Abstract
Current research on the angiotensin-converting-enzyme (ACE) gene has yielded controversial results on whether different ACE polymorphisms are linked with human longevity. ACE polymorphisms are a risk factor for Alzheimer’s disease and age-onset diseases that may contribute to the mortality of older people. Our goal is to consolidate existing studies, using artificial intelligence-assisted software to come to a more precise understanding of the role of the ACE gene in human longevity. The I (insertion) and D (deletion) polymorphisms in the intron are correlated with the levels of circulating ACE; homozygous D (DD) is high, and homozygous I (II) is low. Here, we performed a detailed meta-analysis of the I and D polymorphisms using centenarians (100+ years old), long-lived subjects (85+ years old), and control groups. ACE genotype distribution was analyzed across a total of 2054 centenarians and 12,074 controls, as well as 1367 long-lived subjects between the ages of 85–99, using the inverse variance and random effects methods. The ACE DD genotype was found to be favored in centenarians (OR: 1.41 (95% CI: 1.19–1.67), p < 0.0001) with a heterogeneity of 32%, and the II genotype slightly favored the control groups (OR: 0.81 (95% CI: 0.66–0.98), p = 0.03) with a heterogeneity of 28%, corroborating results from previous meta-analyses. Novel to our meta-analysis, the ID genotype was found to be favored in control groups (OR: 0.86 (95% CI: 0.76–0.97), p = 0.01) with a heterogeneity of 0%. The long-lived group showed a similar positive association between the DD genotype and longevity (OR: 1.34 (95% CI: 1.21–1.48), p < 0.0001) and a negative association between the II genotype and longevity (OR: 0.79 (95% CI: 0.70–0.88), p < 0.0001). The long-lived ID genotype did not show significant findings (OR: 0.93 (95% CI: 0.84–1.02), p = 0.79). In conclusion, the results suggest a significant positive association of the DD genotype with human longevity. However, despite the previous study, the results do not confirm a positive association of the ID genotype with human longevity. We suggest a few important paradoxical implications: (1) inhibition of ACE can increase longevity in model systems from nematodes to mammals, seemingly opposite to the finding in humans; (2) exceptional longevity associated with homozygous DD is also associated with age-related diseases with higher mortality risks in homozygous DD. We discuss ACE, longevity, and age-related diseases.
1. Introduction
Human life expectancy has increased in the past millennia since the studies of longevity in ancient Greek and Roman populations [1,2,3]. The study of aging, including the biology of aging and populations of older people, has suggested that longevity is influenced by a combination of genetic, lifestyle, and environmental factors [4]. The search for genetic causes of longevity has implicated several possible candidate genes, including SIRT1, APOE, FOXO3A, ACE, ATM, NOS1, NOS2, KLOTHO, and IL6 [5,6]. Of these, ACE encodes an angiotensin-converting enzyme, which has two functions with opposing effects on health [7,8]. First, ACE is an essential enzyme for hemodynamic control via the renin-angiotensin-aldosterone system and the kinin-kallikrein system. ACE converts the inactive angiotensin I to the active angiotensin II, a potent vasoconstrictor, and inactivates bradykinin, a potent vasodilator. ACE inhibitors are one of the first-line treatments for hypertension and congestive heart failure. In this function, ACE inhibition is beneficial to health through controlling hypertension. Secondly, ACE is an amyloid-degrading enzyme that can reduce the proteo-toxicity of amyloid. Importantly, ACE, when altered, is a risk factor gene in Alzheimer’s disease [9,10]. ACE alterations are associated with Alzheimer’s disease [8]. In this function, ACE inhibition may be deleterious to the health through amyloid toxicity. Thus, the effect of ACE on longevity may differ depending on the health conditions of advanced aging [8]. Thus, we reason that long-lived groups should be included in this study.
The ACE gene is located on chromosome 17q23. The ACE I/D polymorphisms were first identified in 1990; they are characterized by the presence (insertion, I) or absence (deletion, D) of a 287-bp (base pair) Alu repeat sequence in intron 16 [11]. The I/D polymorphisms are correlated with the serum levels of circulating ACE; the D-allele is higher, and the I-allele is lower [12]. DD individuals have approximately twice the plasma ACE levels as II individuals do [11,13]. Other meta-analyses have also shown a positive association between the D-allele and increased risk of essential hypertension [14], ischemic stroke [15], coronary artery disease [16], left ventricular hypertrophy [17], and pneumonia [18]. In addition, the D-allele has been linked with higher mortality rates from COVID-19 [19].
Paradoxically, though the ACE D allele is associated with the risk of cardiovascular disease (CVD), hypertension, Alzheimer’s disease, and other common causes of death in older people, a previous study noted an increased frequency of the DD genotype among a group of centenarians from France [20]. Since then, centenarian studies have been performed across different cohorts, resulting in conflicting results. A more recent study suggests that the DD genotype and the D allele are enriched in centenarians [21], whereas another study suggests that there is no significant difference among the I and D alleles [22,23]. In this meta-analysis, we further consolidated these findings to more precisely determine whether the D polymorphism is enriched in centenarians and long-lived individuals.
2. Results
2.1. AI-Assisted Software and Centenarian Results
We tested the following AI-assisted programs for meta-analysis: ASReview, Colandr, Parsifal, and Rayyan. Of the programs, Colandr was selected for its accessibility and intuitive user interface. It is assisted by machine learning and natural language processing, which provide an integrated data extraction function and seven additional functions (set-up review, piloting or scoping, literature search, duplicate check, article screen, data coding, appraisal, and documentation) [24]. It uses machine-learning algorithms to determine the relevance of a given study based on other studies deemed relevant by the researcher and orders the list of unscreened studies based on calculated relevance, though the final decision to include or exclude a study requires a critical appraisal by the researcher. Compared with other software we tested, Colandr had the quickest machine-learning algorithm, providing feedback on study relevance after approximately five studies marked for inclusion by the researcher, compared with eight or more studies for other programs. Overall, Colandr provides a good framework for the systematic review of the literature and offers helpful feedback to expedite the process.
