gut seem promising, since a match between blood EVs and gut microbiota have been repeatedly reported [219]. Interestingly, different bacterial composition and abundances characterize GERD and EoE patients [215,223], suggesting the potential of bacterial-derived EVs in discriminating between these two commonly misdiagnosed diseases.
5. Conclusions and Perspectives
Although blood is the source of choice in the majority of studies seeking non-invasive biomarkers, a robust candidate for EoE still has not been found [12]. In this review we show that 18 circulating molecules suggested as disease biomarkers are common for either BA and AD (7), BA and EoE (6) or BA, EoE and AD (5), demonstrating that the co-existence of EoE with other eosinophilic disorders hinders the finding of specific biomarkers. Although our review is limited to concomitancy of EoE with BA and AD, the existence of other common biomarkers for EoE and concomitant allergies such as allergic rhinitis or food allergy is very likely and should be considered for further research. Such findings indicate the need for alternative sampling methods. Other minimally invasive methods employing esophageal mucus seem promising [224,225] and potentially more specific in the context of EoE; however, they are more or less limited to the site of collection. In contrast, blood-based biomarkers have the potential to provide a more complete picture of the state of the disease. To overcome current limitations, further studies should consider either the detailed characterization of patient comorbidities or, when possible, the inclusion of appropriate atopic controls. Additionally, a more refined manipulation of blood samples might improve specificity, such as the enrichment of EVs from patient´s blood. Despite being in its infancy, a considerable number of studies have explored the potential of EVs as biomarkers of disease. In regard to the malignancies covered in this review, BA constitutes the highest number of EV-related studies, doubling those dedicated to AD using blood exclusively. Indeed, up to 19 potential EV-biomarkers were described for BA, five of which had been previously uncovered in circulation including Il-4, IL-5, TNF-α, CCL17/TARC, and miR-21 (Figure 2), thus indicating the specificity and robustness of these biomarkers. In contrast, no coincidences were found between circulating and EV biomarkers described for AD. This is most likely due to the still limited amount of research for this disease. We also noticed a great deal of heterogeneity in the EV-isolation methods employed, as well as an uneven technical accuracy for EV characterization [226,227], highlighting room for improvement, still, within translational research in EVs. Table 2 summarizes all studies involving EVs as potential biomarkers included in this work.

Table 2.
Studies exploring the potential as biomarkers of circulating EVs included in this work. TEM: transmission electron microscopy, UC: ultracentrifugation, NTA: nanoparticle tracking analysis, BCA: bicinchoninic acid assay, DLS: dynamic light scattering, SEM: scanning electron microscopy, SEC: size exclusion chromatography, PBMC: peripheral blood mononuclear cells.
In summary, common circulating biomarkers have been described for BA, AD, and EoE diseases. The suggestion of lack of specificity highlights the need to include an exhaustive control of concomitant atopies. EVs are a promising source that could increase biomarker specificity; however, rigorous characterization and method homogeneity are key to ensuring robust and reproducible results.
Author Contributions
Conceptualization, E.G.-N. and A.J.L.; writing—original draft preparation, E.G.-N.; writing—review and editing, A.J.L., L.A.-G., E.J.L.-M. and P.N. All authors have read and agreed to the published version of the manuscript.
Funding
This work was supported in part by grant from the Instituto de Salud Carlos III (ISCIII) (PI18/01252 III), and co-funded by the European Union, and by Association of Biomedical Research La Mancha Centro. E.J.L.-M. is in receipt of a Juan Rodes grant (JR19/00005) from the ISCIII, Spanish Ministry of Health-Social Services and Equality, which is partly funded by the European Social Fund (period 2014–2020). P.N. is in receipt of a Sara Borrell grant (CD19/00102), both from the ISCIII, Spanish Ministry of Health, Social Services and Equality, which is partly funded by the European Social Fund (period 2014–2020).
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
Not applicable.
Acknowledgments
We are grateful to Melanie Radcliff for English language revision.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Lucendo, A.J.; Molina-Infante, J.; Arias, Á.; von Arnim, U.; Bredenoord, A.J.; Bussmann, C.; Dias, J.A.; Bove, M.; González-Cervera, J.; Larsson, H.; et al. Guidelines on eosinophilic esophagitis: Evidence-based statements and recommendations for diagnosis and management in children and adults. United Eur. Gastroenterol. J. 2017, 5, 335–358. [Google Scholar] [CrossRef] [PubMed]
- Arias, Á.; Lucendo, A.J. Incidence and prevalence of eosinophilic oesophagitis increase continiously in adults and children in Central Spain: A 12-year population-based study. Dig. Liver Dis. 2019, 51, 55–62. [Google Scholar] [CrossRef] [PubMed]
- Straumann, A.; Bauer, M.; Fischer, B.; Blaser, K.; Simon, H.-U. Idiopathic eosinophilic esophagitis is associated with a TH2-type allergic inflammatory response. J. Allergy Clin. Immunol. 2001, 108, 954–961. [Google Scholar] [CrossRef]
- Capucilli, P.; Hill, D.A. Allergic Comorbidity in Eosinophilic Esophagitis: Mechanistic Relevance and Clinical Implications. Clin. Rev. Allergy Immunol. 2019, 57, 111–127. [Google Scholar] [CrossRef] [PubMed]
- Arias, Á.; Lucendo, A.J. Epidemiology and risk factors for eosinophilic esophagitis: Lessons for clinicians. Expert Rev. Gastroenterol. Hepatol. 2020, 14, 1069–1082. [Google Scholar] [CrossRef] [PubMed]
- Navarro, P.; Arias, Á.; Arias-González, L.; Laserna-Mendieta, E.J.; Ruiz-Ponce, M.; Lucendo, A.J. Systematic review with meta-analysis: The growing incidence and prevalence of eosinophilic oesophagitis in children and adults in population-based studies. Aliment. Pharmacol. Ther. 2019, 49, 1116–1125. [Google Scholar] [CrossRef]
- Navarro, P.; Laserna-Mendieta, E.J.; Casabona, S.; Savarino, E.; Pérez-Fernández, M.T.; Ghisa, M.; Pérez-Martínez, I.; Guagnozzi, D.; Perelló, A.; Guardiola-Arévalo, A.; et al. Accurate and timely diagnosis of Eosinophilic Esophagitis improves over time in Europe. An analysis of the EoE CONNECT Registry. United Eur. Gastroenterol. J. 2022, 10, 507–517. [Google Scholar] [CrossRef]
- Rossi, C.M.; Lenti, M.V.; di Sabatino, A. The need for a reliable non-invasive diagnostic biomarker for eosinophilic oesophagitis. Lancet Gastroenterol. Hepatol. 2022, 7, 202–203. [Google Scholar] [CrossRef]
- González-Cervera, J.; Arias, Á.; Redondo-González, O.; Cano-Mollinedo, M.M.; Terreehorst, I.; Lucendo, A.J. Association between atopic manifestations and eosinophilic esophagitis. Ann. Allergy Asthma Immunol. 2017, 118, 582–590.e2. [Google Scholar] [CrossRef]
- McCormick, J.P.; Lee, J.T. Insights into the Implications of Coexisting Type 2 Inflammatory Diseases. J. Inflamm. Res. 2021, 14, 4259–4266. [Google Scholar] [CrossRef]
- Hill, D.A.; Grundmeier, R.W.; Ramos, M.; Spergel, J.M. Eosinophilic Esophagitis Is a Late Manifestation of the Allergic March. J. Allergy Clin. Immunol. Pract. 2018, 6, 1528–1533. [Google Scholar] [CrossRef]
- Hines, B.T.; Rank, M.A.; Wright, B.L.; Marks, L.; Hagan, J.B.; Straumann, A.; Greenhawt, M.; Dellon, E.S. Minimally invasive biomarker studies in eosinophilic esophagitis. Ann. Allergy Asthma Immunol. 2018, 121, 218–228. [Google Scholar] [CrossRef] [PubMed]
- Bullock, J.Z.; Villanueva, J.M.; Blanchard, C.; Filipovich, A.H.; Putnam, P.E.; Collins, M.H.; Risma, K.A.; Akers, R.M.; Kirby, C.L.; Buckmeier, B.K.; et al. Interplay of Adaptive Th2 Immunity with Eotaxin-3/C-C Chemokine Receptor 3 in Eosinophilic Esophagitis. J. Craniofacial Surg. 2007, 45, 22–31. [Google Scholar] [CrossRef]
