Genome-Wide Characterization and Analysis of bHLH Transcription Factors Related to Anthocyanin Biosynthesis in Cinnamomum camphora (‘Gantong 1’)
Abstract
1. Introduction
2. Results
2.1. Identification and Physiochemical Characteristics of CcbHLHs
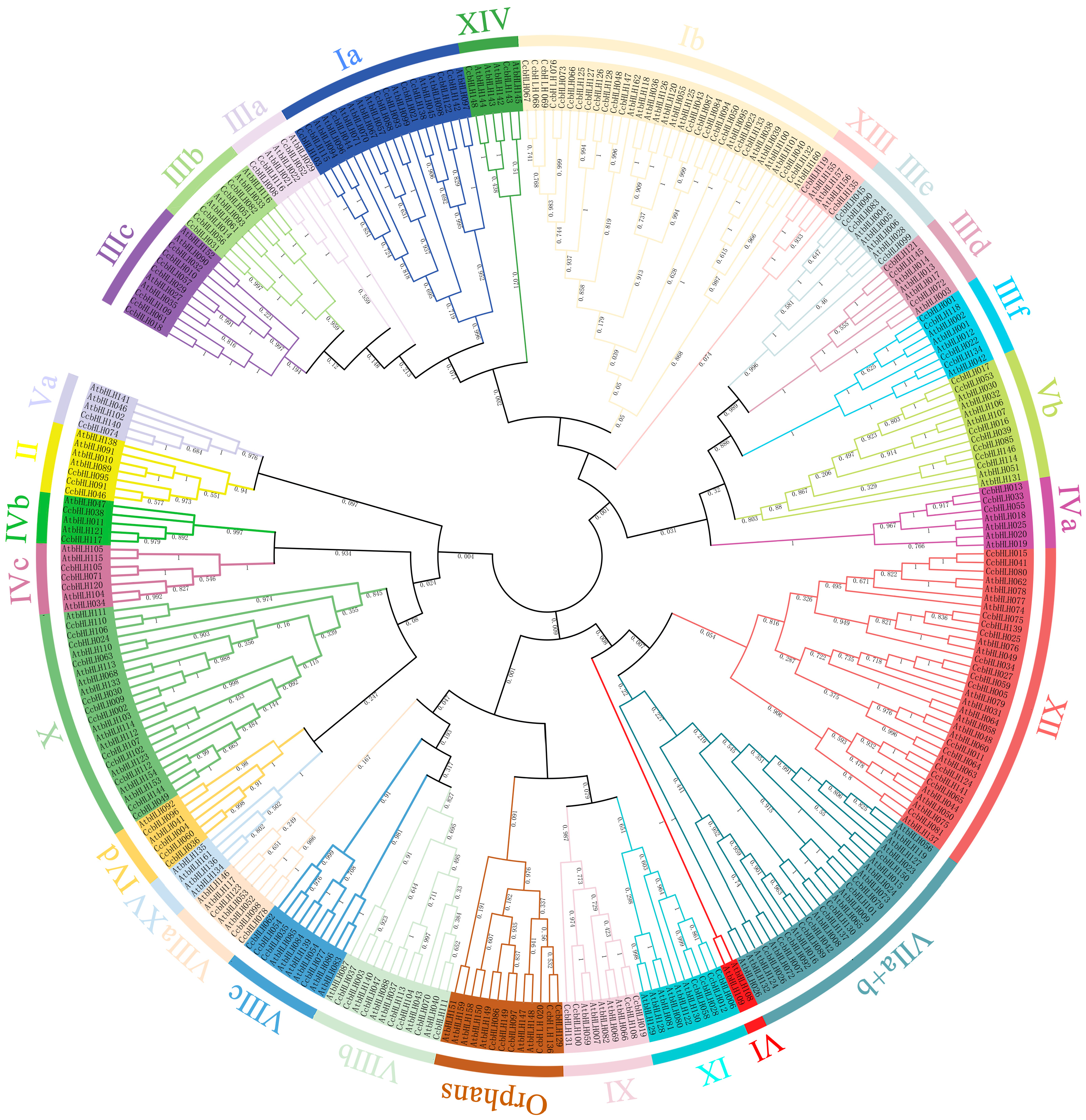
2.2. Phylogenetic Analysis of CcbHLHs
2.3. Analysis of Structure and Conserved Motifs of CcbHLHs
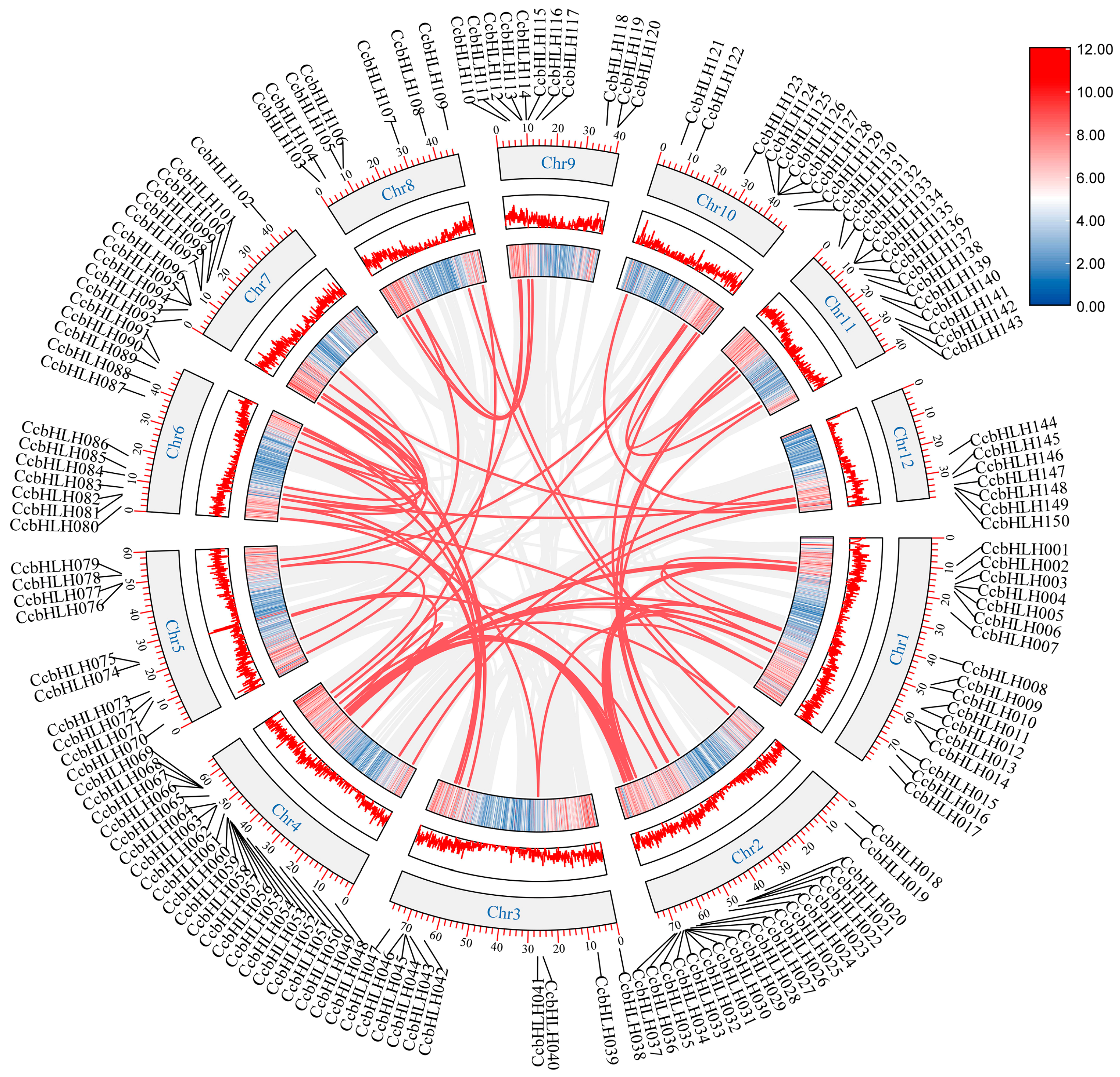
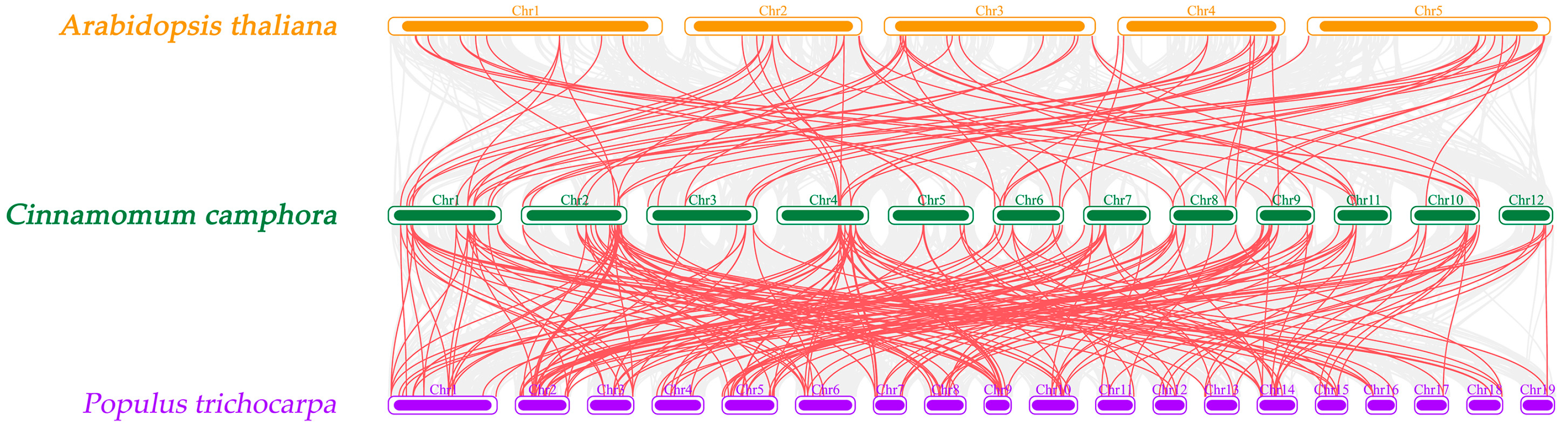
2.4. Chromosomal Localization and Collinearity Analysis of CcbHLHs
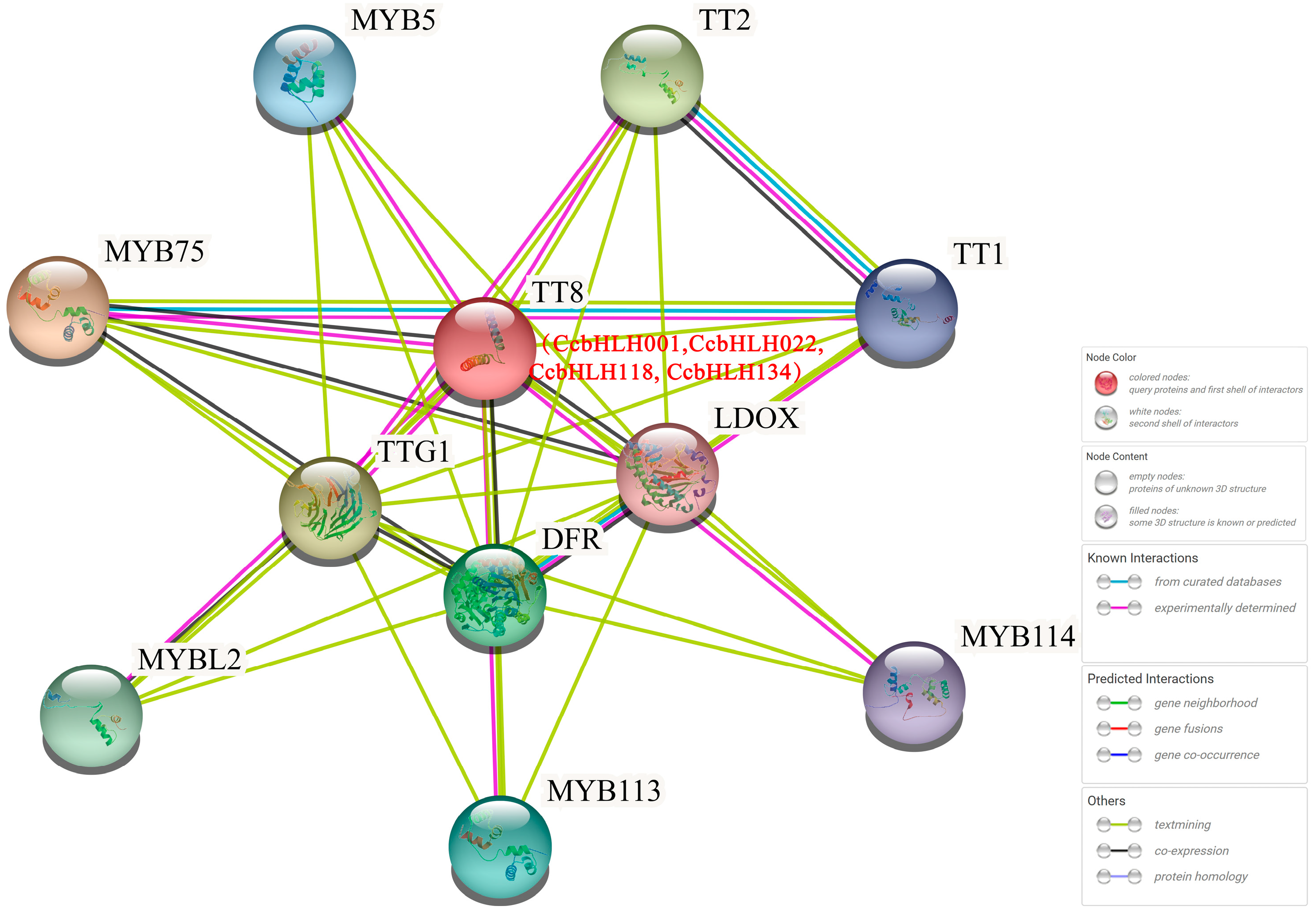
2.5. Predicted Protein–Protein Interaction Network of CcbHLHs
2.6. Expression Levels of CcbHLHs in Different Tissues
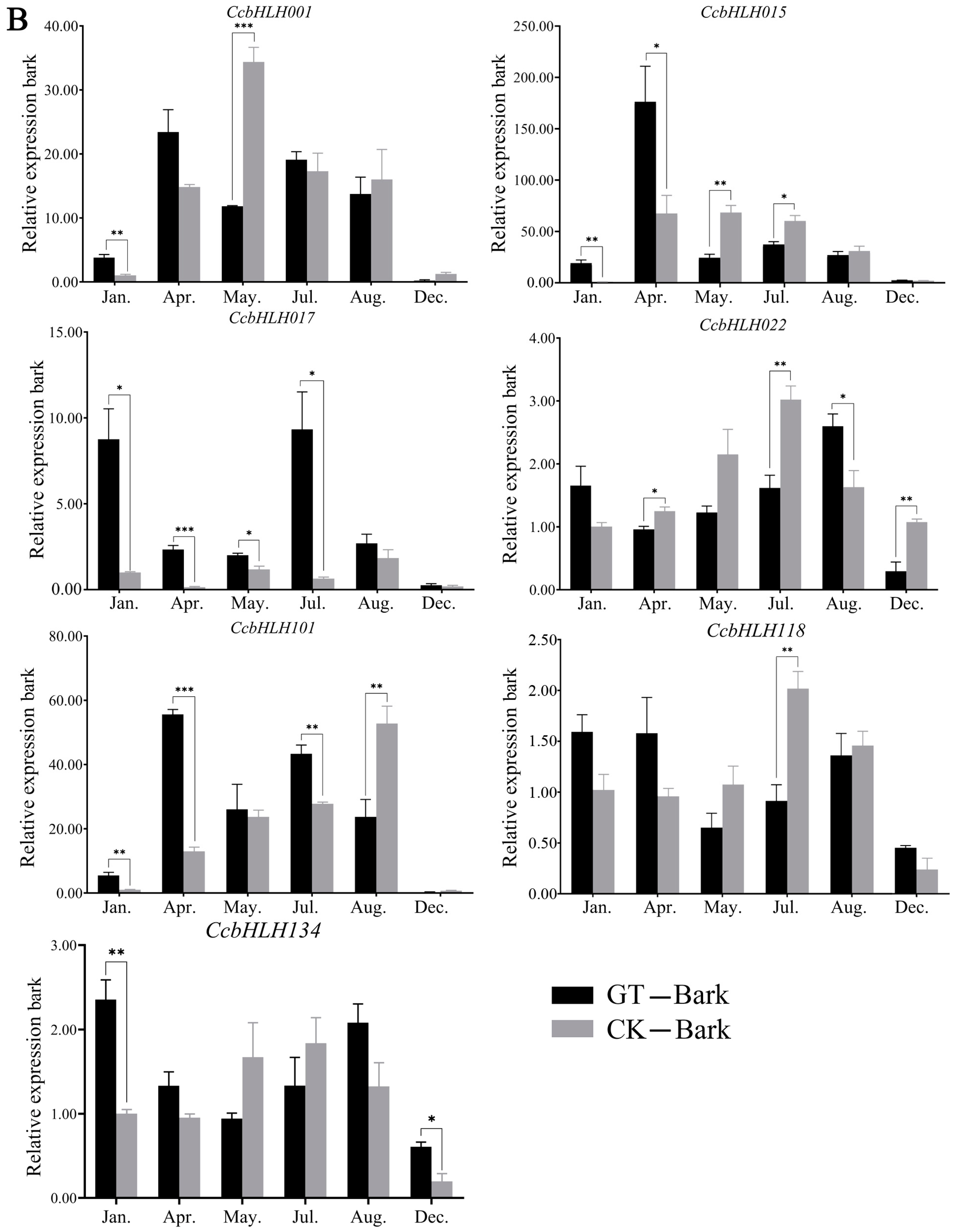
2.7. Analysis of Expression Levels of Candidate TFs for Anthocyanin Biosynthesis
3. Discussion
4. Materials and Methods
4.1. Plant Materials
4.2. Identification and Physicochemical Properties Analysis of CcbHLHs
4.3. Phylogenetic Analysis of CcbHLHs
4.4. Analysis of the Structure and Conserved Motifs in CcbHLHs
4.5. Chromosomal Localization and Collinearity Analysis of CcbHLHs
4.6. Protein–Protein Interaction Network Prediction
4.7. Expression Levels of CcbHLHs in Different Tissues
4.8. Analysis of Expression Levels of Candidate TFs Regulating Anthocyanin Biosynthesis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Zhong, Y.; Yang, A.; Li, Z.; Zhang, H.; Liu, L.; Wu, Z.; Li, Y.; Liu, T.; Xu, M.; Yu, F. Genetic Diversity and Population Genetic Structure of Cinnamomum camphora in South China Revealed by EST-SSR Markers. Forests 2019, 10, 1019. [Google Scholar] [CrossRef]
- Miller, R.; Owens, S.J.; Rørslett, B. Plants and colour: Flowers and pollination. Opt. Laser Technol. 2011, 43, 282–294. [Google Scholar] [CrossRef]
- Zhong, Y.; Chen, C.; Gong, X.; Luan, X.; Wu, Z.; Li, H.; Liu, Q.; Xu, M.; Yu, F. Transcriptome and metabolome analyses reveal a key role of the anthocyanin biosynthetic pathway cascade in the pigmentation of a Cinnamomum camphora red bark mutant (‘Gantong 1’). Ind. Crops Prod. 2022, 175, 114236. [Google Scholar] [CrossRef]
