An Angiogenic Gene Signature for Prediction of the Prognosis and Therapeutic Responses of Hepatocellular Carcinoma
Abstract
1. Introduction
2. Results
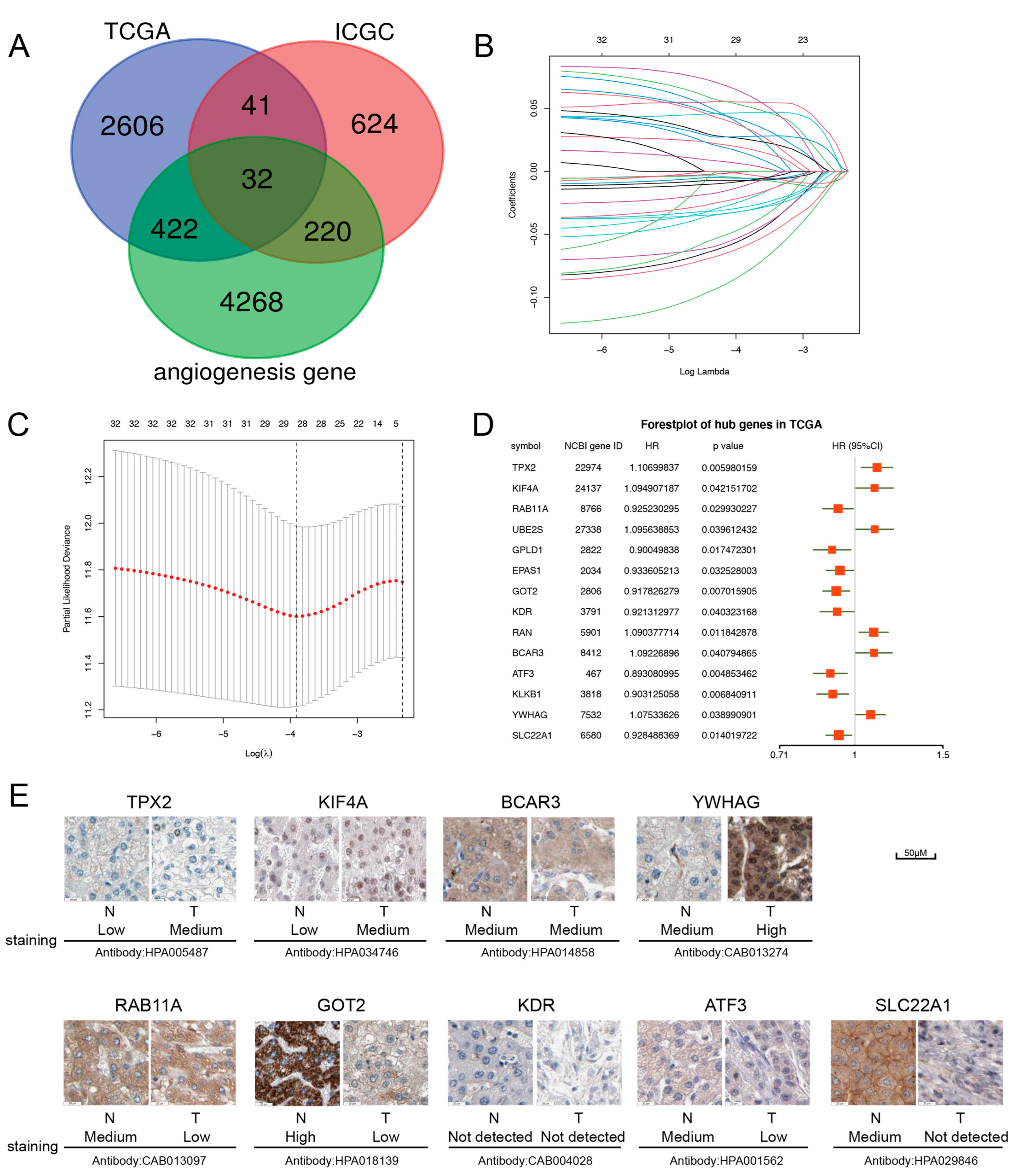
2.1. Construction of the Prognostic Model with the