Our literature search is summarized in Figure 1. The extracted data of centenarians from the identified studies are summarized in Table 1. The meta-analysis included 13 studies [20,22,25,26,27,28,29,30,31,32,33,34,35] involving a total of 2054 centenarians and 10,986 controls. Ethnicities of the studied populations were Caucasian (n = 10), Korean (n = 1), Uyghur (n = 1), and Han Chinese (n = 1). This meta-analysis investigated the age groups that could be analyzed. We found three age groups that consistently showed up among the previous studies, resulting in centenarians (100+ years old), long-lived groups (85+ years old), and control groups. The range of the control groups was diverse among previous studies. We set the ages of control groups as 18–85 years old, which was judged to be inclusive of most of the studies.
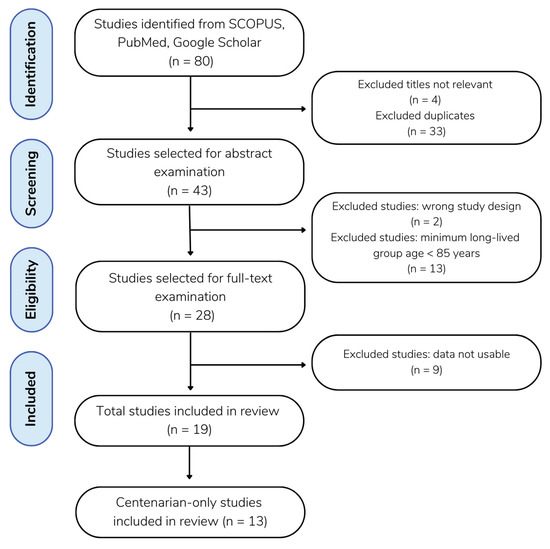
Figure 1.
Flow diagram of the literature search. A total of 80 studies were gathered from SCOPUS (n = 39), PubMed (n = 26), and Google Scholar (n = 15). Excluding 33 duplicates and 4 studies with irrelevant titles, 43 remaining studies were selected for abstract examination. Following the abstract screening, 2 studies were excluded for having the wrong study design, and 13 studies were excluded for defining their minimum long-lived population age to be < 85 years of age. A full-text review was performed on the remaining 28 studies, and 9 of those were excluded, as the data was not usable, meaning that the study collected and presented data in a form other than the frequency of individuals’ genotypes, and that the frequency of genotypes was not able to be calculated from the data available.

Table 1.
Characteristics of studies on ACE I/D polymorphisms in centenarians. A total of 13 studies were included that looked at the frequency of different ACE I/D polymorphisms in centenarians.
The major differences from the previous meta-analysis were as follows. First, we used different age groups for a more precise understanding of the effect of ACE on longevity. The previous meta-analysis compared the frequencies of various combinations of I and D (i.e., DD vs. II; DD vs. ID; ID vs. II; DD + ID vs. II; and DD vs. ID + II) [21] and lacked comparisons among different age groups. Secondly, we excluded a study [36] that was used in the previous meta-analysis [21] for the following reason: the excluded study used 62–88 years old, which overlapped with the age range for the long-lived group (85+ years old) commonly used in previous studies (see II. Long-Lived Results). Finally, we incorporated AI-assisted software to reduce the burden of the meta-analysis studies.
As shown in Figure 2, the association between the DD genotype and exceptional longevity was analyzed in 13 studies [20,22,25,26,27,28,29,30,31,32,33,34,35]. The DD genotype was more frequent among centenarians than in controls, with an OR of 1.41 (95% CI: 1.19–1.67, p < 0.0001) with mild heterogeneity (32%), indicating a significant positive association between the DD genotype and longevity.
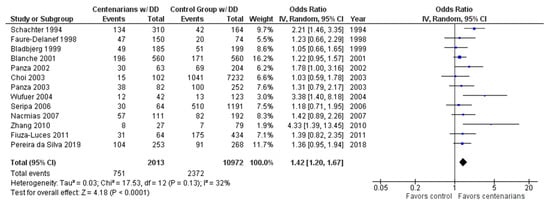
Figure 2.
Comparison of DD genotype frequency in centenarians and control groups. The ages of centenarians and control groups are as described in Table 1. The p-value is indicated by P in the Figure.
As shown in Figure 3, the association between the ID genotype and exceptional longevity was analyzed in 13 studies [20,22,25,26,27,28,29,30,31,32,33,34,35]. The ID genotype was found to have an OR of 0.86 (95% CI: 0.76–0.97, p = 0.01) with no evidence of heterogeneity (0%), indicating a significant negative association between the ID genotype and longevity.
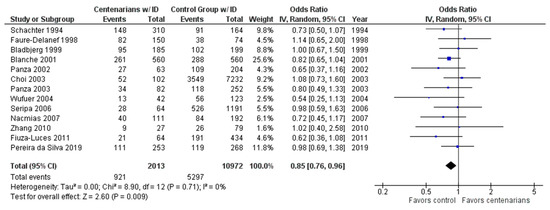
Figure 3.
Comparison of ID genotype frequency in centenarians and control groups. The ages of centenarians and control groups are described in Table 1. The p-value is indicated by P in the Figure.
As shown in Figure 4, the association between the II genotype and exceptional longevity was analyzed in 13 studies [20,22,25,26,27,28,29,30,31,32,33,34,35]. The II genotype was found to have an OR of 0.81 (95% CI: 0.66–0.98, p = 0.03) with mild heterogeneity (28%), indicating a significant negative association between the II genotype and longevity.
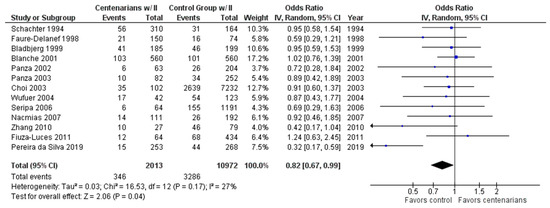
Figure 4.
Comparison of II genotype frequency in centenarians and control groups. The ages of centenarians and control groups are as described in Table 1. The p-value is indicated by P in the Figure.