- Pang, Q.; Yu, W.; Ehsan, S.; Chen, X.; Hong, P.; Tariq, P.; Ren, Y.; Zhang, Y.; Dong, T.; Jia, H.; et al. Omic analysis of anthocyanin synthesis in wine grape leaves under low-temperature. Sci. Hortic. 2022, 307, 111483. [Google Scholar] [CrossRef]
- Lin, Y.; Fan, L.; He, J.; Wang, Z.; Yin, Y.; Cheng, Y.; Li, Z. Anthocyanins contribute to fruit defense against postharvest green mold. Postharvest Biol. Technol. 2021, 181, 111661. [Google Scholar] [CrossRef]
- Zhu, Z.; Wang, H.; Wang, Y.; Guan, S.; Wang, F.; Tang, J.; Zhang, R.; Xie, L.; Lu, Y. Characterization of the cis elements in the proximal promoter regions of the anthocyanin pathway genes reveals a common regulatory logic that governs pathway regulation. J. Exp. Bot. 2015, 66, 3775–3789. [Google Scholar] [CrossRef] [PubMed]
- Xu, W.; Dubos, C.; Lepiniec, L. Transcriptional control of flavonoid biosynthesis by MYB-bHLH-WDR complexes. Trends Plant Sci. 2015, 20, 1360–1385. [Google Scholar] [CrossRef]
- Davies, K.M.; Albert, N.W.; Schwinn, K.E. From landing lights to mimicry: The molecular regulation of flower colouration and mechanisms for pigmentation patterning. Funct. Plant Biol. 2012, 39, 619–638. [Google Scholar] [CrossRef] [PubMed]
- Perez-Diaz, J.R.; Perez-Diaz, J.; Madrid-Espinoza, J.; Gonzalez-Villanueva, E.; Moreno, Y.; Ruiz-Lara, S. New member of the R2R3-MYB transcription factors family in grapevine suppresses the anthocyanin accumulation in the flowers of transgenic tobacco. Plant Mol. Biol. 2016, 90, 63–76. [Google Scholar] [CrossRef] [PubMed]
- Duek, P.D.; Fankhauser, C. bHLH class transcription factors take centre stage in phytochrome signalling. Trends Plant Sci. 2005, 10, 51–54. [Google Scholar] [CrossRef] [PubMed]
- Feller, A.; Machemer, K.; Braun, E.L.; Grotewold, E. Evolutionary and comparative analysis of MYB and bHLH plant transcription factors. Plant J. 2011, 66, 94–116. [Google Scholar] [CrossRef] [PubMed]