Angiogenesis-Related Gene in HCC
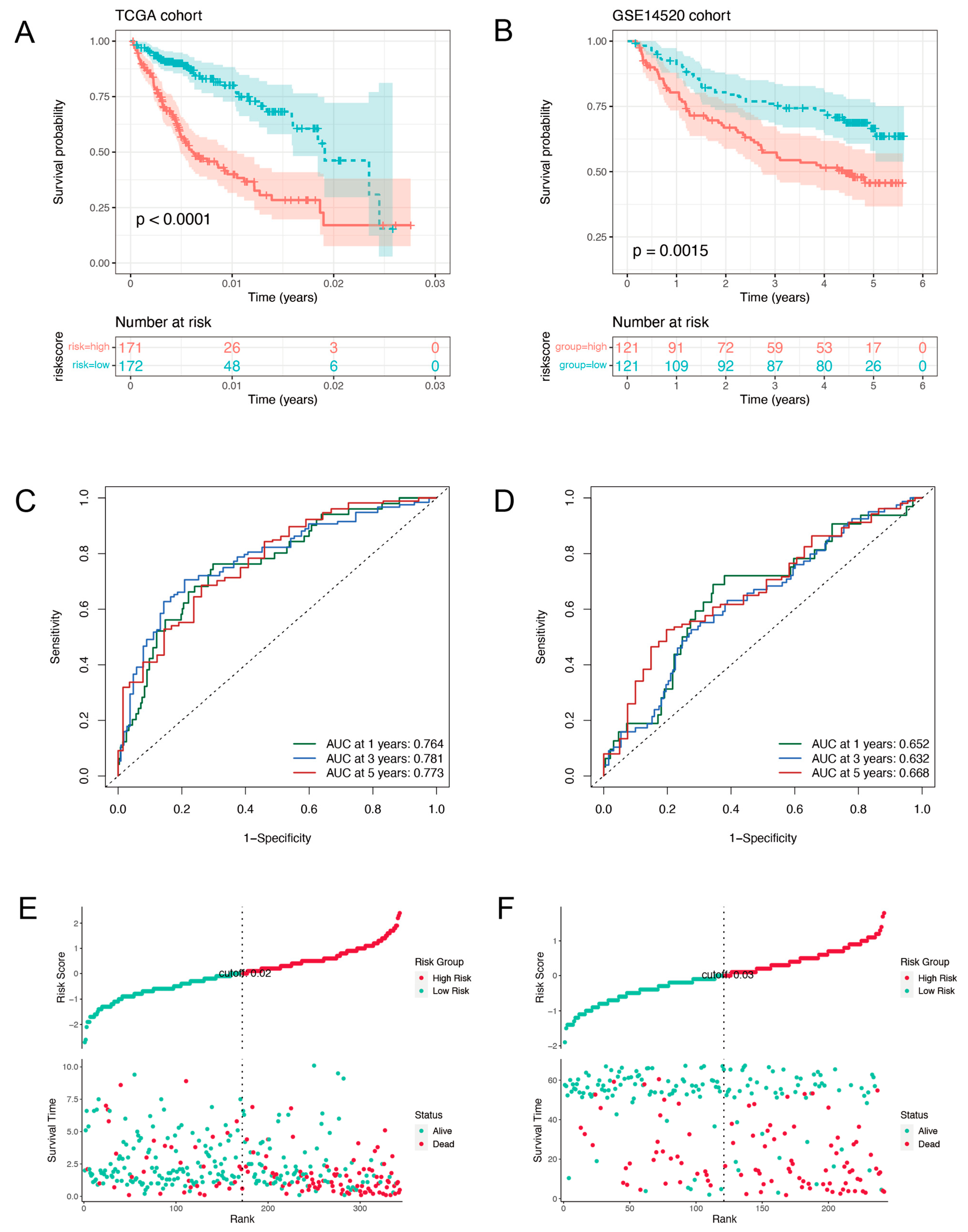
2.2. Validation of the Prognostic Signature