- Cunnion, K.M.; Willis, L.K.; Minto, H.B.; Burch, T.C.; Werner, A.L.; Shah, T.A.; Krishna, N.K.; Nyalwidhe, J.O.; Maples, K.M. Eosinophil Quantitated Urine Kinetic. Ann. Allergy Asthma Immunol. 2016, 116, 435–439. [Google Scholar] [CrossRef] [PubMed]
- Johnsson, M.; Bove, M.; Bergquist, H.; Olsson, M.; Fornwall, S.; Hassel, K.; Wold, A.E.; Wennerås, C. Distinctive Blood Eosinophilic Phenotypes and Cytokine Patterns in Eosinophilic Esophagitis, Inflammatory Bowel Disease and Airway Allergy. J. Innate Immun. 2011, 3, 594–604. [Google Scholar] [CrossRef] [PubMed]
- Konikoff, M.R.; Blanchard, C.; Kirby, C.; Buckmeier, B.K.; Cohen, M.; Heubi, J.E.; Putnam, P.E.; Rothenberg, M.E. Potential of Blood Eosinophils, Eosinophil-Derived Neurotoxin, and Eotaxin-3 as Biomarkers of Eosinophilic Esophagitis. Clin. Gastroenterol. Hepatol. 2006, 4, 1328–1336. [Google Scholar] [CrossRef]
- Morris, D.W.; Stucke, E.M.; Martin, L.J.; Abonia, J.P.; Mukkada, V.A.; Putnam, P.E.; Rothenberg, M.E.; Fulkerson, P.C. Eosinophil progenitor levels are increased in patients with active pediatric eosinophilic esophagitis. J. Allergy Clin. Immunol. 2016, 138, 915–918.e5. [Google Scholar] [CrossRef]
- Nguyen, T.; Gernez, Y.; Fuentebella, J.; Patel, A.; Tirouvanziam, R.; Reshamwala, N.; Bass, D.; Berquist, W.E.; Cox, K.L.; Kerner, J.A.; et al. Immunophenotyping of Peripheral Eosinophils Demonstrates Activation in Eosinophilic Esophagitis. J. Craniofacial Surg. 2011, 53, 40–47. [Google Scholar] [CrossRef]
- Sawant, D.V.; Yao, W.; Wright, Z.; Sawyers, C.; Tepper, R.S.; Gupta, S.K.; Kaplan, M.H.; Dent, A.L. Serum MicroRNA-21 as a Biomarker for Allergic Inflammatory Disease in Children. MicroRNA 2015, 4, 36–40. [Google Scholar] [CrossRef] [PubMed]
- Schlag, C.; Miehlke, S.; Heiseke, A.; Brockow, K.; Krug, A.; von Arnim, U.; Straumann, A.; Vieth, M.; Bussmann, C.; Mueller, R.; et al. Peripheral blood eosinophils and other non-invasive biomarkers can monitor treatment response in eosinophilic oesophagitis. Aliment. Pharmacol. Ther. 2015, 42, 1122–1130. [Google Scholar] [CrossRef]
- Margolis, L.; Sadovsky, Y. The biology of extracellular vesicles: The known unknowns. PLoS Biol. 2019, 17, e3000363. [Google Scholar] [CrossRef]
- Couch, Y.; Buzàs, E.I.; di Vizio, D.; Gho, Y.S.; Harrison, P.; Hill, A.F.; Lötvall, J.; Raposo, G.; Stahl, P.D.; Théry, C.; et al. A brief history of nearly EV-erything—The rise and rise of extracellular vesicles. J. Extracell. Vesicles 2021, 10. [Google Scholar] [CrossRef]
- Alhamwe, B.A.; Potaczek, D.; Miethe, S.; Alhamdan, F.; Hintz, L.; Magomedov, A.; Garn, H. Extracellular Vesicles and Asthma—More Than Just a Co-Existence. Int. J. Mol. Sci. 2021, 22, 4984. [Google Scholar] [CrossRef] [PubMed]
- Nazimek, K.; Bryniarski, K.; Askenase, P.W. Functions of Exosomes and Microbial Extracellular Vesicles in Allergy and Contact and Delayed-Type Hypersensitivity. Int. Arch. Allergy Immunol. 2016, 171, 1–26. [Google Scholar] [CrossRef] [PubMed]
- Buzas, E.I. The roles of extracellular vesicles in the immune system. Nat. Rev. Immunol. 2022, 1–15. [Google Scholar] [CrossRef] [PubMed]
- Zhou, B.; Xu, K.; Zheng, X.; Chen, T.; Wang, J.; Song, Y.; Shao, Y.; Zheng, S. Application of exosomes as liquid biopsy in clinical diagnosis. Signal Transduct. Target. Ther. 2020, 5, 144. [Google Scholar] [CrossRef]
- Wolf, M.; Poupardin, R.W.; Ebner-Peking, P.; Andrade, A.C.; Blöchl, C.; Obermayer, A.; Gomes, F.G.; Vari, B.; Maeding, N.; Eminger, E.; et al. A functional corona around extracellular vesicles enhances angiogenesis, skin regeneration and immunomodulation. J. Extracell. Vesicles 2022, 11, e12207. [Google Scholar] [CrossRef]
- Hendrix, A. The nature of blood(y) extracellular vesicles. Nat. Rev. Mol. Cell Biol. 2021, 22, 243. [Google Scholar] [CrossRef]
- Ghezzi, M.; Pozzi, E.; Abbattista, L.; Lonoce, L.; Zuccotti, G.V.; D’Auria, E. Barrier Impairment and Type 2 Inflammation in Allergic Diseases: The Pediatric Perspective. Children 2021, 8, 1165. [Google Scholar] [CrossRef]
- Sugita, K.; Kabashima, K. Tight junctions in the development of asthma, chronic rhinosinusitis, atopic dermatitis, eosinophilic esophagitis, and inflammatory bowel diseases. J. Leukoc. Biol. 2020, 107, 749–762. [Google Scholar] [CrossRef]
- Cianferoni, A.; Spergel, J. The importance of TSLP in allergic disease and its role as a potential therapeutic target. Expert Rev. Clin. Immunol. 2014, 10, 1463–1474. [Google Scholar] [CrossRef]
- Hammad, H.; Lambrecht, B.N. The basic immunology of asthma. Cell 2021, 184, 1469–1485. [Google Scholar] [CrossRef]
- Lucendo, A.J. Cellular and molecular immunological mechanisms in eosinophilic esophagitis: An updated overview of their clinical implications. Expert Rev. Gastroenterol. Hepatol. 2014, 8, 669–685. [Google Scholar] [CrossRef]
- Sroka-Tomaszewska, J.; Trzeciak, M. Molecular Mechanisms of Atopic Dermatitis Pathogenesis. Int. J. Mol. Sci. 2021, 22, 4130. [Google Scholar] [CrossRef]
- Zhernov, Y.V.; Vysochanskaya, S.O.; Sukhov, V.A.; Zaostrovtseva, O.K.; Gorshenin, D.S.; Sidorova, E.A.; Mitrokhin, O.V. Molecular Mechanisms of Eosinophilic Esophagitis. Int. J. Mol. Sci. 2021, 22, 13183. [Google Scholar] [CrossRef] [PubMed]
- Radonjic-Hoesli, S.; Brüggen, M.-C.; Feldmeyer, L.; Simon, H.-U.; Simon, D. Eosinophils in skin diseases. Semin. Immunopathol. 2021, 43, 393–409. [Google Scholar] [CrossRef] [PubMed]
- Bakakos, A.; Loukides, S. Severe Eosinophilic Asthma. J. Clin. Med. 2019, 8, 1375. [Google Scholar] [CrossRef]
- Doyle, A.D.; Masuda, M.Y.; Kita, H.; Wright, B.L. Eosinophils in Eosinophilic Esophagitis: The Road to Fibrostenosis is Paved with Good Intentions. Front. Immunol. 2020, 11, 603295. [Google Scholar] [CrossRef]
- Doucet-Ladevèze, R.; Holvoet, S.; Raymond, F.; Foata, F.; Hershey, G.K.K.; Sherrill, J.D.; Rothenberg, M.E.; Blanchard, C. Transcriptomic Analysis Links Eosinophilic Esophagitis and Atopic Dermatitis. Front. Pediatr. 2019, 7, 467. [Google Scholar] [CrossRef] [PubMed]
- Simon, D.; Cianferoni, A.; Spergel, J.M.; Aceves, S.; Holbreich, M.; Venter, C.; Rothenberg, M.E.; Terreehorst, I.; Muraro, A.; Lucendo, A.J.; et al. Eosinophilic esophagitis is characterized by a non-IgE-mediated food hypersensitivity. Allergy 2016, 71, 611–620. [Google Scholar] [CrossRef]
- Mocanu, M.; Vâță, D.; Alexa, A.-I.; Trandafir, L.; Patrașcu, A.-I.; Hâncu, M.F.; Gheucă-Solovăstru, L. Atopic Dermatitis—Beyond the Skin. Diagnostics 2021, 11, 1553. [Google Scholar] [CrossRef]
- Liang, Z.; Liu, L.; Zhao, H.; Xia, Y.; Zhang, W.; Ye, Y.; Jiang, M.; Cai, S. A Systemic Inflammatory Endotype of Asthma with More Severe Disease Identified by Unbiased Clustering of the Serum Cytokine Profile. Medicine 2016, 95, e3774. [Google Scholar] [CrossRef] [PubMed]
- Godwin, B.; Wilkins, B.; Muir, A.B. EoE disease monitoring. Ann. Allergy Asthma Immunol. 2019, 124, 240–247. [Google Scholar] [CrossRef] [PubMed]
- Porpodis, K.; Tsiouprou, I.; Apostolopoulos, A.; Ntontsi, P.; Fouka, E.; Papakosta, D.; Vliagoftis, H.; Domvri, K. Eosinophilic Asthma, Phenotypes-Endotypes and Current Biomarkers of Choice. J. Pers. Med. 2022, 12, 1093. [Google Scholar] [CrossRef] [PubMed]
- Renert-Yuval, Y.; Thyssen, J.P.; Bissonnette, R.; Bieber, T.; Kabashima, K.; Hijnen, D.; Guttman-Yassky, E. Biomarkers in atopic dermatitis—A review on behalf of the International Eczema Council. J. Allergy Clin. Immunol. 2021, 147, 1174–1190.e1. [Google Scholar] [CrossRef]
- Matsumoto, H. Roles of Periostin in Asthma. Adv. Exp. Med. Biol. 2019, 1132, 145–159. [Google Scholar] [CrossRef] [PubMed]
- Matsumoto, H. Role of serum periostin in the management of asthma and its comorbidities. Respir. Investig. 2020, 58, 144–154. [Google Scholar] [CrossRef]