- Hichri, I.; Barrieu, F.; Bogs, J.; Kappel, C.; Delrot, S.; Lauvergeat, V. Recent advances in the transcriptional regulation of the flavonoid biosynthetic pathway. J. Exp. Bot. 2011, 62, 2465–2483. [Google Scholar] [CrossRef] [PubMed]
- Murre, C.; Page-McCaw, P.S.; Baltimore, D. A new DNA binding and dimerization motif in immunoglobulin enhancer binding, daughterless, MyoD, and myc proteins. Cell 1989, 56, 777–783. [Google Scholar] [CrossRef]
- Sun, H.; Fan, H.J.; Ling, H.Q. Genome-wide identification and characterization of the bHLH gene family in tomato. BMC Genom. 2015, 16, 9. [Google Scholar] [CrossRef] [PubMed]
- Bailey, P.C.; Martin, C.; Toledo-Ortiz, G.; Quail, P.H.; Huq, E.; Heim, M.A.; Jakoby, M.; Werber, M.; Weisshaar, B. Update on the Basic Helix-Loop-Helix Transcription Factor Gene Family in Arabidopsis thaliana. Plant Cell 2003, 15, 2497–2501. [Google Scholar] [CrossRef]
- Carretero-Paulet, L.; Galstyan, A.; Roig-Villanova, I.; Martínez-García, J.F.; Bilbao-Castro, J.R.; Robertson, D.L. Genome-wide classification and evolutionary analysis of the bHLH family of transcription factors in Arabidopsis, poplar, rice, moss, and algae. Plant Physiol. 2010, 153, 1398–1412. [Google Scholar] [CrossRef]
- Dong, H.; Chen, Q.; Dai, Y.; Hu, W.; Zhang, S.; Huang, X. Genome-wide identification of PbrbHLH family genes, and expression analysis in response to drought and cold stresses in pear (Pyrus bretschneideri). BMC Plant Biol. 2021, 21, 86. [Google Scholar] [CrossRef]
- Li, M.; Sun, L.; Gu, H.; Cheng, D.; Guo, X.; Chen, R.; Wu, Z.; Jiang, J.; Fan, X.; Chen, J. Genome-wide characterization and analysis of bHLH transcription factors related to anthocyanin biosynthesis in spine grapes (Vitis davidii). Sci. Rep. 2021, 11, 6863. [Google Scholar] [CrossRef]
- Li, R.; Ahmad, B.; Hwarari, D.; Li, D.A.; Lu, Y.; Gao, M.; Chen, J.; Yang, L. Genomic Survey and Cold-Induced Expression Patterns of bHLH Transcription Factors in Liriodendron chinense (Hemsl) Sarg. Forests 2012, 13, 518. [Google Scholar] [CrossRef]