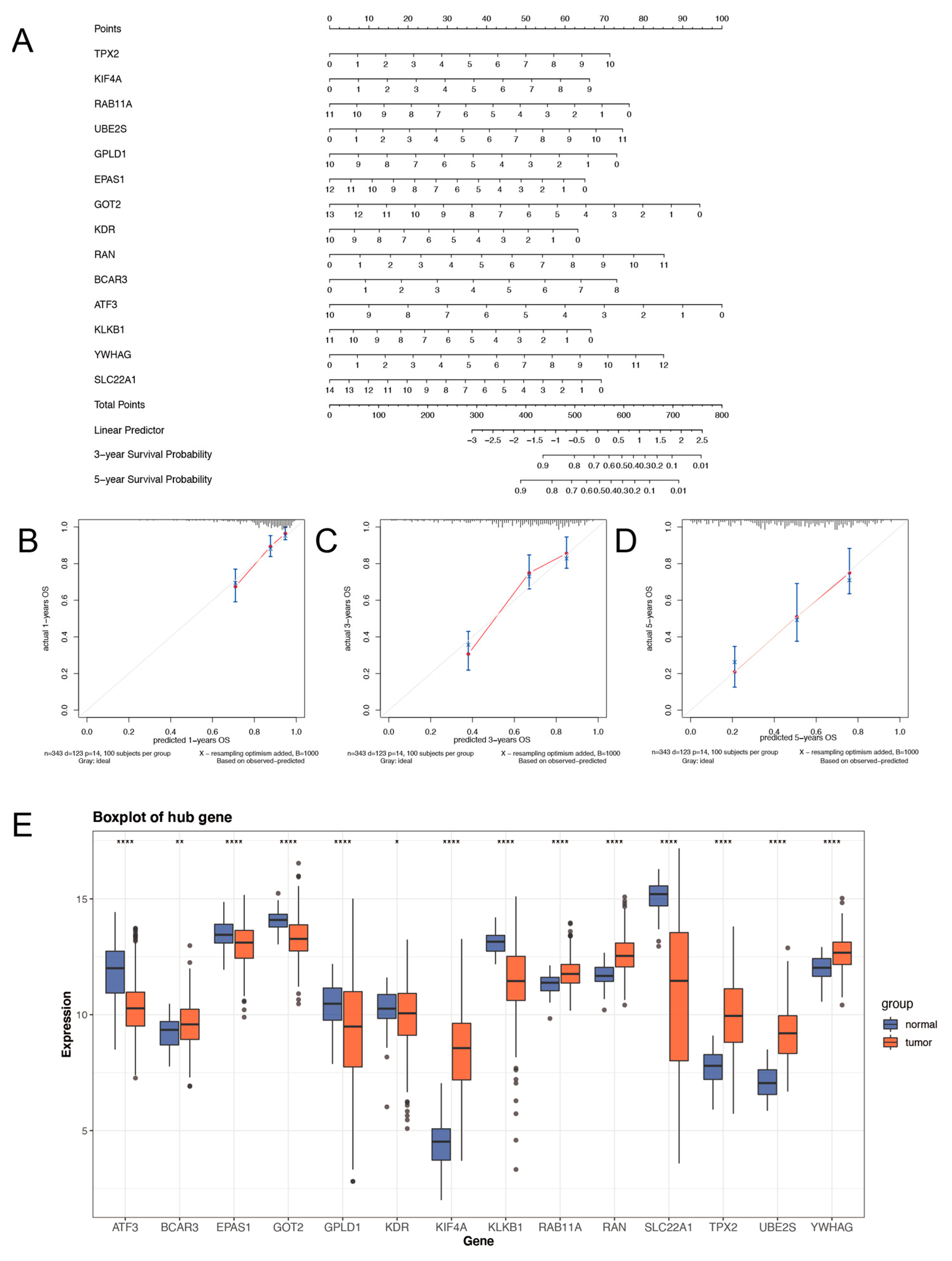
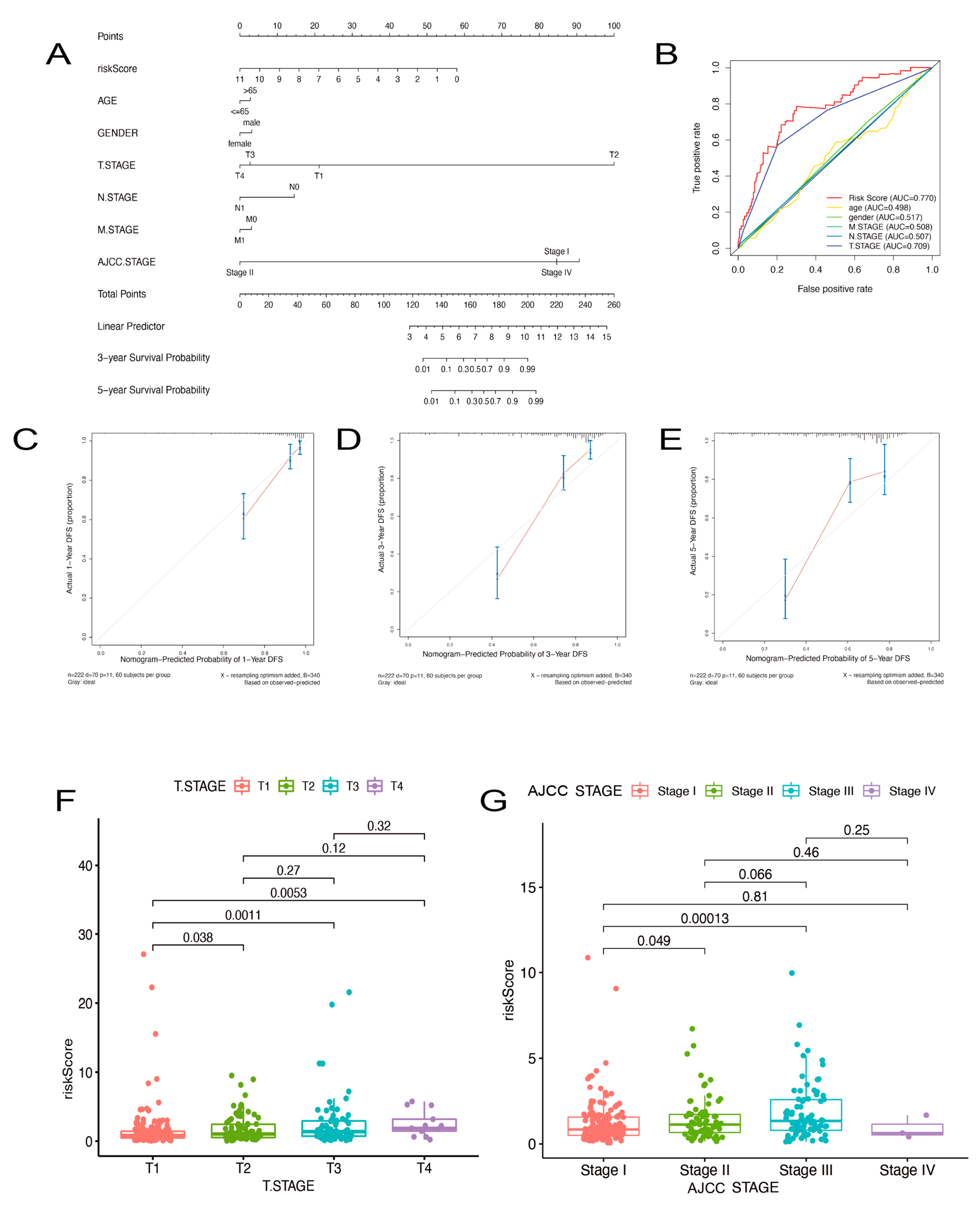
2.3. Prognostic Nomogram of Risk Model by This Gene Set