2.2. Long-Lived Results
Table 2 summarizes the data of long-lived groups. The meta-analysis included 19 studies, with 13 centenarian-only studies, 5 studies that included both nonagenarians and centenarians [23,37,38,39,40], and 1 study with individuals 85 years and older [41], involving a total of 3421 long-lived individuals (85+ years old) and 11,959 controls. Ethnicities of the studied populations were Caucasian (n = 13), Korean (n = 1), Uyghur (n = 1), Han Chinese (n = 2), Russian/Yakut (n = 1), and Peruvian (n = 1).

Table 2.
Characteristics of studies on ACE I/D polymorphisms in long-lived individuals (85+ years old).
The association between the DD genotype and exceptional longevity was analyzed in 18 studies [20,22,23,25,26,27,28,29,30,31,32,33,34,35,38,39,40,41] (Figure 5). The DD genotype was found to have an OR of 1.32 (95% CI: 1.19–1.47, p < 0.00001) with mild heterogeneity (30%), indicating a significant positive association between the DD genotype and longevity.
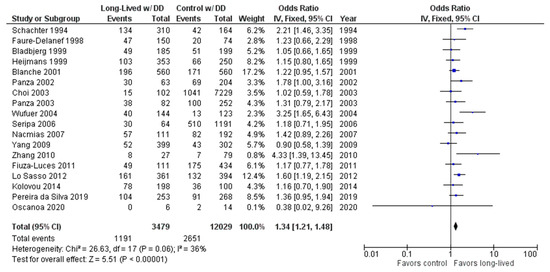
Figure 5.
Comparison of DD genotype frequency in long-lived and control groups. The ages of long-lived and control groups are described in Table 1. The p-value is indicated by P in the Figure.
As shown in Figure 6, the association between the ID genotype and exceptional longevity was analyzed in 18 studies [20,22,23,25,26,27,28,29,30,31,32,33,34,35,38,39,40,41].
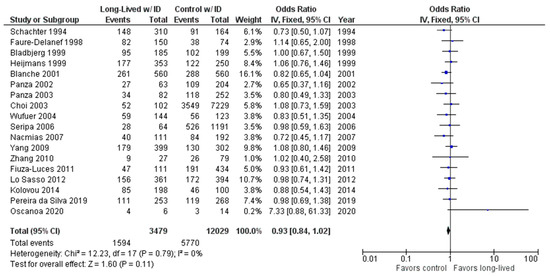
Figure 6.
Comparison of ID genotype frequency in long-lived and control groups. The ages of long-lived and control groups are described in Table 1. The p-value is indicated by P in the Figure.
The ID genotype was found to have an OR of 0.92 (95% CI: 0.84–1.02, p = 0.11) with no evidence of heterogeneity (0%). There was no significant difference in the frequency of the ID genotype between the long-lived (85+ years old) and control groups.
As shown in Figure 7, the association between the II genotype and exceptional longevity was analyzed in 19 studies [20,22,23,25,26,27,28,29,30,31,32,33,34,35,38,39,40,41,42]. Arkhipova et al. (2014) only provided raw frequency data for the II genotype and thus was not included in the DD or ID analyses. The II genotype was found to have an OR of 0.80 (95% CI: 0.71–0.90, p = 0.002) with mild heterogeneity (30%), indicating a significant negative association between the II genotype and longevity. The risk of bias of the studies was assessed using ROBINS-1 (method). As shown in Figure 8, the result suggests an overall low risk of bias.
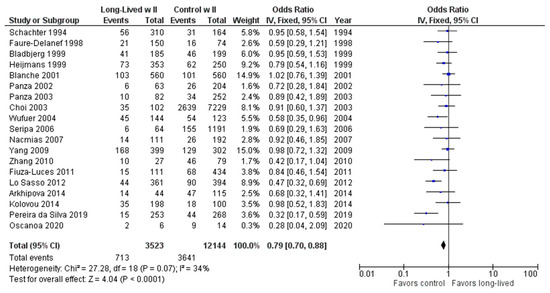
Figure 7.
Comparison of II genotype frequency in long-lived and control groups. The ages of long-lived and control groups are described in Table 1. The p-value is indicated by P in the Figure.
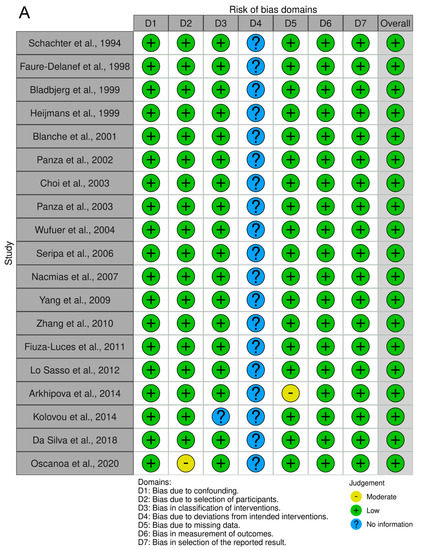
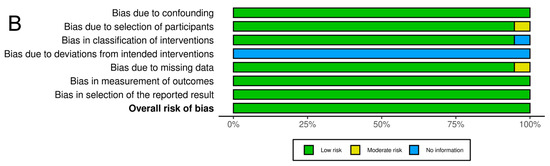
Figure 8.
Risk assessment of the studies. (A) Traffic light plot showing the assessment of the risk of bias using ROBINS-1. (B) Summary plot showing the assessment of the risk of bias using ROBINS-1.
3. Discussion
Centenarians are the fastest-growing demographic group of the world’s population, having roughly doubled every decade since 1950 [42]. It has been nearly a decade since the most recent meta-analysis was published in 2013 [21], and additional studies on centenarians and nonagenarians have been conducted that further elucidate the association between ACE genotype and longevity. In addition, the use of AI and machine-learning software has aided in the search and screening process to compile even more studies and process the associated data. Our meta-analysis suggests, with increased confidence, that there is a significant positive association of the DD genotype with exceptional longevity across populations of different ethnicities. It has been reported that serum ACE levels correlate in the order of DD > ID > II, in which DD is the highest [11,12,13]. The higher levels of ACE were correlated with increased lifespans, especially more in centenarians than in long-lived controls.