- Jia, G.; Erickson, R.W.; Choy, D.F.; Mosesova, S.; Wu, L.C.; Solberg, O.D.; Shikotra, A.; Carter, R.; Audusseau, S.; Hamid, Q.; et al. Periostin is a systemic biomarker of eosinophilic airway inflammation in asthmatic patients. J. Allergy Clin. Immunol. 2012, 130, 647–654.e10. [Google Scholar] [CrossRef] [PubMed]
- Anderson, H.M.; Lemanske, R.F.; Arron, J.R.; Holweg, C.T.J.; Rajamanickam, V.; Gangnon, R.E.; Gern, J.E.; Jackson, D.J. Relationships among aeroallergen sensitization, peripheral blood eosinophils, and periostin in pediatric asthma development. J. Allergy Clin. Immunol. 2017, 139, 790–796. [Google Scholar] [CrossRef]
- Yavuz, S.T.; Bagci, S.; Bolat, A.; Akin, O.; Ganschow, R. Association of serum periostin levels with clinical features in children with asthma. Pediatr. Allergy Immunol. 2021, 32, 937–944. [Google Scholar] [CrossRef] [PubMed]
- Samitas, K.; Zervas, E.; Vittorakis, S.; Semitekolou, M.; Alissafi, T.; Bossios, A.; Gogos, H.; Economidou, E.; Lotvall, J.; Xanthou, G.; et al. Osteopontin expression and relation to disease severity in human asthma. Eur. Respir. J. 2010, 37, 331–341. [Google Scholar] [CrossRef] [PubMed]
- Trinh, H.K.T.; Nguyen, T.V.T.; Kim, S.-H.; Cao, T.B.T.; Luu, Q.Q.; Kim, S.-H.; Park, H.-S. Osteopontin contributes to late-onset asthma phenotypes in adult asthma patients. Exp. Mol. Med. 2020, 52, 253–265. [Google Scholar] [CrossRef]
- Xu, H.; Lou, W.; Fu, F. Association between osteopontin expression and asthma: A meta-analysis. J. Int. Med. Res. 2019, 47, 3513–3521. [Google Scholar] [CrossRef] [PubMed]
- McBrien, C.N.; Menzies-Gow, A. The Biology of Eosinophils and Their Role in Asthma. Front. Med. 2017, 4, 93. [Google Scholar] [CrossRef] [PubMed]
- Kunc, P.; Fabry, J.; Lucanska, M.; Pecova, R. Biomarkers of Bronchial Asthma. Physiol. Res. 2020, 69, S29–S34. [Google Scholar] [CrossRef]
- Peona, V.; de Amici, M.; Quaglini, S.; Bellaviti, G.; Castellazzi, A.M.; Marseglia, G.; Ciprandi, G. Serum Eosinophilic Cationic Protein: Is There a Role in Respiratory Disorders. J. Asthma 2010, 47, 131–134. [Google Scholar] [CrossRef]
- Koh, G.C.-H.; Shek, L.P.-C.; Goh, D.Y.-T.; van Bever, H.; Koh, D.S.-Q. Eosinophil cationic protein: Is it useful in asthma? A systematic review. Respir. Med. 2007, 101, 696–705. [Google Scholar] [CrossRef] [PubMed]
- Zorampari, C.; Prakash, A.; Rehan, H.S.; Gupta, L.K. Serum dipeptidyl peptidase-4 and eosinophil cationic protein levels in patients of bronchial asthma. Pulm. Pharmacol. Ther. 2021, 72, 102109. [Google Scholar] [CrossRef]
- del Rio, P.R.; Liu, A.H.; Borres, M.P.; Södergren, E.; Iachetti, F.; Casale, T.B. Asthma and Allergy: Unravelling a Tangled Relationship with a Focus on New Biomarkers and Treatment. Int. J. Mol. Sci. 2022, 23, 3881. [Google Scholar] [CrossRef] [PubMed]
- Rutten, B.; Young, S.; Rhedin, M.; Olsson, M.; Kurian, N.; Syed, F.; Beech, A.; Fidock, M.; Newbold, P.; Singh, D.; et al. Eosinophil-derived neurotoxin: A biologically and analytically attractive asthma biomarker. PLoS ONE 2021, 16, e0246627. [Google Scholar] [CrossRef]
- Shimoda, T.; Obase, Y.; Kishikawa, R.; Iwanaga, T. Serum high-sensitivity C-reactive protein can be an airway inflammation predictor in bronchial asthma. Allergy Asthma Proc. 2015, 36, 167. [Google Scholar] [CrossRef]
- Jin, Y.; Song, J.; Xu, F.; Zhang, D.; He, J.; Zheng, J.; Zhang, Y.; Li, J.; Guo, Y.; Xu, M.; et al. Association between YKL-40 and asthma: A systematic meta-analysis. Sleep Breath. 2022, 26, 1011–1022. [Google Scholar] [CrossRef] [PubMed]
- Naik, S.P.; Mahesh, P.A.; Jayaraj, B.S.; Madhunapantula, S.V.; Jahromi, S.R.; Yadav, M.K. Evaluation of inflammatory markers interleukin-6 (IL-6) and matrix metalloproteinase-9 (MMP-9) in asthma. J. Asthma 2017, 54, 584–593. [Google Scholar] [CrossRef]
- Milara, J.; Morell, A.; de Diego, A.; Artigues, E.; Morcillo, E.; Cortijo, J. Mucin 1 deficiency mediates corticosteroid insensitivity in asthma. Allergy 2018, 74, 111–121. [Google Scholar] [CrossRef]
- Hur, G.-Y.; Ye, Y.-M.; Yang, E.; Park, H.-S. Serum potential biomarkers according to sputum inflammatory cell profiles in adult asthmatics. Korean J. Intern. Med. 2020, 35, 988–997. [Google Scholar] [CrossRef]
- Radonjic-Hoesli, S.; Pavlov, N.; Simon, H.-U.; Simon, D. Are blood cytokines reliable biomarkers of allergic disease diagnosis and treatment responses. J. Allergy Clin. Immunol. 2022, 150, 251–258. [Google Scholar] [CrossRef] [PubMed]
- Silkoff, P.E.; Laviolette, M.; Singh, D.; FitzGerald, J.M.; Kelsen, S.; Backer, V.; Porsbjerg, C.; Girodet, P.-O.; Berger, P.; Kline, J.; et al. Identification of airway mucosal type 2 inflammation by using clinical biomarkers in asthmatic patients. J. Allergy Clin. Immunol. 2017, 140, 710–719. [Google Scholar] [CrossRef] [PubMed]
- Quoc, Q.L.; Moon, J.-Y.; Lee, D.-H.; Ban, G.-Y.; Kim, S.-H.; Park, H.-S. Role of Thymus and Activation-Regulated Chemokine in Allergic Asthma. J. Asthma Allergy 2022, 15, 157–167. [Google Scholar] [CrossRef]
- Jiang, X.-G.; Yang, X.-D.; Lv, Z.; Zhuang, P.-H. Elevated serum levels of TNF-α, IL-8, and ECP can be involved in the development and progression of bronchial asthma. J. Asthma 2017, 55, 111–118. [Google Scholar] [CrossRef]
- Sobkowiak, P.; Banaszak, I.W.; Kowalewska, M.; Wasilewska, E.; Msc, W.L.; Kycler, Z.; Skibinska, M.; Bręborowicz, A.; Jassem, E.; Szczepankiewicz, A. Interleukin 1β polymorphism and serum level are associated with pediatric asthma. Pediatr. Pulmonol. 2017, 52, 1565–1571. [Google Scholar] [CrossRef]
- Wang, C.; Jiang, S.; Zhang, S.; Ouyang, Z.; Wang, G.; Wang, F. Research Progress of Metabolomics in Asthma. Metabolites 2021, 11, 567. [Google Scholar] [CrossRef]
- Gautam, Y.; Johansson, E.; Mersha, T.B. Multi-Omics Profiling Approach to Asthma: An Evolving Paradigm. J. Pers. Med. 2022, 12, 66. [Google Scholar] [CrossRef] [PubMed]
- Ghosh, N.; Choudhury, P.; Kaushik, S.R.; Arya, R.; Nanda, R.; Bhattacharyya, P.; Roychowdhury, S.; Banerjee, R.; Chaudhury, K. Metabolomic fingerprinting and systemic inflammatory profiling of asthma COPD overlap (ACO). Respir. Res. 2020, 21, 126. [Google Scholar] [CrossRef]
- Reinke, S.N.; Gallart-Ayala, H.; Gómez, C.; Checa, A.; Fauland, A.; Naz, S.; Kamleh, M.A.; Djukanović, R.; Hinks, T.S.; Wheelock, C.E. Metabolomics analysis identifies different metabotypes of asthma severity. Eur. Respir. J. 2017, 49, 1601740. [Google Scholar] [CrossRef] [PubMed]
- Liang, Y.; Gai, X.Y.; Chang, C.; Zhang, X.; Wang, J.; Li, T.T. Metabolomic Profiling Differences among Asthma, COPD, and Healthy Subjects: A LC-MS-based Metabolomic Analysis. Biomed. Environ. Sci. 2019, 32, 659–672. [Google Scholar] [CrossRef] [PubMed]
- Matysiak, J.; Klupczynska, A.; Packi, K.; Mackowiak-Jakubowska, A.; Bręborowicz, A.; Pawlicka, O.; Olejniczak, K.; Kokot, Z.; Matysiak, J. Alterations in Serum-Free Amino Acid Profiles in Childhood Asthma. Int. J. Environ. Res. Public Health 2020, 17, 4758. [Google Scholar] [CrossRef] [PubMed]
- Pang, Z.; Wang, G.; Wang, C.; Zhang, W.; Liu, J.; Wang, F. Serum Metabolomics Analysis of Asthma in Different Inflammatory Phenotypes: A Cross-Sectional Study in Northeast China. BioMed Res. Int. 2018, 2018, 2860521. [Google Scholar] [CrossRef]
- Turi, K.N.; McKennan, C.; Gebretsadik, T.; Snyder, B.; Seroogy, C.M.; Lemanske, R.F.; Zoratti, E.; Havstad, S.; Ober, C.; Lynch, S.; et al. Unconjugated bilirubin is associated with protection from early-life wheeze and childhood asthma. J. Allergy Clin. Immunol. 2021, 148, 128–138. [Google Scholar] [CrossRef]
- Bian, X.; Sun, B.; Zheng, P.; Li, N.; Wu, J.-L. Derivatization enhanced separation and sensitivity of long chain-free fatty acids: Application to asthma using targeted and non-targeted liquid chromatography-mass spectrometry approach. Anal. Chim. Acta 2017, 989, 59–70. [Google Scholar] [CrossRef]
- Jung, J.; Kim, S.-H.; Lee, H.-S.; Choi, G.S.; Jung, Y.-S.; Ryu, D.H.; Park, H.-S.; Hwang, G.-S. Serum metabolomics reveals pathways and biomarkers associated with asthma pathogenesis. Clin. Exp. Allergy 2013, 43, 425–433. [Google Scholar] [CrossRef]