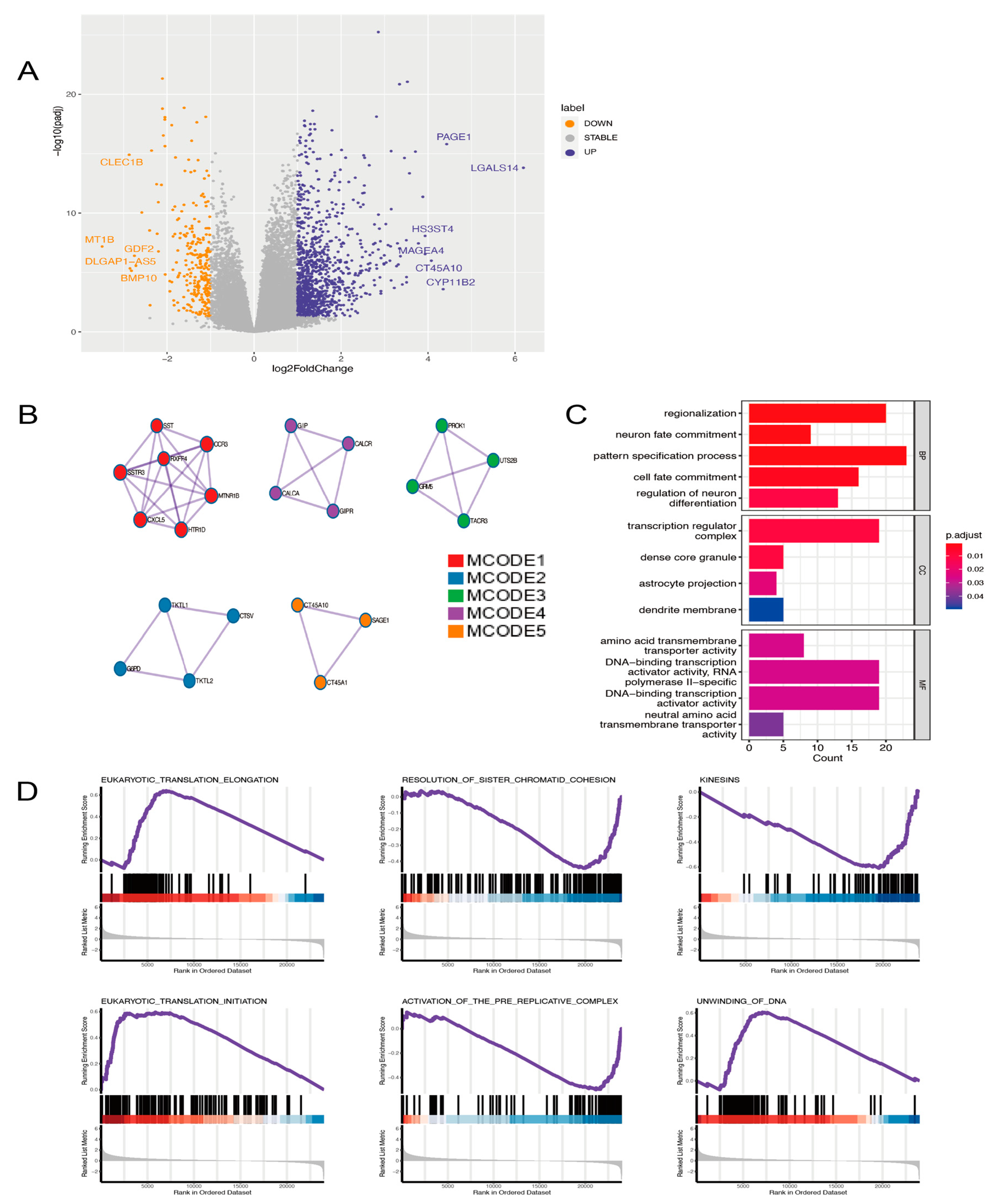
2.4. Comparison of High- and Low-Risk Groups for Somatic Variants