The results present a couple of paradoxes in which the D-allele has been reported as a risk factor for hypertension [14], CVD [16], pneumonia [18], and COVID-19 [19], all of which are major causes of death in older people. However, the DD genotype has been associated with a decreased risk of Alzheimer’s disease, potentially due to ACE’s ability to inhibit amyloid beta aggregation and degrade A beta-(1-40) [43,44,45]. Although the protective effect of the D-allele against Alzheimer’s disease does not fully explain the higher proportion of DD individuals in centenarian cohorts, the D-allele or DD genotype may impart a health risk while also providing a long-term longevity advantage. The middle-life crisis theory of aging [8,46,47,48], in which midlife events are the catalyst for the transition from normal aging to a more advanced pathological state, can be used to explain ACE’s dual functions as an angiotensin-converting enzyme and an amyloid-degrading enzyme [8]; low levels of ACE plasma activity, associated with the II genotype, may initially confer stress resistance before midlife, but will be insufficient to counter the abnormal accumulation of beta-amyloid when it begins and lead to higher mortality rates earlier in the aging process due to AD, whereas DD individuals who have survived midlife comorbidities would now have lower mortality rates from AD. However, this does not fully explain the ACE paradox, as Alzheimer’s disease is only one of many diseases that affect older people. As the full role of ACE in the human body has not been determined, the D-allele may have some neuroendocrine or immunomodulatory functions that have yet to be discovered [49] that may further explain ACE’s role in longevity. This raises an unanswered key question as to how exceptional longevity is associated with positive risks of age-onset diseases that may increase the risk of mortality. Interestingly, the study of two traits (IP6K3 and IPMK) suggests that gene–gene interactions may be more meaningful than single polymorphisms [50]. Thus, studies of interaction networks are needed [51].
Another paradox is that the inhibition of ACE by mutations or drugs can extend lifespans in model systems from nematodes, fruit flies, and mammals [8,52]. This observation raises awareness that life-extension interventions in model systems may be different from exceptional longevity in humans. Thus, it raises another unanswered key question of whether exceptional longevity in humans shares common underlying mechanisms with life extension in model systems.
Our meta-analysis includes a comprehensive review of studies performed on centenarians, from Schachter’s initial study in 1994 up to the present, for a total of 2054 centenarians. We did not limit the literature search to only studies written in English, thus our data represent all of the current ACE centenarian studies in the databases searched. The results corroborated the positive association between the DD genotype and exceptional longevity and the negative association between the II genotype and exceptional longevity that have been noted in previous meta-analyses. A novel finding was that the ID genotype, which had previously shown no significant associations with longevity, was found to have a significant negative association with longevity in the centenarian-only analysis.
It is implicated that the effect of ACE on health may differ among different age groups [8]. Thus, we compared the centenarian-only data with the long-lived (85+ years old) data. We found that the distributions of ACE DD and II genotypes were consistent across both age groups, with strong associations in the DD genotype with exceptional longevity and with negative associations in the II genotype. Interestingly, the ID genotype had a significant negative association among centenarians but did not show a significant association among long-lived groups. The straightforward interpretations are as follows: (1) The DD genotype is associated with centenarians and the long-lived groups, and the D alleles follow the inheritance pattern of autosomal recessive. (2) The II genotype is negatively associated with longevity in both age groups. (3) The ID genotype shows a weakly negative association with centenarians, but the trend was not significant in the long-lived control. Thus, more detailed studies with a series of ages will be needed.
The limitations of this study are as follows. First, one issue can be summarized as a general limitation of the meta-analysis method. Each search engine lists the papers that are indexed by their criteria, which may mean missing relevant studies. We included three search engines to reduce this risk of missing studies. Second, related to the first limitation is that the criteria to analyze the data rely on the criteria used in each study. For example, control groups of the past studies ranged broadly and were diverse (18–85 years old). There was a lack of detailed age groups to compare the effect of ACE. Thus, we were not able to have a series of ages to define age-dependent enrichment of the ACE D and I alleles. Third, another limitation is that the vast majority of the centenarian studies on ACE used Caucasian populations. Selectively analyzing the studies of Caucasian populations gives the same results as the pooled data, indicating that there were no major differences in associations across ethnicities within this present review. However, there were not enough Asian studies to sufficiently analyze the strength of association within Asian populations only. We also did not find any studies on ACE polymorphisms in Hispanic/Latino or Black centenarian cohorts in our literature search. For future meta-analyses, a greater diversity of such studies should be taken to better illuminate the role of ACE in aging populations across all different environmental or lifestyle risk factors.
4. Materials and Methods
4.1. Nomenclature, PRISMA, and Database Search
Due to nomenclature inconsistency among studies, we used I (insertion) and D (deletion) alleles as follows: The genotypes were referred to as homozygous insertion (II), heterozygous insertion/deletion (ID), and homozygous deletion (DD). The I and D polymorphisms were referred to as I/D. We followed the Preferred Reporting Items for Systematic Reviews and Meta-Analyses (PRISMA) [53] and performed a meta-analysis as described [54] with updates described in PRISMA 2020 [55]. We performed a literature search to collect publications on ACE I/D polymorphisms in centenarians published before 25 May 2022. A total of 80 studies were obtained from the Human Genomic Aging Resource’s Longevity Map database, Scopus, PubMed, and Google Scholar using the keywords “ACE” OR “angiotensin-converting enzyme” AND “longevity” OR “centenarian” OR “nonagenarian” (subjects 90+ years). Studies of all languages were included. The titles and abstracts of each article were imported to Colandr [24], which uses AI and machine-learning algorithms to determine an article’s relevance based on text-based evidence. Of the 80 studies initially identified, 37 were excluded as duplicates. The remaining 43 articles were then screened in Colandr according to specified inclusion criteria.