- Chiu, C.-Y.; Cheng, M.-L.; Chiang, M.-H.; Wang, C.-J.; Tsai, M.-H.; Lin, G. Metabolomic Analysis Reveals Distinct Profiles in the Plasma and Urine Associated with IgE Reactions in Childhood Asthma. J. Clin. Med. 2020, 9, 887. [Google Scholar] [CrossRef] [PubMed]
- Bigler, J.; Boedigheimer, M.; Schofield, J.P.R.; Skipp, P.J.; Corfield, J.; Rowe, A.; Sousa, A.R.; Timour, M.; Twehues, L.; Hu, X.; et al. A Severe Asthma Disease Signature from Gene Expression Profiling of Peripheral Blood from U-BIOPRED Cohorts. Am. J. Respir. Crit. Care Med. 2017, 195, 1311–1320. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.; Mao, Z.-D.; Shi, Y.-J.; Qian, Y.; Liu, Z.-G.; Yin, X.-W.; Zhang, Q. Comprehensive analysis of miRNA–mRNA–lncRNA networks in severe asthma. Epigenomics 2019, 11, 115–131. [Google Scholar] [CrossRef] [PubMed]
- Kyyaly, M.A.; Vorobeva, E.V.; Kothalawala, D.M.; Fong, W.C.G.; He, P.; Sones, C.L.; Al-Zahrani, M.; Sanchez-Elsner, T.; Arshad, S.H.; Kurukulaaratchy, R.J. MicroRNAs—A Promising Tool for Asthma Diagnosis and Severity Assessment: A Systematic Review. J. Pers. Med. 2022, 12, 543. [Google Scholar] [CrossRef]
- Panganiban, R.P.; Wang, Y.; Howrylak, J.; Chinchilli, V.M.; Craig, T.J.; August, A.; Ishmael, F.T. Circulating microRNAs as biomarkers in patients with allergic rhinitis and asthma. J. Allergy Clin. Immunol. 2016, 137, 1423–1432. [Google Scholar] [CrossRef] [PubMed]
- Xu, L.; Yi, M.; Tan, Y.; Yi, Z.; Zhang, Y. A comprehensive analysis of microRNAs as diagnostic biomarkers for asthma. Ther. Adv. Respir. Dis. 2020, 14, 1753466620981863. [Google Scholar] [CrossRef] [PubMed]
- Ye, S.; Zhu, S.; Feng, L. LncRNA ANRIL/miR-125a axis exhibits potential as a biomarker for disease exacerbation, severity, and inflammation in bronchial asthma. J. Clin. Lab. Anal. 2019, 34, e23092. [Google Scholar] [CrossRef]
- Zhu, Y.; Mao, D.; Gao, W.; Han, G.; Hu, H. Analysis of lncRNA Expression in Patients with Eosinophilic and Neutrophilic Asthma Focusing on LNC_000127. Front. Genet. 2019, 10, 141. [Google Scholar] [CrossRef]
- Dai, B.; Sun, F.; Cai, X.; Li, C.; Liu, F.; Shang, Y. Long noncoding RNA PTTG3P/miR-192-3p/CCNB1 axis is a potential biomarker of childhood asthma. Int. Immunopharmacol. 2021, 101, 108229. [Google Scholar] [CrossRef]
- Yang, Y.; Sun, Z.; Ren, T.; Lei, W. Differential Expression of lncRNA CASC2 in the Serum of Childhood Asthma and Its Role in Airway Smooth Muscle Cells Proliferation and Migration. J. Asthma Allergy 2022, 15, 197–207. [Google Scholar] [CrossRef]
- Yu, H.; Qi, N.; Zhou, Q. LncRNA H19 Inhibits Proliferation and Migration of Airway Smooth Muscle Cells Induced by PDGF-BB Through miR-21/PTEN/Akt Axis. J. Asthma Allergy 2021, 14, 71–80. [Google Scholar] [CrossRef]
- García-Sánchez, A.; Estravís, M.; Martin, M.J.; Pérez-Pazos, J.; Martín-García, C.; Gil-Melcón, M.; Ramos-González, J.; Eguiluz-Gracia, I.; Triviño, J.C.; Isidoro-García, M.; et al. PTGDR2 Expression in Peripheral Blood as a Potential Biomarker in Adult Patients with Asthma. J. Pers. Med. 2021, 11, 827. [Google Scholar] [CrossRef] [PubMed]
- Czech, W.; Krutmann, J.; Schopf, E.; Kapp, A. Serum eosinophil cationic protein (ECP) is a sensitive measure for disease activity in atopic dermatitis. Br. J. Dermatol. 1992, 126, 351–355. [Google Scholar] [CrossRef] [PubMed]
- Kägi, M.; Joller-Jemelka, H.; Wüthrich, B. Correlation of Eosinophils, Eosinophil Cationic Protein and Soluble lnterleukin-2 Receptor with the Clinical Activity of Atopic Dermatitis. Dermatology 1992, 185, 88–92. [Google Scholar] [CrossRef] [PubMed]
- Kim, H.S.; Kim, J.H.; Seo, Y.M.; Chun, Y.H.; Yoon, J.-S.; Kim, H.H.; Lee, J.S.; Kim, J.T. Eosinophil-derived neurotoxin as a biomarker for disease severity and relapse in recalcitrant atopic dermatitis. Ann. Allergy Asthma Immunol. 2017, 119, 441–445. [Google Scholar] [CrossRef]
- Murat-Susić, S.; Lipozencić, J.; Zizić, V.; Husar, K.; Marinović, B. Serum eosinophil cationic protein in children with atopic dermatitis. Int. J. Dermatol. 2006, 45, 1156–1160. [Google Scholar] [CrossRef]
- Selnes, A.; Dotterud, L. No association between serum eosinophil cationic protein and atopic dermatitis or allergic rhinitis in an unselected population of children. J. Eur. Acad. Dermatol. Venereol. 2005, 19, 61–65. [Google Scholar] [CrossRef]
- Jung, K.; Linse, F.; Heller, R.; Moths, C.; Goebel, R.; Neumann, C. Adhesion Molecules in Atopic Dermatitis: VCAM-1 and ICAM-1 Expression Is Increased in Healthy-Appearing Skin. Allergy 1996, 51, 452–460. [Google Scholar] [CrossRef]
- Kou, K.; Okawa, T.; Yamaguchi, Y.; Ono, J.; Inoue, Y.; Kohno, M.; Matsukura, S.; Kambara, T.; Ohta, S.; Izuhara, K.; et al. Periostin levels correlate with disease severity and chronicity in patients with atopic dermatitis. Br. J. Dermatol. 2014, 171, 283–291. [Google Scholar] [CrossRef] [PubMed]
- Ariëns, L.F.M.; van der Schaft, J.; Bakker, D.S.; Balak, D.; Romeijn, M.L.E.; Kouwenhoven, T.; Kamsteeg, M.; Giovannone, B.; Drylewicz, J.; van Amerongen, C.C.A.; et al. Dupilumab is very effective in a large cohort of difficult-to-treat adult atopic dermatitis patients: First clinical and biomarker results from the BioDay registry. Allergy 2019, 75, 116–126. [Google Scholar] [CrossRef] [PubMed]
- Brunner, P.M.; He, H.; Pavel, A.B.; Czarnowicki, T.; Lefferdink, R.; Erickson, T.; Canter, T.; Puar, N.; Rangel, S.M.; Malik, K.; et al. The blood proteomic signature of early-onset pediatric atopic dermatitis shows systemic inflammation and is distinct from adult long-standing disease. J. Am. Acad. Dermatol. 2019, 81, 510–519. [Google Scholar] [CrossRef]
- Kakinuma, T.; Nakamura, K.; Wakugawa, M.; Mitsui, H.; Tada, Y.; Saeki, H.; Torii, H.; Asahina, A.; Onai, N.; Matsushima, K.; et al. Thymus and activation-regulated chemokine in atopic dermatitis: Serum thymus and activation-regulated chemokine level is closely related with disease activity. J. Allergy Clin. Immunol. 2001, 107, 535–541. [Google Scholar] [CrossRef]
- Kagami, S.; Kakinuma, T.; Saeki, H.; Tsunemi, Y.; Fujita, H.; Nakamura, K.; Takekoshi, T.; Kishimoto, M.; Mitsui, H.; Torii, H.; et al. Significant elevation of serum levels of eotaxin-3/CCL26, but not of eotaxin-2/CCL24, in patients with atopic dermatitis: Serum eotaxin-3/CCL26 levels reflect the disease activity of atopic dermatitis. Clin. Exp. Immunol. 2003, 134, 309–313. [Google Scholar] [CrossRef]
- Wen, H.-C.; Czarnowicki, T.; Noda, S.; Malik, K.; Pavel, A.B.; Nakajima, S.; Honda, T.; Shin, J.U.; Lee, H.; Chou, M.; et al. Serum from Asian patients with atopic dermatitis is characterized by TH2/TH22 activation, which is highly correlated with nonlesional skin measures. J. Allergy Clin. Immunol. 2018, 142, 324–328.e11. [Google Scholar] [CrossRef] [PubMed]
- McAleer, M.; Jakasa, I.; Hurault, G.; Sarvari, P.; McLean, W.; Tanaka, R.; Kezic, S.; Irvine, A. Systemic and stratum corneum biomarkers of severity in infant atopic dermatitis include markers of innate and T helper cell-related immunity and angiogenesis. Br. J. Dermatol. 2018, 180, 586–596. [Google Scholar] [CrossRef] [PubMed]
- Hon, K.L.; Ching, G.K.; Ng, P.C.; Leung, T.F. Exploring CCL18, eczema severity and atopy. Pediatr. Allergy Immunol. 2011, 22, 704–707. [Google Scholar] [CrossRef] [PubMed]
- Machura, E.; Rusek-Zychma, M.; Jachimowicz, M.; Wrzask, M.; Mazur, B.; Kasperska-Zajac, A. Serum TARC and CTACK concentrations in children with atopic dermatitis, allergic asthma, and urticaria. Pediatr. Allergy Immunol. 2011, 23, 278–284. [Google Scholar] [CrossRef]
- He, H.; Li, R.; Choi, S.; Zhou, L.; Pavel, A.; Estrada, Y.D.; Krueger, J.G.; Guttman-Yassky, E. Increased cardiovascular and atherosclerosis markers in blood of older patients with atopic dermatitis. Ann. Allergy Asthma Immunol. 2019, 124, 70–78. [Google Scholar] [CrossRef]