2.5. Analyzing the Model’s Functional Enrichment
2.6. The Gene Signature for Nomogram Construction
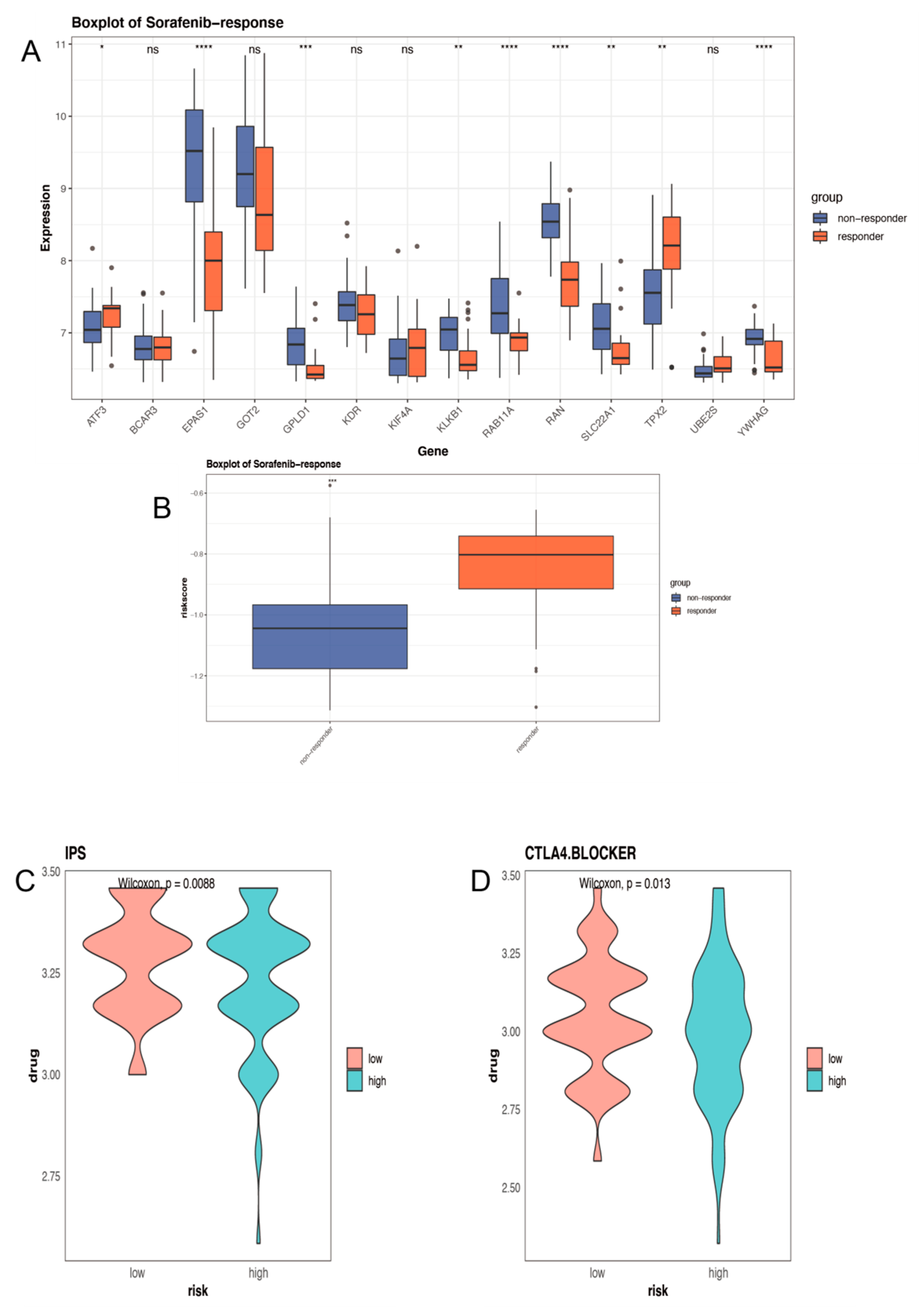