4.2. Inclusion Criteria
Studies were included if they met all of the following conditions: (1) evaluation of the ACE I/D polymorphisms and longevity; (2) case-control study design; (3) study provides sample size, genotype data, odds ratio (OR), and 95% confidence interval (CI) or the data necessary to calculate the results. The exclusion criteria of the studies were as follows: (1) not relevant to ACE I/D polymorphisms or longevity; (2) review or meta-analysis; (3) minimum long-lived population age <85 years or unspecified; (4) no control group or maximum control group age >85 years. Data were then separated into centenarian-only and long-lived (85+ years old) sections. Studies were included in the centenarian-only section if the mean population age was >100. In total, we obtained 13 papers that met all our inclusion criteria for centenarians and 19 papers for the long-lived group.
4.3. Data Extraction
Data were extracted independently by the author and an assistant, and any discrepancies were settled by consensus. The data extracted included the author’s name, year of publication, ethnicity of the study population, sample size, and genotype numbers in long-lived and control groups. Allele frequencies were calculated if not directly reported. If the study had multiple control groups of varying age ranges, the control group was defined as all individuals ≤85 years of age, and data were extracted accordingly.
4.4. Data Synthesis, Statistics, and Assessment of the Risk of Bias
To determine the association between ACE genotype and longevity, the odds ratio (OR) with the corresponding 95% confidence interval (CI) was calculated for each genotype (DD, ID, and II). All statistical analyses were performed using The Cochrane Collaboration’s Review Manager (RevMan) Version 5.4 using the random effects statistical model and inverse variance methods to determine the odds ratio, p-value, and heterogeneity I2, among others, as described previously [54]. We used a threshold of p-value < 0.05 to judge statistical significance. The risk of bias in the studies was evaluated as described previously [54]. The result was visualized with Risk-of-bias VISualization (robvis) ROBINS-1 [56,57]. Bias due to deviations from intended interventions did not apply to this meta-analysis, as participants were categorized by age and could not be blinded to the group they were assigned.
5. Conclusions
Our meta-analysis suggests a significant positive association between the ACE DD genotype and centenarians and long-lived groups. Although ACE polymorphisms are a risk factor for Alzheimer’s disease and age-onset diseases that may contribute to mortality, the ACE DD genotype but not the ID genotype was in favor of exceptional longevity over the mortality expected in older people. The DD genotype is associated with a reduced risk of Alzheimer’s disease and increased longevity. It is consistent with an ACE function as an amyloid degradation enzyme, which may be important in exceptional longevity in humans. Additionally, the DD genotype causes a high circulating ACE. In contrast, the DD genotype is also associated with an increased risk of various other diseases. These include hypertension [14], ischemic stroke [15], coronary artery disease [16], left ventricular hypertrophy [17], and pneumonia [18]. Some of these diseases, including hypertension and cardiovascular conditions (stroke and coronary artery disease), are associated with the risks of COVID-19 mortality [54]. The DD genotype is associated with higher COVID-19 mortality [19]. This may imply disease interactions mediated by the D allele. The underlying mechanisms of human longevity remain unclear, especially when considering the D-allele’s role as a risk factor for many age-related comorbidities. Further studies on nonagenarians and centenarians of different backgrounds may provide better insight into the role of ACE as a longevity gene.
Author Contributions
L.L. was involved in data collection, the organization of AI components, data analysis, and writing of the manuscript. S.M. directed and guided the entire research process. All authors have read and agreed to the published version of the manuscript.
Funding
This research received no external funding.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
The data supporting the reported results have been made available within the manuscript.
Acknowledgments
We thank all the members of the Murakami Laboratory at Touro University-California for useful discussion. The work was initiated at the Master of Science in Medical Health Sciences, Touro University California.
Conflicts of Interest
The authors declare no conflict of interest.
Abbreviations
| ACE | angiotensin-converting enzyme |
| I | insertion |
| D | deletion |
| CVD | cardiovascular disease |
| COVID-19 | coronavirus 2019 |
| OR | odds ratio |
| CI | confidence interval |
References
- Montagu, J.D. Length of life in the ancient world: A controlled study. J. R. Soc. Med. 1994, 87, 25–26. [Google Scholar] [CrossRef] [PubMed]
- Batrinos, M.L. The length of life and eugeria in classical Greece. Hormones 2008, 7, 82–83. [Google Scholar] [CrossRef] [PubMed]
- Roser, M.; Ortiz-Ospina, E.; Ritchie, H. Life Expectancy. OurWorldInData.org. 2013. Available online: https://ourworldindata.org/life-expectancy (accessed on 29 November 2022).