- Ungar, B.; Garcet, S.; Gonzalez, J.; Dhingra, N.; da Rosa, J.C.; Shemer, A.; Krueger, J.G.; Suarez-Farinas, M.; Guttman-Yassky, E. An Integrated Model of Atopic Dermatitis Biomarkers Highlights the Systemic Nature of the Disease. J. Investig. Dermatol. 2016, 137, 603–613. [Google Scholar] [CrossRef]
- Kou, K.; Aihara, M.; Matsunaga, T.; Chen, H.; Taguri, M.; Morita, S.; Fujita, H.; Yamaguchi, Y.; Kambara, T.; Ikezawa, Z. Association of serum interleukin-18 and other biomarkers with disease severity in adults with atopic dermatitis. Arch. Dermatol. Res. 2011, 304, 305–312. [Google Scholar] [CrossRef]
- Ahrens, B.; Schulz, G.; Bellach, J.; Niggemann, B.; Beyer, K. Chemokine levels in serum of children with atopic dermatitis with regard to severity and sensitization status. Pediatr. Allergy Immunol. 2015, 26, 634–640. [Google Scholar] [CrossRef] [PubMed]
- Nakazato, J.; Kishida, M.; Kuroiwa, R.; Fujiwara, J.; Shimoda, M.; Shinomiya, N. Serum levels of Th2 chemokines, CCL17, CCL22, and CCL27, were the important markers of severity in infantile atopic dermatitis. Pediatr. Allergy Immunol. 2008, 19, 605–613. [Google Scholar] [CrossRef]
- Song, T.W.; Sohn, M.H.; Kim, E.S.; Kim, K.W.; Kim, K.-E. Increased serum thymus and activation-regulated chemokine and cutaneous T cell-attracting chemokine levels in children with atopic dermatitis. Clin. Exp. Allergy 2006, 36, 346–351. [Google Scholar] [CrossRef] [PubMed]
- Zedan, K. Immunoglobulin E, Interleukin-18 and Interleukin-12 in Patients with Atopic Dermatitis: Correlation with Disease Activity. J. Clin. Diagn. Res. 2015, 9, WC01–WC05. [Google Scholar] [CrossRef] [PubMed]
- Katoh, N.; Hirano, S.; Suehiro, M.; Ikenaga, K.; Yamashita, T.; Sugawara, N.; Yasuno, H. Soluble CD30 is more relevant to disease activity of atopic dermatitis than soluble CD26. Clin. Exp. Immunol. 2000, 121, 187–192. [Google Scholar] [CrossRef]
- Bock, O.; Kreiselmeyer, I.; Mrowietz, U. Expression of dipeptidyl-peptidase IV (CD26) on CD8+ T cells is significantly decreased in patients with psoriasis vulgaris and atopic dermatitis. Exp. Dermatol. 2001, 10, 414–419. [Google Scholar] [CrossRef]
- Miyagaki, T.; Sugaya, M.; Suga, H.; Morimura, S.; Kamata, M.; Ohmatsu, H.; Fujita, H.; Asano, Y.; Tada, Y.; Kadono, T.; et al. Serum soluble CD26 levels: Diagnostic efficiency for atopic dermatitis, cutaneous T-cell lymphoma and psoriasis in combination with serum thymus and activation-regulated chemokine levels. J. Eur. Acad. Dermatol. Venereol. 2011, 27, 19–24. [Google Scholar] [CrossRef]
- Lv, Y.; Qi, R.; Xu, J.; Di, Z.; Zheng, H.; Huo, W.; Zhang, L.; Chen, H.; Gao, X. Profiling of Serum and Urinary MicroRNAs in Children with Atopic Dermatitis. PLoS ONE 2014, 9, e115448. [Google Scholar] [CrossRef]
- Vekaria, A.S.; Brunner, P.M.; Aleisa, A.I.; Bonomo, L.; Lebwohl, M.G.; Israel, A.; Guttman-Yassky, E. Moderate-to-severe atopic dermatitis patients show increases in serum C-reactive protein levels, correlating with skin disease activity. F1000Research 2017, 6, 1712. [Google Scholar] [CrossRef]
- Morishima, Y.; Kawashima, H.; Takekuma, K.; Hoshika, A. Changes in serum lactate dehydrogenase activity in children with atopic dermatitis. Pediatr. Int. 2010, 52, 171–174. [Google Scholar] [CrossRef]
- Mancuso, P. The role of adipokines in chronic inflammation. ImmunoTargets Ther. 2016, 5, 47–56. [Google Scholar] [CrossRef]
- Jaworek, A.K.; Szepietowski, J.C.; Szafraniec, K.; Jaworek, M.; Hałubiec, P.; Wojas-Pelc, A.; Pokorski, M. Adipokines as Biomarkers of Atopic Dermatitis in Adults. J. Clin. Med. 2020, 9, 2858. [Google Scholar] [CrossRef]
- Salomon, J.; Matusiak, Ł.; Nowicka-Suszko, D.; Szepietowski, J.C. Chitinase-3-Like Protein 1 (YKL-40) Reflects the Severity of Symptoms in Atopic Dermatitis. J. Immunol. Res. 2017, 2017, 5746031. [Google Scholar] [CrossRef] [PubMed]
- Izuhara, K.; Yamaguchi, Y.; Ohta, S.; Nunomura, S.; Nanri, Y.; Azuma, Y.; Nomura, N.; Noguchi, Y.; Aihara, M. Squamous Cell Carcinoma Antigen 2 (SCCA2, SERPINB4): An Emerging Biomarker for Skin Inflammatory Diseases. Int. J. Mol. Sci. 2018, 19, 1102. [Google Scholar] [CrossRef] [PubMed]
- Afghani, J.; Traidl-Hoffmann, C.; Schmitt-Kopplin, P.; Reiger, M.; Mueller, C. An Overview of the Latest Metabolomics Studies on Atopic Eczema with New Directions for Study. Int. J. Mol. Sci. 2022, 23, 8791. [Google Scholar] [CrossRef]
- Ilves, L.; Ottas, A.; Kaldvee, B.; Abram, K.; Soomets, U.; Zilmer, M.; Jaks, V.; Kingo, K. Metabolomic Differences between the Skin and Blood Sera of Atopic Dermatitis and Psoriasis. Int. J. Mol. Sci. 2022, 23, 13001. [Google Scholar] [CrossRef]
- Mihály, J.; Gamlieli, A.; Worm, M.; Rühl, R. Decreased retinoid concentration and retinoid signalling pathways in human atopic dermatitis. Exp. Dermatol. 2011, 20, 326–330. [Google Scholar] [CrossRef]
- Ottas, A.; Fishman, D.; Okas, T.-L.; Püssa, T.; Toomik, P.; Martson, A.; Kingo, K.; Soomets, U. Blood serum metabolome of atopic dermatitis: Altered energy cycle and the markers of systemic inflammation. PLoS ONE 2017, 12, e0188580. [Google Scholar] [CrossRef] [PubMed]
- Töröcsik, D.; Weise, C.; Gericke, J.; Szegedi, A.; Lucas, R.; Mihaly, J.; Worm, M.; Rühl, R. Transcriptomic and lipidomic profiling of eicosanoid/docosanoid signalling in affected and non-affected skin of human atopic dermatitis patients. Exp. Dermatol. 2018, 28, 177–189. [Google Scholar] [CrossRef] [PubMed]
- Chiu, C.; Lin, G.; Wang, C.; Hung, S.; Chung, W. Metabolomics reveals microbial-derived metabolites associated with immunoglobulin E responses in filaggrin -related atopic dermatitis. Pediatr. Allergy Immunol. 2021, 32, 1709–1717. [Google Scholar] [CrossRef]
- Huang, Y.; Chen, G.; Liu, X.; Shao, Y.; Gao, P.; Xin, C.; Cui, Z.; Zhao, X.; Xu, G. Serum Metabolomics Study and Eicosanoid Analysis of Childhood Atopic Dermatitis Based on Liquid Chromatography–Mass Spectrometry. J. Proteome Res. 2014, 13, 5715–5723. [Google Scholar] [CrossRef]
- Hotze, M.; Baurecht, H.; Rodríguez, E.; Chapman-Rothe, N.; Ollert, M.; Fölster-Holst, R.; Adamski, J.; Illig, T.; Ring, J.; Weidinger, S. Increased efficacy of omalizumab in atopic dermatitis patients with wild-type filaggrin status and higher serum levels of phosphatidylcholines. Allergy 2013, 69, 132–135. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L.; Wen, X.; Hou, Y.; Yang, Y.; Song, W.; Zeng, Y.; Sun, J. Integrated metabolomics and lipidomics study of patients with atopic dermatitis in response to dupilumab. Front. Immunol. 2022, 13, 1002536. [Google Scholar] [CrossRef]
- Lucas, R.; Szklenar, M.; Mihály, J.; Szegedi, A.; Töröcsik, D.; Rühl, R. Plasma Levels of Bioactive Vitamin D and A5 Ligands Positively Correlate with Clinical Atopic Dermatitis Markers. Dermatology 2022, 238, 1076–1083. [Google Scholar] [CrossRef] [PubMed]
- Schneider, M. MicroRNAs as novel players in skin development, homeostasis and disease. Br. J. Dermatol. 2011, 166, 22–28. [Google Scholar] [CrossRef]
- Bélanger, É.; Madore, A.-M.; Boucher-Lafleur, A.-M.; Simon, M.-M.; Kwan, T.; Pastinen, T.; Laprise, C. Eosinophil microRNAs Play a Regulatory Role in Allergic Diseases Included in the Atopic March. Int. J. Mol. Sci. 2020, 21, 9011. [Google Scholar] [CrossRef]
- Sonkoly, E.; Janson, P.; Majuri, M.-L.; Savinko, T.; Fyhrquist, N.; Eidsmo, L.; Xu, N.; Meisgen, F.; Wei, T.; Bradley, M.; et al. MiR-155 is overexpressed in patients with atopic dermatitis and modulates T-cell proliferative responses by targeting cytotoxic T lymphocyte–associated antigen 4. J. Allergy Clin. Immunol. 2010, 126, 581–589.e20. [Google Scholar] [CrossRef]
- Massimiliano, B.; Martina, A.; Ilaria, G.; Paola, C.; Paola, M.; Pietro, Q.; Francesco, S. Expression of miRNA 155, FOXP3 and ROR gamma, in children with moderate and severe atopic dermatitis. G. Ital. Dermatol. Venereol. 2020, 155. [Google Scholar] [CrossRef]
- Ma, L.; Xue, H.-B.; Wang, F.; Shu, C.-M.; Zhang, J.-H. MicroRNA-155 may be involved in the pathogenesis of atopic dermatitis by modulating the differentiation and function of T helper type 17 (Th17) cells. Clin. Exp. Immunol. 2015, 181, 142–149. [Google Scholar] [CrossRef]