- Liu, R.; Wang, Y.; Tang, S.; Cai, J.; Liu, S.; Zheng, P.; Sun, B. Genome-wide identification of the tea plant bHLH transcription factor family and discovery of candidate regulators of trichome formation. Sci. Rep. 2021, 11, 10764. [Google Scholar] [CrossRef]
- Song, M.; Wang, H.; Wang, Z.; Huang, H.; Chen, S.; Ma, H. Genome-Wide Characterization and Analysis of bHLH Transcription Factors Related to Anthocyanin Biosynthesis in Fig (Ficus carica L.). Front. Plant Sci. 2021, 12, 730692. [Google Scholar] [CrossRef] [PubMed]
- Tian, Y.; Pu, X.; Yu, H.; Ji, A.; Gao, R.; Hu, Y.; Xu, Z.; Wang, H. Genome-Wide Characterization and Analysis of bHLH Transcription Factors Related to Crocin Biosynthesis in Gardenia jasminoides Ellis (Rubiaceae). BioMed Res. Int. 2020, 1, 1–11. [Google Scholar] [CrossRef]
- Zhao, W.; Liu, Y.; Li, L.; Meng, H.; Yang, Y.; Dong, Z.; Wang, L.; Wu, G. Genome-Wide Identification and Characterization of bHLH Transcription Factors Related to Anthocyanin Biosynthesis in Red Walnut (Juglans regia L.). Front. Genet. 2021, 12, 632509. [Google Scholar] [CrossRef] [PubMed]
- Zhao, M.; Li, J.; Zhu, L.; Chang, P.; Li, L.; Zhang, L. Identification and characterization of MYB-bHLH-WD40 regulatory complex members controlling anthocyanidin biosynthesis in blueberry fruits development. Genes 2019, 10, 496. [Google Scholar] [CrossRef] [PubMed]
- Baudry, A.; Heim, M.A.; Dubreucq, B.; Caboche, M.; Weisshaar, B.; Lepiniec, L. TT2, TT8, and TTG1 synergistically specify the expression of BANYULS and proanthocyanidin biosynthesis in Arabidopsis thaliana. Plant J. 2004, 39, 366–380. [Google Scholar] [CrossRef]
- Li, C.; Qiu, J.; Ding, L.; Huang, M.; Huang, S.; Yang, G.; Yin, J. Anthocyanin biosynthesis regulation of DhMYB2 and DhbHLH1 in Dendrobium hybrids petals. Plant Physiol. Biochem. 2017, 112, 335–345. [Google Scholar] [CrossRef]
- Yuan, Y.; Shi, Y.; Tang, D. Isolation and characterization of R2R3-MYB and basic helix–loop–helix (bHLH) transcription factors involved in anthocyanin biosynthesis in tulip tepals. Acta Physiol. Plant. 2020, 42, 32. [Google Scholar] [CrossRef]