- Weinert, B.T.; Timiras, P.S. Invited Review: Theories of aging. J. Appl. Physiol. 2003, 95, 1706–1716. [Google Scholar] [CrossRef]
- Revelas, M.; Thalamuthu, A.; Oldmeadow, C.; Evans, T.-J.; Armstrong, N.J.; Kwok, J.B.; Brodaty, H.; Schofield, P.R.; Scott, R.J.; Sachdev, P.S.; et al. Review and meta-analysis of genetic polymorphisms associated with exceptional human longevity. Mech. Ageing Dev. 2018, 175, 24–34. [Google Scholar] [CrossRef] [PubMed]
- Costa, D.; Scognamiglio, M.; Fiorito, C.; Benincasa, G.; Napoli, C. Genetic background, epigenetic factors and dietary interventions which influence human longevity. Biogerontology 2019, 20, 605–626. [Google Scholar] [CrossRef] [PubMed]
- Le, D.; Crouch, N.; Villanueva, A.; Ta, P.; Dmitriyev, R.; Tunzi, M.; Murakami, S. Evidence-Based Genetics and Identification of Key Human Alzheimer’s Disease Alleles with Co-morbidities. J. Neurol. Exp. Neurosci. 2020, 6, 20. [Google Scholar] [CrossRef]
- Le, D.; Brown, L.; Malik, K.; Murakami, S. Two Opposing Functions of Angiotensin-Converting Enzyme (ACE) That Links Hypertension, Dementia, and Aging. Int. J. Mol. Sci. 2021, 22, 13178. [Google Scholar] [CrossRef] [PubMed]
- Nia, B.V.; Kang, C.; Tran, M.G.; Lee, D.; Murakami, S. Meta Analysis of Human AlzGene Database: Benefits and Limitations of Using C. elegans for the Study of Alzheimer’s Disease and Co-morbid Conditions. Front. Genet. 2017, 8, 55. [Google Scholar] [CrossRef]
- Murakami, S.; Lacayo, P. Biological and disease hallmarks of Alzheimer’s disease defined by Alzheimer’s disease genes. Front. Aging Neurosci. 2022, 14, 996030. [Google Scholar] [CrossRef]
- Rigat, B.; Hubert, C.; Alhenc-Gelas, F.; Cambien, F.; Corvol, P.; Soubrier, F. An insertion/deletion polymorphism in the angiotensin I-converting enzyme gene accounting for half the variance of serum enzyme levels. J. Clin. Investig. 1990, 86, 1343–1346. [Google Scholar] [CrossRef]
- Tiret, L.; Rigat, B.; Visvikis, S.; Breda, C.; Corvol, P.; Cambien, F.; Soubrier, F. Evidence, from combined segregation and linkage analysis, that a variant of the angiotensin I-converting enzyme (ACE) gene controls plasma ACE levels. Am. J. Hum. Genet. 1992, 51, 197–205. [Google Scholar] [PubMed]
- Tomita, H.; Ina, Y.; Sugiura, Y.; Sato, S.; Kawaguchi, H.; Morishita, M.; Yamamoto, M.; Ueda, R. Polymorphism in the Angiotensin-Converting Enzyme (ACE) Gene and Sarcoidosis. Am. J. Respir. Crit. Care Med. 1997, 156, 255–259. [Google Scholar] [CrossRef] [PubMed]
- Li, Y. Angiotensin-converting enzyme gene insertion/deletion polymorphism and essential hypertension in the Chinese population: A meta-analysis including 21,058 participants. Intern. Med. J. 2012, 42, 439–444. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Z.; Xu, G.; Liu, D.; Fan, X.; Zhu, W.; Liu, X. Angiotensin-Converting Enzyme Insertion/Deletion Polymorphism Contributes to Ischemic Stroke Risk: A Meta-Analysis of 50 Case-Control Studies. PLoS ONE 2012, 7, e46495. [Google Scholar] [CrossRef]
- Zintzaras, E.; Raman, G.; Kitsios, G.; Lau, J. Angiotensin-Converting Enzyme Insertion/Deletion Gene Polymorphic Variant as a Marker of Coronary Artery DiseaseA Meta-analysis. Arch. Intern. Med. 2008, 168, 1077–1089. [Google Scholar] [CrossRef]
- Li, X.; Li, Y.; Jia, N.; Guo, S.; Chu, S.; Niu, W. Angiotensin-converting enzyme gene deletion allele increases the risk of left ventricular hypertrophy: Evidence from a meta-analysis. Mol. Biol. Rep. 2012, 39, 10063–10075. [Google Scholar] [CrossRef]
- Nie, W.; Zang, Y.; Chen, J.; Liu, T.; Xiao, L.; Xiu, Q. Angiotensin-converting enzyme I/D polymorphism is associated with pneumonia risk: A meta-analysis. J. Renin-Angiotensin-Aldosterone Syst. 2013, 15, 585–592. [Google Scholar] [CrossRef]
- Yamamoto, N.; Ariumi, Y.; Nishida, N.; Yamamoto, R.; Bauer, G.; Gojobori, T.; Shimotohno, K.; Mizokami, M. SARS-CoV-2 infections and COVID-19 mortalities strongly correlate with ACE1 I/D genotype. Gene 2020, 758, 144944. [Google Scholar] [CrossRef]
- Schächter, F.; Faure-Delanef, L.; Guénot, F.; Rouger, H.; Froguel, P.; Lesueur-Ginot, L.; Cohen, D. Genetic associations with human longevity at the APOE and ACE loci. Nat. Genet. 1994, 6, 29–32. [Google Scholar] [CrossRef]
- Garatachea, N.; Marín, P.J.; Lucia, A. The ACE DD genotype and D-allele are associated with exceptional longevity: A meta-analysis. Ageing Res. Rev. 2013, 12, 1079–1087. [Google Scholar] [CrossRef]
- Choi, Y.-H.; Kim, J.-H.; Kim, D.K.; Lee, M.S.; Kim, C.H.; Park, S.C. Distributions of ACE and APOE Polymorphisms and Their Relations with Dementia Status in Korean Centenarians. J. Gerontol. Ser. A Biol. Sci. Med. Sci. 2003, 58, M227–M231. [Google Scholar] [CrossRef] [PubMed]
- Oscanoa, T.J.; Cieza, E.C.; Lizaraso-Soto, F.A.; Guevara, M.L.; Fujita, R.M.; Romero-Ortuño, R. Angiotensin-Converting Enzyme (ACE) genetic variation and longevity in Peruvian older people: A cross-sectional study. Ann. Hum. Biol. 2020, 47, 309–312. [Google Scholar] [CrossRef] [PubMed]