- Koga, Y.; Jinnin, M.; Ichihara, A.; Fujisawa, A.; Moriya, C.; Sakai, K.; Fukushima, S.; Inoue, Y.; Ihn, H. Analysis of expression pattern of serum microRNA levels in patients with psoriasis. J. Dermatol. Sci. 2014, 74, 170–171. [Google Scholar] [CrossRef] [PubMed]
- Yan, F.; Meng, W.; Ye, S.; Zhang, X.; Mo, X.; Liu, J.; Chen, D.; Lin, Y. MicroRNA-146a as a potential regulator involved in the pathogenesis of atopic dermatitis. Mol. Med. Rep. 2019, 20, 4645–4653. [Google Scholar] [CrossRef]
- Gu, C.; Li, Y.; Wu, J.; Xu, J. IFN-γ-induced microRNA-29b up-regulation contributes tokeratinocyte apoptosis in atopic dermatitis through inhibiting Bcl2L2. Int. J. Clin. Exp. Pathol. 2017, 10, 10117–10126. [Google Scholar] [PubMed]
- Chen, X.-F.; Zhang, L.-J.; Zhang, J.; Dou, X.; Shao, Y.; Jia, X.-J.; Zhang, W.; Yu, B. MiR-151a is involved in the pathogenesis of atopic dermatitis by regulating interleukin-12 receptor β2. Exp. Dermatol. 2017, 27, 427–432. [Google Scholar] [CrossRef]
- Meng, L.; Li, M.; Gao, Z.; Ren, H.; Chen, J.; Liu, X.; Cai, Q.; Jiang, L.; Ren, X.; Yu, Q.; et al. Possible role of hsa-miR-194-5p, via regulation of HS3ST2, in the pathogenesis of atopic dermatitis in children. Eur. J. Dermatol. 2019, 29, 603–613. [Google Scholar] [CrossRef] [PubMed]
- Blanchard, C.; Mingler, M.K.; McBride, M.; Putnam, P.E.; Collins, M.H.; Chang, G.; Stringer, K.; Abonia, J.P.; Molkentin, J.D.; Rothenberg, M.E. Periostin facilitates eosinophil tissue infiltration in allergic lung and esophageal responses. Mucosal Immunol. 2008, 1, 289–296. [Google Scholar] [CrossRef] [PubMed]
- Dellon, E.S.; Higgins, L.L.; Beitia, R.; Rusin, S.; Woosley, J.T.; Veerappan, R.; Selitsky, S.R.; Parker, J.S.; Genta, R.M.; Lash, R.H.; et al. Prospective assessment of serum periostin as a biomarker for diagnosis and monitoring of eosinophilic oesophagitis. Aliment. Pharmacol. Ther. 2016, 44, 189–197. [Google Scholar] [CrossRef] [PubMed]
- Dellon, E.S.; Lin, L.; Beitia, R.; Moran, T.P.; Qian, Y. Serum autoantibodies against epithelial cell adhesion molecules as disease biomarkers of eosinophilic esophagitis. Clin. Exp. Allergy 2017, 48, 343–346. [Google Scholar] [CrossRef] [PubMed]
- Dellon, E.S.; Rusin, S.; Gebhart, J.H.; Covey, S.; Higgins, L.L.; Beitia, R.; Speck, O.; Woodward, K.; Woosley, J.T.; Shaheen, N.J. Utility of a Noninvasive Serum Biomarker Panel for Diagnosis and Monitoring of Eosinophilic Esophagitis: A Prospective Study. Am. J. Gastroenterol. 2015, 110, 821–827. [Google Scholar] [CrossRef]
- Wright, B.L.; Ochkur, S.I.; Olson, N.S.; Shim, K.P.; Jacobsen, E.A.; Rank, M.A.; Dellon, E.S.; Lee, J.J. Normalized serum eosinophil peroxidase levels are inversely correlated with esophageal eosinophilia in eosinophilic esophagitis. Dis. Esophagus 2017, 31. [Google Scholar] [CrossRef] [PubMed]
- Min, S.B.; Nylund, C.M.; Baker, T.P.; Ally, M.; Reinhardt, B.; Chen, Y.-J.; Nazareno, L.; Moawad, F.J. Longitudinal Evaluation of Noninvasive Biomarkers for Eosinophilic Esophagitis. J. Clin. Gastroenterol. 2017, 51, 127–135. [Google Scholar] [CrossRef]
- Subbarao, G.; Rosenman, M.B.; Ohnuki, L.; Georgelas, A.; Davis, M.; Fitzgerald, J.F.; Molleston, J.P.; Croffie, J.M.; Pfefferkorn, M.D.; Corkins, M.R.; et al. Exploring Potential Noninvasive Biomarkers in Eosinophilic Esophagitis in Children. J. Craniofacial Surg. 2011, 53, 651–658. [Google Scholar] [CrossRef]
- Cengiz, C. Serum eosinophilic cationic protein is correlated with food impaction and endoscopic severity in eosinophilic esophagitis. Turk. J. Gastroenterol. 2019, 30, 345–349. [Google Scholar] [CrossRef]
- Wechsler, J.B.; Ackerman, S.J.; Chehade, M.; Amsden, K.; Riffle, M.E.; Wang, M.; Du, J.; Kleinjan, M.L.; Alumkal, P.; Gray, E.; et al. Noninvasive biomarkers identify eosinophilic esophagitis: A prospective longitudinal study in children. Allergy 2021, 76, 3755–3765. [Google Scholar] [CrossRef]
- Straumann, A.; Conus, S.; Grzonka, P.; Kita, H.; Kephart, G.; Bussmann, C.; Beglinger, C.; Smith, D.A.; Patel, J.; Byrne, M.; et al. Anti-interleukin-5 antibody treatment (mepolizumab) in active eosinophilic oesophagitis: A randomised, placebo-controlled, double-blind trial. Gut 2009, 59, 21–30. [Google Scholar] [CrossRef] [PubMed]
- Schlag, C.; Pfefferkorn, S.; Brockow, K.; Haller, B.; Slotta-Huspenia, J.; Schulz, S.; von Werder, A.; Ring, J.; Schmid, R.M.; Bajbouj, M. Serum Eosinophil Cationic Protein is Superior to Mast Cell Tryptase as Marker for Response to Topical Corticosteroid Therapy in Eosinophilic Esophagitis. J. Clin. Gastroenterol. 2014, 48, 600–606. [Google Scholar] [CrossRef] [PubMed]
- Witek, J.D.; Cerdà, V.J.; Gil Guillén, V.; Clar, J.B.D.; Pacheco, R.R. Assessing eosinophilic cationic protein as a biomarker for monitoring patients with eosinophilic esophagitis treated with specific exclusion diets. World Allergy Organ. J. 2017, 10, 12. [Google Scholar] [CrossRef] [PubMed]
- Rodríguez-Sánchez, J.; Gómez-Torrijos, E.; Santa-Belda, E.D.-L.; López-Viedma, B.; Martín-Dávila, F.; Pilkington-Woll, J.P.; Donado-Palencia, P.; Sánchez-Miranda, P.; Olmedo-Camacho, J. Effectiveness of serological markers of eosinophil activity in monitoring eosinophilic esophagitis. Rev. Esp. Enferm. Dig. 2013, 105, 462–468. [Google Scholar] [CrossRef]
- Sarbinowska, J.; Wiatrak, B.; Waśko-Czopnik, D. Searching for Noninvasive Predictors of the Diagnosis and Monitoring of Eosinophilic Esophagitis—The Importance of Biomarkers of the Inflammatory Reaction Involving Eosinophils. Biomolecules 2021, 11, 890. [Google Scholar] [CrossRef]
- Blanchard, C.; Stucke, E.M.; Rodriguez-Jimenez, B.; Burwinkel, K.; Collins, M.H.; Ahrens, A.; Alexander, E.S.; Butz, B.K.B.; Jameson, S.C.; Kaul, A.; et al. A striking local esophageal cytokine expression profile in eosinophilic esophagitis. J. Allergy Clin. Immunol. 2011, 127, 208–217.e7. [Google Scholar] [CrossRef]
- Kouro, T.; Takatsu, K. IL-5- and eosinophil-mediated inflammation: From discovery to therapy. Int. Immunol. 2009, 21, 1303–1309. [Google Scholar] [CrossRef]
- Kinoshita, Y.; Furuta, K.; Ishimura, N.; Ishihara, S. Elevated Plasma Cytokines in Japanese Patients with Eosinophilic Esophagitis and Gastroenteritis. Digestion 2012, 86, 238–243. [Google Scholar] [CrossRef]
- Lu, S.; Herzlinger, M.; Cao, W.; Noble, L.; Yang, D.; Shapiro, J.; Kurtis, J.; LeLeiko, N.; Resnick, M. Utility of 15(S)-HETE as a Serological Marker for Eosinophilic Esophagitis. Sci. Rep. 2018, 8, 14498. [Google Scholar] [CrossRef]
- Bhattacharya, B.; Carlsten, J.; Sabo, E.; Kethu, S.; Meitner, P.; Tavares, R.; Jakate, S.; Mangray, S.; Aswad, B.; Resnick, M.B. Increased expression of eotaxin-3 distinguishes between eosinophilic esophagitis and gastroesophageal reflux disease. Hum. Pathol. 2007, 38, 1744–1753. [Google Scholar] [CrossRef]
- Wen, T.; Stucke, E.M.; Grotjan, T.M.; Kemme, K.A.; Abonia, J.P.; Putnam, P.E.; Franciosi, J.P.; Garza, J.M.; Kaul, A.; King, E.C.; et al. Molecular Diagnosis of Eosinophilic Esophagitis by Gene Expression Profiling. Gastroenterology 2013, 145, 1289–1299. [Google Scholar] [CrossRef] [PubMed]
- Matoso, A.; Mukkada, V.A.; Lu, S.; Monahan, R.; Cleveland, K.; Noble, L.; Mangray, S.; Resnick, M.B. Expression microarray analysis identifies novel epithelial-derived protein markers in eosinophilic esophagitis. Mod. Pathol. 2013, 26, 665–676. [Google Scholar] [CrossRef] [PubMed]
- Moye, L.M.; Liu, Y.; Coarfa, C.; Putluri, N.; Rhoads, J.M. Plasma Urea Cycle Metabolites May Be Useful Biomarkers in Children with Eosinophilic Esophagitis. Front. Pediatr. 2019, 6, 423. [Google Scholar] [CrossRef]
- Lu, T.; Sherrill, J.D.; Wen, T.; Plassard, A.J.; Besse, J.A.; Abonia, J.; Franciosi, J.P.; Putnam, P.E.; Eby, M.; Martin, L.; et al. MicroRNA signature in patients with eosinophilic esophagitis, reversibility with glucocorticoids, and assessment as disease biomarkers. J. Allergy Clin. Immunol. 2012, 129, 1064–1075.e9. [Google Scholar] [CrossRef] [PubMed]