- Arlotta, C.; Puglia, G.D.; Genovese, C.; Toscano, V.; Karlova, R.; Beekwilder, J.; De Vos, R.C.H.; Raccuia, S.A. MYB5-like and bHLH influence flavonoid composition in pomegranate. Plant Sci. 2020, 298, 110563. [Google Scholar] [CrossRef]
- Shen, T.; Qi, H.; Luan, X.; Xu, W.; Yu, F.; Zhong, Y.; Xu, M. The chromosome-level genome sequence of the camphor tree provides insights into Lauraceae evolution and terpene biosynthesis. Plant Biotechnol. J. 2022, 20, 244–246. [Google Scholar] [CrossRef]
- Buck, M.J.; Atchley, W.R. Phylogenetic analysis of plant basic helix-loop-helix proteins. J. Mol. Evol. 2003, 56, 742–750. [Google Scholar] [CrossRef]
- Pires, N.; Dolan, L. Origin and diversification of basic-helix-loop-helix proteins in plants. Mol. Biol. Evol. 2010, 27, 862–874. [Google Scholar] [CrossRef] [PubMed]
- Heim, M.A.; Jakoby, M.; Werber, M.; Martin, C.; Weisshaar, B.; Bailey, P.C. The Basic Helix-Loop-Helix Transcription Factor Family in Plants: A Genome-Wide Study of Protein Structure and Functional Diversity. Mol. Biol. Evol. 2003, 20, 735–747. [Google Scholar] [CrossRef] [PubMed]
- Abrouk, M.; Murat, F.; Pont, C.; Messing, J.; Jackson, S.; Faraut, T.; Tannier, E.; Plomion, C.; Cooke, R.; Feuillet, C.; et al. Palaeogenomics of plants: Synteny-based modelling of extinct ancestors. Trends Plant Sci. 2010, 15, 479–487. [Google Scholar] [CrossRef] [PubMed]
- Tiley, G.P.; Ane, C.; Burleigh, J.G. Evaluating and Characterizing Ancient Whole-Genome Duplications in Plants with Gene Count Data. Genome Biol. Evol. 2016, 8, 1023–1237. [Google Scholar] [CrossRef]
- Maher, C.; Stein, L.; Ware, D. Evolution of Arabidopsis microRNA families through duplication events. Genome Res. 2006, 16, 510–519. [Google Scholar] [CrossRef]
- Yang, Z.; Bielawski, J.P. Statistical methods for detecting molecular adaptation. Trends Ecol. Evol. 2000, 15, 496–503. [Google Scholar] [CrossRef]