- Cheng, S.; Augustin, C.; Bethel, A.; Gill, D.; Anzaroot, S.; Brun, J.; DeWilde, B.; Minnich, R.; Garside, R.; Masuda, Y.; et al. Using machine learning to advance synthesis and use of conservation and environmental evidence. Conserv. Biol. 2018, 32, 762–764. [Google Scholar] [CrossRef] [PubMed]
- Faure-Delanef, L.; Baudin, B.; Bénézteau-Burnat, B.; Beaudoin, J.-C.; Giboudeau, J.; Cohen, D. Plasma concentration, kinetic constants, and gene polymorphism of angiotensin I-converting enzyme in centenarians. Clin. Chem. 1998, 44, 2083–2087. [Google Scholar] [CrossRef] [PubMed]
- Bladbjerg, E.M.; Andersen-Ranberg, K.; de Maat, M.P.; Kristensen, S.R.; Jeune, B.; Gram, J.; Jespersen, J. Longevity is independent of common variations in genes associated with cardiovascular risk. Thromb. Haemost. 1999, 82, 1100–1105. [Google Scholar] [PubMed]
- Blanché, H.; Cabanne, L.; Sahbatou, M.; Thomas, G. A study of French centenarians: Are ACE and APOE associated with longevity? Comptes Rendus L’académie Sci. Ser. III-Sci. De La Vie 2001, 324, 129–135. [Google Scholar] [CrossRef] [PubMed]
- Panza, F.; Solfrizzi, V.; D‘Introno, A.; Capurso, C.; Basile, A.M.; Colacicco, A.M.; Sabba, M.; Noya, R.; Capurso, A. Apolipo-protein and angiotensin converting enzyme genes: Regional differences and extreme longevity in Europe. Arch. Gerontol. Geriatr. 2002, 8, 247–251. [Google Scholar] [CrossRef]
- Panza, F.; Solfrizzi, V.; D’Introno, A.; Colacicco, A.M.; Capurso, C.; Kehoe, P.G.; Capurso, A. Angiotensin I converting enzyme (ACE) gene polymorphism in centenarians: Different allele frequencies between the North and South of Europe. Exp. Gerontol. 2003, 38, 1015–1020. [Google Scholar] [CrossRef]
- Wufuer, M.; Fang, M.-W.; Cheng, Z.-H.; Qiu, C.-C. Polymorphism of angiotensin converting enzyme gene and natural longevity in the Xinjiang Uygur people: An association study. Zhonghua Yi Xue Za Zhi 2004, 84, 1603–1606. (In Chinese) [Google Scholar]
- Seripa, D.; Franceschi, M.; Matera, M.G.; Panza, F.; Kehoe, P.; Gravina, C.; Orsitto, G.; Solfrizzi, V.; Di Minno, G.; Dallapiccola, B.; et al. Sex Differences in the Association of Apolipoprotein E and Angiotensin-Converting Enzyme Gene Polymorphisms With Healthy Aging and Longevity: A Population-Based Study From Southern Italy. J. Gerontol. Ser. A Biol. Sci. Med. Sci. 2006, 61, 918–923. [Google Scholar] [CrossRef]
- Nacmias, B.; Bagnoli, S.; Tedde, A.; Cellini, E.; Bessi, V.; Guarnieri, B.; Ortensi, L.; Piacentini, S.; Bracco, L.; Sorbi, S. Angiotensin converting enzyme insertion/deletion polymorphism in sporadic and familial Alzheimer’s disease and longevity. Arch. Gerontol. Geriatr. 2007, 45, 201–206. [Google Scholar] [CrossRef] [PubMed]
- Zhang, W.-F.; Ling, J.-H.; Wang, A.-J. Relationship between shen-deficiency syndrome and polymorphism of angiotensin converting enzyme gene in longevous elders in Bama area of Guangxi municipality. Zhongguo Zhong Xi Yi Jie He Za Zhi Zhongguo Zhongxiyi Jiehe Zazhi 2010, 30, 259–263. (In Chinese) [Google Scholar] [PubMed]
- Fiuza-Luces, C.; Ruiz, J.R.; Rodríguez-Romo, G.; Santiago, C.; Gómez-Gallego, F.; Cano-Nieto, A.; Garatachea, N.; Rodríguez-Moreno, I.; Morán, M.; Lucia, A. Is the ACE I/D polymorphism associated with extreme longevity? A study on a Spanish cohort. J. Renin-Angiotensin-Aldosterone Syst. 2011, 12, 202–207. [Google Scholar] [CrossRef] [PubMed]
- Pereira da Silva, A.; Matos, A.; Aguiar, L.; Ramos-Marques, N.; Ribeiro, R.; Gil, Â.; Gorjão-Clara, J.; Bicho, M. Hypertension and longevity: Role of genetic polymorphisms in renin-angiotensin-aldosterone system and endothelial nitric oxide synthase. Mol. Cell. Biochem. 2018, 455, 61–71. [Google Scholar] [CrossRef] [PubMed]
- Paolisso, G.; Tagliamonte, M.R.; De Lucia, D.; Palmieri, F.; Manzella, D.; Rinaldi, C.; Bossone, A.; Colaizzo, D.; Margaglione, M.; Varricchio, M. ACE Gene Polymorphism and Insulin Action in Older Subjects and Healthy Centenarians. J. Am. Geriatr. Soc. 2001, 49, 610–614. [Google Scholar] [CrossRef]
- Arkhipova, N.; Popova, E.; Grigorieva, L. The angiotension converting enzyme (ACE) gene I/D polymorphism in different ethnic groups of geriatric age living in the Far North. Health 2014, 6, 426–431. [Google Scholar] [CrossRef]
- Kolovou, G.; Kolovou, V.; Vasiliadis, I.; Giannakopoulou, V.; Mihas, C.; Bilianou, H.; Kollia, A.; Papadopoulou, E.; Marvaki, A.; Goumas, G.; et al. The Frequency of 4 Common Gene Polymorphisms in Nonagenarians, Centenarians, and Average Life Span Individuals. Angiology 2014, 65, 210–215. [Google Scholar] [CrossRef]
- Lo Sasso, B.; Bellia, C.; Tomaiuolo, R.; Zarrilli, F.; Scorza, M.; Caruso, A.; Agnello, L.; Bazza, F.; Carru, C.; Zinellu, A.; et al. Polimorfismo I/D del gene per l’enzima di conversione dell’angiotensina (ACE): Gene della longevità o fattore di rischio nella patologia ipertensiva? Biochim. Clin. 2013, 37, 461–464. [Google Scholar]