- Cañas, J.A.; Tabares, A.; Barbero, C.; García-Sánchez, D.; Sastre, B.; Rodrigo-Muñoz, J.M.; Mahíllo-Fernández, I.; Rayo, A.; Borrell, B.; Cilleruelo, M.L.; et al. Proton-pump Inhibitor Response Prediction Using Esophageal microRNAs in Children with Eosinophilic Esophagitis. J. Craniofacial Surg. 2020, 71, 755–763. [Google Scholar] [CrossRef]
- Venkateshaiah, S.U.; Rayapudi, M.; Kandikattu, H.K.; Yadavalli, C.S.; Mishra, A. Blood mRNA levels of T cells and IgE receptors are novel non-invasive biomarkers for eosinophilic esophagitis (EoE). Clin. Immunol. 2021, 227, 108752. [Google Scholar] [CrossRef]
- Venkateshaiah, S.U.; Manohar, M.; Verma, A.K.; Blecker, U.; Mishra, A. Possible Noninvasive Biomarker of Eosinophilic Esophagitis: Clinical and Experimental Evidence. Case Rep. Gastroenterol. 2016, 10, 685–692. [Google Scholar] [CrossRef]
- Venkateshaiah, S.U.; Kandikattu, H.K.; Yadavalli, C.S.; Mishra, A. Eosinophils and T Cell Surface Molecule Transcript Levels in the Blood Differentiate Eosinophilic Esophagitis (EoE) from GERD. Int. J. Basic Clin. Immunol. 2021, 4, 1–8. [Google Scholar]
- Zhu, X.; Wang, M.; Mavi, P.; Rayapudi, M.; Pandey, A.K.; Kaul, A.; Putnam, P.E.; Rothenberg, M.E.; Mishra, A. Interleukin-15 Expression Is Increased in Human Eosinophilic Esophagitis and Mediates Pathogenesis in Mice. Gastroenterology 2010, 139, 182–193.e7. [Google Scholar] [CrossRef]
- Bardou, P.; Mariette, J.; Escudié, F.; Djemiel, C.; Klopp, C. jvenn: An interactive Venn diagram viewer. BMC Bioinform. 2014, 15, 293. [Google Scholar] [CrossRef]
- György, B.; Szabó, T.G.; Pásztói, M.; Pál, Z.; Misják, P.; Aradi, B.; László, V.; Pállinger, É.; Pap, E.; Kittel, Á.; et al. Membrane vesicles, current state-of-the-art: Emerging role of extracellular vesicles. Cell. Mol. Life Sci. 2011, 68, 2667–2688. [Google Scholar] [CrossRef] [PubMed]
- van Niel, G.; Carter, D.R.F.; Clayton, A.; Lambert, D.W.; Raposo, G.; Vader, P. Challenges and directions in studying cell–cell communication by extracellular vesicles. Nat. Rev. Mol. Cell Biol. 2022, 23, 369–382. [Google Scholar] [CrossRef]
- Rossaint, J.; Kühne, K.; Skupski, J.; van Aken, H.; Looney, M.R.; Hidalgo, A.; Zarbock, A. Directed transport of neutrophil-derived extracellular vesicles enables platelet-mediated innate immune response. Nat. Commun. 2016, 7, 13464. [Google Scholar] [CrossRef] [PubMed]
- Fitzgerald, W.; Freeman, M.L.; Lederman, M.M.; Vasilieva, E.; Romero, R.; Margolis, L. A System of Cytokines Encapsulated in ExtraCellular Vesicles. Sci. Rep. 2018, 8, 8973. [Google Scholar] [CrossRef] [PubMed]
- Picca, A.; Guerra, F.; Calvani, R.; Coelho-Júnior, H.J.; Landi, F.; Bernabei, R.; Romano, R.; Bucci, C.; Marzetti, E. Extracellular Vesicles and Damage-Associated Molecular Patterns: A Pandora’s Box in Health and Disease. Front. Immunol. 2020, 11, 601740. [Google Scholar] [CrossRef] [PubMed]
- Wozniak, A.L.; Adams, A.; King, K.E.; Dunn, W.; Christenson, L.K.; Hung, W.-T.; Weinman, S.A. The RNA binding protein FMR1 controls selective exosomal miRNA cargo loading during inflammation. J. Cell Biol. 2020, 219, e201912074. [Google Scholar] [CrossRef] [PubMed]
- Fendl, B.; Weiss, R.; Eichhorn, T.; Linsberger, I.; Afonyushkin, T.; Puhm, F.; Binder, C.J.; Fischer, M.B.; Weber, V. Extracellular vesicles are associated with C-reactive protein in sepsis. Sci. Rep. 2021, 11, 6996. [Google Scholar] [CrossRef]
- Calvo, V.; Izquierdo, M. Inducible Polarized Secretion of Exosomes in T and B Lymphocytes. Int. J. Mol. Sci. 2020, 21, 2631. [Google Scholar] [CrossRef] [PubMed]
- Raposo, G.; Nijman, H.W.; Stoorvogel, W.; Liejendekker, R.; Harding, C.V.; Melief, C.J.; Geuze, H.J. B lymphocytes secrete antigen-presenting vesicles. J. Exp. Med. 1996, 183, 1161–1172. [Google Scholar] [CrossRef]
- Duarte, D.; Taveira-Gomes, T.; Sokhatska, O.; Palmares, C.; Costa, R.; Negrão, R.; Guimarães, J.T.; Delgado, L.; Soares, R.; Moreira, A. Increased circulating platelet microparticles as a potential biomarker in asthma. Allergy 2013, 68, 1073–1075. [Google Scholar] [CrossRef] [PubMed]
- Gao, J.; Xu, X.; Ying, Z.; Jiang, L.; Zhong, M.; Wang, A.; Chen, L.-C.; Lu, B.; Sun, Q. Post-Effect of Air Quality Improvement on Biomarkers for Systemic Inflammation and Microparticles in Asthma Patients After the 2008 Beijing Olympic Games: A Pilot Study. Inflammation 2017, 40, 1214–1224. [Google Scholar] [CrossRef] [PubMed]
- Zheng, R.; Du, M.; Tian, M.; Zhu, Z.; Wei, C.; Chu, H.; Gan, C.; Liang, J.; Xue, R.; Gao, F.; et al. Fine Particulate Matter Induces Childhood Asthma Attacks via Extracellular Vesicle-Packaged Let-7i-5p-Mediated Modulation of the MAPK Signaling Pathway. Adv. Sci. 2021, 9, e2102460. [Google Scholar] [CrossRef] [PubMed]
- Wagner, N.; Eberhardt, M.; Vera, J.; Cuomo, F.; Blume, K.; Galster, S.; Achenbach, S.; Laffert, B.; Kahlert, H.; Schuler, G.; et al. Plasma-derived extracellular vesicles discriminate type-1 allergy subjects from non-allergic controls. World Allergy Organ. J. 2021, 14, 100583. [Google Scholar] [CrossRef] [PubMed]
- Zhao, M.; Li, Y.-P.; Geng, X.-R.; Zhao, M.; Ma, S.-B.; Yang, Y.-H.; Deng, Z.-H.; Luo, L.-M.; Pan, X.-Q. Expression Level of MiRNA-126 in Serum Exosomes of Allergic Asthma Patients and Lung Tissues of Asthmatic Mice. Curr. Drug Metab. 2019, 20, 799–803. [Google Scholar] [CrossRef] [PubMed]
- Zhao, M.; Juanjuan, L.; Weijia, F.; Jing, X.; Qiuhua, H.; Hua, Z.; Fuhe, L.; Hao, P. Expression Levels of MicroRNA-125b in Serum Exosomes of Patients with Asthma of Different Severity and its Diagnostic Significance. Curr. Drug Metab. 2019, 20, 781–784. [Google Scholar] [CrossRef]
- Atashbasteh, M.; Mortaz, E.; Mahdaviani, S.A.; Jamaati, H.; Allameh, A. Expression levels of plasma exosomal miR-124, miR-125b, miR-133b, miR-130a and miR-125b-1-3p in severe asthma patients and normal individuals with emphasis on inflammatory factors. Allergy Asthma Clin. Immunol. 2021, 17, 51. [Google Scholar] [CrossRef]
- Hir, S.R.; Alizadeh, Z.; Mazinani, M.; Rad, M.M.; Fazlollahi, M.R.; Kazemnejad, A.; Hosseini, A.Z.; Moin, M. Exosomal MicroRNAs as Biomarkers in Allergic Asthma. Iran. J. Allergy Asthma Immunol. 2021, 20, 160–168. [Google Scholar] [CrossRef]
- Bahmer, T.; Krauss-Etschmann, S.; Buschmann, D.; Behrends, J.; Watz, H.; Kirsten, A.; Pedersen, F.; Waschki, B.; Fuchs, O.; Pfaffl, M.W.; et al. RNA-seq–based profiling of extracellular vesicles in plasma reveals a potential role of miR-122-5p in asthma. Allergy 2020, 76, 366–371. [Google Scholar] [CrossRef]
- Suzuki, M.; Cole, J.J.; Konno, S.; Makita, H.; Kimura, H.; Nishimura, M.; Maciewicz, R.A. Large-scale plasma proteomics can reveal distinct endotypes in chronic obstructive pulmonary disease and severe asthma. Clin. Transl. Allergy 2021, 11, e12091. [Google Scholar] [CrossRef]
- Fölster-Holst, R. The role of the skin microbiome in atopic dermatitis—Correlations and consequences. J. Dtsch. Dermatol. Ges. 2022, 20, 571–577. [Google Scholar] [CrossRef]
- Kulp, A.; Kuehn, M.J. Biological Functions and Biogenesis of Secreted Bacterial Outer Membrane Vesicles. Annu. Rev. Microbiol. 2010, 64, 163–184. [Google Scholar] [CrossRef] [PubMed]
- Kim, M.-H.; Choi, S.J.; Choi, H.-I.; Choi, J.-P.; Park, H.-K.; Kim, E.K.; Moon, B.S.; Min, T.-K.; Rho, M.; Cho, Y.-J.; et al. Lactobacillus plantarum-derived Extracellular Vesicles Protect Atopic Dermatitis Induced by Staphylococcus aureus-derived Extracellular Vesicles. Allergy Asthma Immunol. Res. 2018, 10, 516–532. [Google Scholar] [CrossRef] [PubMed]
- Yang, J.; McDowell, A.; Seo, H.; Kim, S.; Min, T.K.; Jee, Y.-K.; Choi, Y.; Park, H.-S.; Pyun, B.Y.; Kim, Y.-K. Diagnostic Models for Atopic Dermatitis Based on Serum Microbial Extracellular Vesicle Metagenomic Analysis: A Pilot Study. Allergy Asthma Immunol. Res. 2020, 12, 792–805. [Google Scholar] [CrossRef] [PubMed]
- Oba, R.; Isomura, M.; Igarashi, A.; Nagata, K. Circulating CD3+HLA-DR+Extracellular Vesicles as a Marker for Th1/Tc1-Type Immune Responses. J. Immunol. Res. 2019, 2019, 6720819. [Google Scholar] [CrossRef]