- Chu, Y.; Xiao, S.; Su, H.; Liao, B.; Zhang, J.; Xu, J.; Chen, S. Genome-wide characterization and analysis of bHLH transcription factors in Panax ginseng. Acta Pharm. Sin. B 2018, 8, 666–677. [Google Scholar] [CrossRef]
- Shen, T.; Wen, X.; Wen, Z.; Qiu, Z.; Hou, Q.; Li, Z.; Mei, L.; Yu, H.; Qiao, G. Genome-wide identification and expression analysis of bHLH transcription factor family in response to cold stress in sweet cherry (Prunus avium L.). Sci. Hortic. 2021, 279, 109905. [Google Scholar] [CrossRef]
- Li, J.; Li, X.; Han, P.; Liu, H.; Gong, J.; Zhou, W.; Shi, B.; Liu, A.; Xu, L. Genome-wide investigation of bHLH genes and expression analysis under different biotic and abiotic stresses in Helianthus annuus L. Int. J. Biol. Macromol. 2021, 189, 72–83. [Google Scholar] [CrossRef]
- Pani, A.; Mahapatra, R.K. Computational identification of microRNAs and their targets in Catharanthus roseus expressed sequence tags. Genom Data 2013, 1, 2–6. [Google Scholar] [CrossRef]
- Hong, G.J.; Xue, X.Y.; Mao, Y.B.; Wang, L.J.; Chen, X.Y. Arabidopsis MYC2 interacts with DELLA proteins in regulating sesquiterpene synthase gene expression. Plant Cell 2012, 24, 2635–2648. [Google Scholar] [CrossRef] [PubMed]
- Xu, J.; Xu, H.; Zhao, H.; Liu, H.; Xu, L.; Liang, Z. Genome-wide investigation of bHLH genes and expression analysis under salt and hormonal treatments in Andrographis paniculata. Ind. Crops Prod. 2022, 183, 114928. [Google Scholar] [CrossRef]
- Li, Y.; Li, L.; Ding, W.; Li, H.; Shi, T.; Yang, X.; Wang, L.; Yue, Y. Genome-wide identification of Osmanthus fragrans bHLH transcription factors and their expression analysis in response to abiotic stress. Environ. Exp. Bot. 2020, 172, 103990. [Google Scholar] [CrossRef]
- Li, Y.; Shan, X.; Gao, R.; Yang, S.; Wang, S.; Gao, X.; Wang, L. Two IIIf Clade-bHLHs from Freesia hybrida Play Divergent Roles in Flavonoid Biosynthesis and Trichome Formation when Ectopically Expressed in Arabidopsis. Sci. Rep. 2016, 6, 30514. [Google Scholar] [CrossRef] [PubMed]