- Yang, J.-K.; Gong, Y.-Y.; Xie, L.; Lian, S.-G.; Yang, J.; Xu, L.-Y.; Gao, S.-J.; Zhang, Y.-P. Lack of genetic association between the angiotensin-converting enzyme gene insertion/deletion polymorphism and longevity in a Han Chinese population. J. Renin-Angiotensin-Aldosterone Syst. 2009, 10, 115–118. [Google Scholar] [CrossRef]
- Heijmans, B.T.; Westendorp, R.G.; Knook, D.L.; Kluft, C.; Slagboom, P. Angiotensin I–converting enzyme and plasminogen activator inhibitor-1 gene variants: Risk of mortality and fatal cardiovascular disease in an elderly population-based cohort. J. Am. Coll. Cardiol. 1999, 34, 1176–1183. [Google Scholar] [CrossRef]
- Willcox, D.C.; Willcox, B.J.; Poon, L.W. Centenarian Studies: Important Contributors to Our Understanding of the Aging Process and Longevity. Curr. Gerontol. Geriatr. Res. 2010, 2010, 484529. [Google Scholar] [CrossRef] [PubMed]
- Hu, J.; Igarashi, A.; Kamata, M.; Nakagawa, H. Angiotensin-converting Enzyme Degrades Alzheimer Amyloid β-Peptide (Aβ); Retards Aβ Aggregation, Deposition, Fibril Formation; and Inhibits Cytotoxicity. J. Biol. Chem. 2001, 276, 47863–47868. [Google Scholar] [CrossRef]
- Kölsch, H.; Jessen, F.; Freymann, N.; Kreis, M.; Hentschel, F.; Maier, W.; Heun, R. ACE I/D polymorphism is a risk factor of Alzheimer’s disease but not of vascular dementia. Neurosci. Lett. 2005, 377, 37–39. [Google Scholar] [CrossRef] [PubMed]
- Lehmann, D.J.; Cortina-Borja, M.; Warden, D.R.; Smith, A.D.; Sleegers, K.; Prince, J.A.; van Duijn, C.M.; Kehoe, P.G. Large Meta-Analysis Establishes the ACE Insertion-Deletion Polymorphism as a Marker of Alzheimer’s Disease. Am. J. Epidemiol. 2005, 162, 305–317. [Google Scholar] [CrossRef]
- Murakami, S.; Cabana, K.; Anderson, D. Chapter 25—Current advances in the study of oxidative stress and age-related memory impairment in C. elegans. In Molecular Aspects of Oxidative Stress on Cell Signaling in Vertebrates and In-Vertebrates; Farooqui, T., Farooqui, A., Eds.; John Wiley & Sons: Hoboken, NJ, USA, 2011; pp. 347–360. [Google Scholar]
- Murakami, S. Chapter 12—Age-dependent modulation of learning and memory in C. elegans. In Handbook of Behavioral Neuroscience; Part of Invertebrate Learning and Memory; Menzel, R., Benjamin, P.R., Eds.; Elsevier: Amsterdam, The Netherlands; Academic Press: Cambridge, MA, USA, 2013; pp. 140–150. [Google Scholar]
- Machino, K.; Link, C.D.; Wang, S.; Murakami, H.; Murakami, S. A semi-automated motion-tracking analysis of locomotion speed in the C. elegans transgenics overexpressing beta-amyloid in neurons. Front. Genet. 2014, 5, 202. [Google Scholar] [CrossRef] [PubMed]
- Costerousse, O.; Allegrini, J.; Lopez, M.; Alhenc-Gelas, F. Angiotensin I-converting enzyme in human circulating mononuclear cells: Genetic polymorphism of expression in T-lymphocytes. Biochem. J. 1993, 290, 33–40. [Google Scholar] [CrossRef]
- Dato, S.; Crocco, P.; De Rango, F.; Iannone, F.; Maletta, R.; Bruni, A.C.; Saiardi, A.; Rose, G.; Passarino, G. IP6K3 and IPMK variations in LOAD and longevity: Evidence for a multifaceted signaling network at the crossroad between neurodegeneration and survival. Mech. Ageing Dev. 2021, 195, 111439. [Google Scholar] [CrossRef]
- Balmorez, T.; Sakazaki, A.; Murakami, S. Genetic Networks of Alzheimer’s Disease, Aging and Longevity in Humans. medRxiv 2023. [Google Scholar] [CrossRef]
- Egan, B.M.; Scharf, A.; Pohl, F.; Kornfeld, K. Control of aging by the renin–angiotensin system: A review of C. elegans, Drosophila, and mammals. Front. Pharmacol. 2022, 13, 938650. [Google Scholar] [CrossRef]
- Moher, D.; Liberati, A.; Tetzlaff, J.; Altman, D.G.; PRISMA Group. Preferred reporting items for systematic reviews and meta-analyses: The PRISMA statement. PLoS Med. 2009, 6, e1000097. [Google Scholar] [CrossRef]
- Antos, A.; Kwong, M.; Balmorez, T.; Villanueva, A.; Murakami, S. Unusually High Risks of COVID-19 Mortality with Age-Related Comorbidities: An Adjusted Meta-Analysis Method to Improve the Risk Assessment of Mortality Using the Comorbid Mortality Data. Infect. Dis. Rep. 2021, 13, 700–711. [Google Scholar] [CrossRef] [PubMed]
- Page, M.J.; McKenzie, J.E.; Bossuyt, P.M.; Boutron, I.; Hoffmann, T.C.; Mulrow, C.D.; Shamseer, L.; Tetzlaff, J.M.; Akl, E.A.; Brennan, S.E.; et al. The PRISMA 2020 Statement: An Updated Guideline for Reporting Systematic Reviews. BMJ 2021, 372, n71. [Google Scholar] [CrossRef] [PubMed]
- Sterne, J.A.C.; Hernán, M.A.; Reeves, B.C.; Savović, J.; Berkman, N.D.; Viswanathan, M.; Henry, D.; Altman, D.G.; Ansari, M.T.; Boutron, I.; et al. ROBINS-I: A tool for assessing risk of bias in non-randomised studies of interventions. BMJ 2016, 355, i4919. [Google Scholar] [CrossRef]
- McGuinness, L.A.; Higgins, J.P.T. Risk-of-bias VISualization (robvis): An R package and Shiny web app for visualizing risk-of-bias assessments. Res. Synth. Methods 2021, 12, 55–61. [Google Scholar] [CrossRef] [PubMed]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).