- Toyoshima, S.; Sakamoto-Sasaki, T.; Kurosawa, Y.; Hayama, K.; Matsuda, A.; Watanabe, Y.; Terui, T.; Gon, Y.; Matsumoto, K.; Okayama, Y. miR103a-3p in extracellular vesicles from FcεRI-aggregated human mast cells enhances IL-5 production by group 2 innate lymphoid cells. J. Allergy Clin. Immunol. 2021, 147, 1878–1891. [Google Scholar] [CrossRef] [PubMed]
- Pandey, K.K.; Madhry, D.; Kumar, Y.R.; Malvankar, S.; Sapra, L.; Srivastava, R.K.; Bhattacharyya, S.; Verma, B. Regulatory roles of tRNA-derived RNA fragments in human pathophysiology. Mol. Ther.-Nucleic Acids 2021, 26, 161–173. [Google Scholar] [CrossRef]
- Meng, L.; Jiang, L.; Chen, J.; Ren, H.; Gao, Z.; Wu, F.; Wen, Y.; Yang, L. Transfer RNA-derived fragment tRF-28-QSZ34KRQ590K in plasma exosomes may be a potential biomarker for atopic dermatitis in pediatric patients. Exp. Ther. Med. 2021, 21, 489. [Google Scholar] [CrossRef]
- Chang, C.-J.; Wang, H.-Q.; Zhang, J.; Zou, Y.; Zhang, Y.-H.; Chen, J.-W.; Chen, C.-B.; Chung, W.-H.; Ji, C. Distinct Proteomic Profiling of Plasma Extracellular Vesicles from Moderate-to-Severe Atopic Dermatitis Patients. Clin. Cosmet. Investig. Dermatol. 2021, 14, 1033–1043. [Google Scholar] [CrossRef] [PubMed]
- Cai, Y.; Sukhova, G.K.; Wong, H.K.; Xu, A.; Tergaonkar, V.; Vanhoutte, P.M.; Tang, E.H.C. Rap1 induces cytokine production in pro-inflammatory macrophages through NFκB signaling and is highly expressed in human atherosclerotic lesions. Cell Cycle 2015, 14, 3580–3592. [Google Scholar] [CrossRef]
- Chan, B.; Wong, W.; Lee, M.L.; Cho, W.C.; Yee, B.K.; Kwan, Y.W.; Tai, W.C.-S. Exosomes in Inflammation and Inflammatory Disease. Proteomics 2019, 19, e1800149. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Q.; Bansal, A. Role of Extracellular Vesicles in the Diagnosis and Pathogenesis of Barrett’s Esophagus: A Mini-Review. Dig. Dis. Sci. 2020, 66, 705–713. [Google Scholar] [CrossRef]
- Spechler, S.J. Gastroesophageal Reflux Disease and Eosinophilic Esophagitis. Gastroenterol. Hepatol. 2019, 15, 111–113. [Google Scholar]
- Uemura, R.; Murakami, Y.; Hashimoto, A.; Sawada, A.; Otani, K.; Taira, K.; Hosomi, S.; Nagami, Y.; Tanaka, F.; Kamata, N.; et al. Expression of Serum Exosomal and Esophageal MicroRNA in Rat Reflux Esophagitis. Int. J. Mol. Sci. 2017, 18, 1611. [Google Scholar] [CrossRef]
- Deretic, V.; Saitoh, T.; Akira, S. Autophagy in infection, inflammation and immunity. Nat. Rev. Immunol. 2013, 13, 722–737. [Google Scholar] [CrossRef]
- Whelan, K.A.; Merves, J.F.; Giroux, V.; Tanaka, K.; Guo, A.; Chandramouleeswaran, P.M.; Benitez, A.J.; Dods, K.; Que, J.; Masterson, J.C.; et al. Autophagy mediates epithelial cytoprotection in eosinophilic oesophagitis. Gut 2016, 66, 1197–1207. [Google Scholar] [CrossRef]
- Salimi, L.; Akbari, A.; Jabbari, N.; Mojarad, B.; Vahhabi, A.; Szafert, S.; Kalashani, S.A.; Soraya, H.; Nawaz, M.; Rezaie, J. Synergies in exosomes and autophagy pathways for cellular homeostasis and metastasis of tumor cells. Cell Biosci. 2020, 10, 64. [Google Scholar] [CrossRef]
- Leidal, A.M.; Debnath, J. Emerging roles for the autophagy machinery in extracellular vesicle biogenesis and secretion. FASEB BioAdvances 2021, 3, 377–386. [Google Scholar] [CrossRef] [PubMed]
- Solvik, T.A.; Nguyen, T.A.; Lin, Y.-H.T.; Marsh, T.; Huang, E.J.; Wiita, A.P.; Debnath, J.; Leidal, A.M. Secretory autophagy maintains proteostasis upon lysosome inhibition. J. Cell Biol. 2022, 221, e202110151. [Google Scholar] [CrossRef]
- Li, Y.; He, X.; Li, Q.; Lai, H.; Zhang, H.; Hu, Z.; Li, Y.; Huang, S. EV-origin: Enumerating the tissue-cellular origin of circulating extracellular vesicles using exLR profile. Comput. Struct. Biotechnol. J. 2020, 18, 2851–2859. [Google Scholar] [CrossRef]
- Akuthota, P.; Carmo, L.A.S.; Bonjour, K.; Murphy, R.O.; Silva, T.P.; Gamalier, J.P.; Capron, K.L.; Tigges, J.; Toxavidis, V.; Camacho, V.; et al. Extracellular Microvesicle Production by Human Eosinophils Activated by “Inflammatory” Stimuli. Front. Cell Dev. Biol. 2016, 4, 117. [Google Scholar] [CrossRef] [PubMed]
- Rodrigo-Muñoz, J.; Gil-Martínez, M.; Sastre, B.; del Pozo, V. Emerging Evidence for Pleiotropism of Eosinophils. Int. J. Mol. Sci. 2021, 22, 7075. [Google Scholar] [CrossRef]
- Grusell, E.N.; Dahlén, G.; Ruth, M.; Bergquist, H.; Bove, M. The cultivable bacterial flora of the esophagus in subjects with esophagitis. Scand. J. Gastroenterol. 2018, 53, 650–656. [Google Scholar] [CrossRef]
- Benitez, A.J.; Hoffmann, C.; Muir, A.B.; Dods, K.K.; Spergel, J.M.; Bushman, F.D.; Wang, M.-L. Inflammation-associated microbiota in pediatric eosinophilic esophagitis. Microbiome 2015, 3, 23. [Google Scholar] [CrossRef] [PubMed]
- Harris, J.; Fang, R.; Wagner, B.; Na Choe, H.; Kelly, C.; Schroeder, S.; Moore, W.; Stevens, M.J.; Yeckes, A.; Amsden, K.; et al. Esophageal Microbiome in Eosinophilic Esophagitis. PLoS ONE 2015, 10, e0128346. [Google Scholar] [CrossRef]
- Laserna-Mendieta, E.J.; FitzGerald, J.A.; Arias-Gonzalez, L.; Ollala, J.M.; Bernardo, D.; Claesson, M.J.; Lucendo, A.J. Esophageal microbiome in active eosinophilic esophagitis and changes induced by different therapies. Sci. Rep. 2021, 11, 7113. [Google Scholar] [CrossRef] [PubMed]
- Yang, J.; Shin, T.-S.; Kim, J.S.; Jee, Y.-K.; Kim, Y.-K. A new horizon of precision medicine: Combination of the microbiome and extracellular vesicles. Exp. Mol. Med. 2022, 54, 466–482. [Google Scholar] [CrossRef]
- Park, J.-Y.; Choi, J.; Lee, Y.; Lee, J.-E.; Lee, E.-H.; Kwon, H.-J.; Yang, J.; Jeong, B.-R.; Kim, Y.-K.; Han, P.-L. Metagenome Analysis of Bodily Microbiota in a Mouse Model of Alzheimer Disease Using Bacteria-derived Membrane Vesicles in Blood. Exp. Neurobiol. 2017, 26, 369–379. [Google Scholar] [CrossRef] [PubMed]
- Samra, M.; Nam, S.K.; Lim, D.H.; Kim, D.H.; Yang, J.; Kim, Y.-K.; Kim, J.H. Urine Bacteria-Derived Extracellular Vesicles and Allergic Airway Diseases in Children. Int. Arch. Allergy Immunol. 2018, 178, 150–158. [Google Scholar] [CrossRef]
- Kim, J.H.; Kim, S.H.; Lim, J.Y.; Kim, D.; Jeong, I.S.; Lee, D.K.; Jang, Y.J. Association between the sinus microbiota with eosinophilic inflammation and prognosis in chronic rhinosinusitis with nasal polyps. Exp. Mol. Med. 2020, 52, 978–987. [Google Scholar] [CrossRef]
- Hasan, A.; Hasan, L.K.; Schnabl, B.; Greytak, M.; Yadlapati, R. Microbiome of the Aerodigestive Tract in Health and Esophageal Disease. Dig. Dis. Sci. 2020, 66, 12–18. [Google Scholar] [CrossRef]
- Furuta, G.T.; Kagalwalla, A.F.; Lee, J.J.; Alumkal, P.; Maybruck, B.T.; Fillon, S.; Masterson, J.C.; Ochkur, S.; Protheroe, C.; Moore, W.; et al. The oesophageal string test: A novel, minimally invasive method measures mucosal inflammation in eosinophilic oesophagitis. Gut 2012, 62, 1395–1405. [Google Scholar] [CrossRef]
- Katzka, D.A.; Geno, D.M.; Ravi, A.; Smyrk, T.C.; Lao-Sirieix, P.; Miramedi, A.; Debiram, I.; O’Donovan, M.; Kita, H.; Kephart, G.M.; et al. Accuracy, Safety, and Tolerability of Tissue Collection by Cytosponge vs Endoscopy for Evaluation of Eosinophilic Esophagitis. Clin. Gastroenterol. Hepatol. 2014, 13, 77–83.e2. [Google Scholar] [CrossRef] [PubMed]
- Coumans, F.A.W.; Brisson, A.R.; Buzas, E.I.; Dignat-George, F.; Drees, E.E.E.; El-Andaloussi, S.; Emanueli, C.; Gasecka, A.; Hendrix, A.; Hill, A.F.; et al. Methodological Guidelines to Study Extracellular Vesicles. Circ. Res. 2017, 120, 1632–1648. [Google Scholar] [CrossRef] [PubMed]
- Théry, C.; Witwer, K.W.; Aikawa, E.; Alcaraz, M.J.; Anderson, J.D.; Andriantsitohaina, R.; Antoniou, A.; Arab, T.; Archer, F.; Atkin-Smith, G.K.; et al. Minimal Information for Studies of Extracellular Vesicles 2018 (MISEV2018): A Position State-ment of the International Society for Extracellular Vesicles and Update of the MISEV2014 Guidelines. J. Extracell. Vesicles 2018, 7, 1535750. [Google Scholar] [CrossRef] [PubMed]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).