- Xiang, L.L.; Liu, X.F.; Li, X.; Yin, X.R.; Grierson, D.; Li, F.; Chen, K.S. A Novel bHLH Transcription Factor Involved in Regulating Anthocyanin Biosynthesis in Chrysanthemums (Chrysanthemum morifolium Ramat.). PLoS ONE 2015, 10, e0143892. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Gao, W.; Li, H.; Wang, Y.; Li, D.; Xue, C.; Liu, Z.; Liu, M.; Zhao, J. Genome-wide analysis of the bZIP gene family in Chinese jujube (Ziziphus jujuba Mill.). BMC Genom. 2020, 21, 483. [Google Scholar] [CrossRef]
- Chandler, V.L.; Radicella, J.P.; Robbins, T.P.; Chen, J.; Turks, D. Two regulatory genes of the maize anthocyanin pathway are homologous: Isolation of B utilizing R genomic sequences. Plant Cell 1989, 1, 1175–1183. [Google Scholar] [CrossRef]
- Ludwig, S.R.; Habera, L.F.; Dellaporta, S.L.; Wessler, S.R. Lc, a member of the maize R gene family responsible for tissue-specific anthocyanin production, encodes a protein similar to transcriptional activators and contains the mychomology region. Proc. Natl. Acad. Sci. USA 1989, 86, 7092–7096. [Google Scholar] [CrossRef]
- Consonni, G.; Viotti, A.; LDellaporta, S.; Tonelli, C. cDNA nucleotide sequence of Sn, a regulatory gene in maize. Nucleic Acids Res. 1991, 20, 373. [Google Scholar] [CrossRef]
- Schneider, M.; Tognolli, M.; Bairoch, A. The Swiss-Prot protein knowledgebase and ExPASy: Providing the plant community with high quality proteomic data and tools. Plant Physiol. Biochem. 2004, 42, 1013–1021. [Google Scholar] [CrossRef]
- Kumar, S.; Stecher, G.; Tamura, K. MEGA7: Molecular evolutionary genetics analysis version 7.0 for bigger datasets brief communication. Mol. Biol. Evol. 2016, 33, 1870–1874. [Google Scholar] [CrossRef] [PubMed]
- Chen, C.; Chen, H.; Zhang, Y.; Thomas, H.R.; Frank, M.H.; He, Y.; Xia, R. TBtools: An Integrative Toolkit Developed for Interactive Analyses of Big Biological Data. Mol. Plant 2020, 13, 1194–1202. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Tang, H.; Jeremy, D., Debarry; Tan, X.; Li, J.; Wang, X.; Tae-ho, L.; Huizhe, J.; Barry, M.; Guo, H.; et al. MCScanX: A toolkit for detection and evolutionary analysis of gene synteny and collinearity. Nucleic Acids Res. 2012, 40, e49. [Google Scholar] [CrossRef] [PubMed]
- Szklarczyk, D.; Gable, A.L.; Lyon, D.; Junge, A.; Wyder, S.; Huerta-Cepas, J.; Simonovic, M.; Doncheva, N.T.; Morris, J.H.; Bork, P.; et al. STRING v11: Protein-protein association networks with increased coverage, supporting functional discovery in genome-wide experimental datasets. Nucleic Acids Res. 2019, 47, D607–D613. [Google Scholar] [CrossRef]



Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Gong, X.; Shen, T.; Li, X.; Lin, H.; Chen, C.; Li, H.; Wu, Z.; Liu, Q.; Xu, M.; Zhang, B.; et al. Genome-Wide Characterization and Analysis of bHLH Transcription Factors Related to Anthocyanin Biosynthesis in Cinnamomum camphora (‘Gantong 1’). Int. J. Mol. Sci. 2023, 24, 3498. https://doi.org/10.3390/ijms24043498
Gong X, Shen T, Li X, Lin H, Chen C, Li H, Wu Z, Liu Q, Xu M, Zhang B, et al. Genome-Wide Characterization and Analysis of bHLH Transcription Factors Related to Anthocyanin Biosynthesis in Cinnamomum camphora (‘Gantong 1’). International Journal of Molecular Sciences. 2023; 24(4):3498. https://doi.org/10.3390/ijms24043498
Chicago/Turabian StyleGong, Xue, Tengfei Shen, Xiuqi Li, Hanbin Lin, Caihui Chen, Huihu Li, Zhaoxiang Wu, Qiaoli Liu, Meng Xu, Bo Zhang, and et al. 2023. "Genome-Wide Characterization and Analysis of bHLH Transcription Factors Related to Anthocyanin Biosynthesis in Cinnamomum camphora (‘Gantong 1’)" International Journal of Molecular Sciences 24, no. 4: 3498. https://doi.org/10.3390/ijms24043498
APA StyleGong, X., Shen, T., Li, X., Lin, H., Chen, C., Li, H., Wu, Z., Liu, Q., Xu, M., Zhang, B., & Zhong, Y. (2023). Genome-Wide Characterization and Analysis of bHLH Transcription Factors Related to Anthocyanin Biosynthesis in Cinnamomum camphora (‘Gantong 1’). International Journal of Molecular Sciences, 24(4), 3498. https://doi.org/10.3390/ijms24043498