2.7. Distinct Sensitivity to Targeted Therapies and Immunotherapy for Different Groups
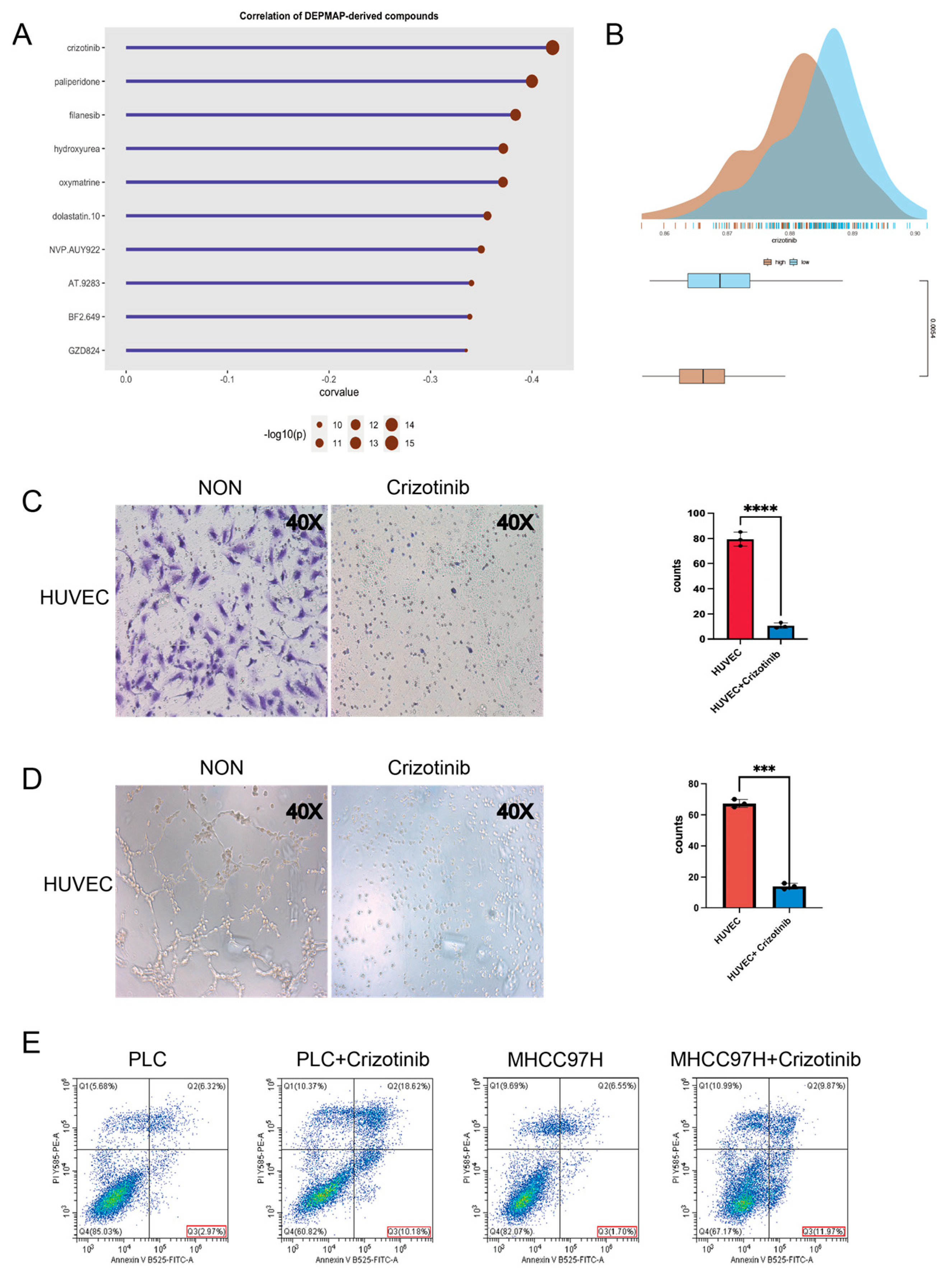
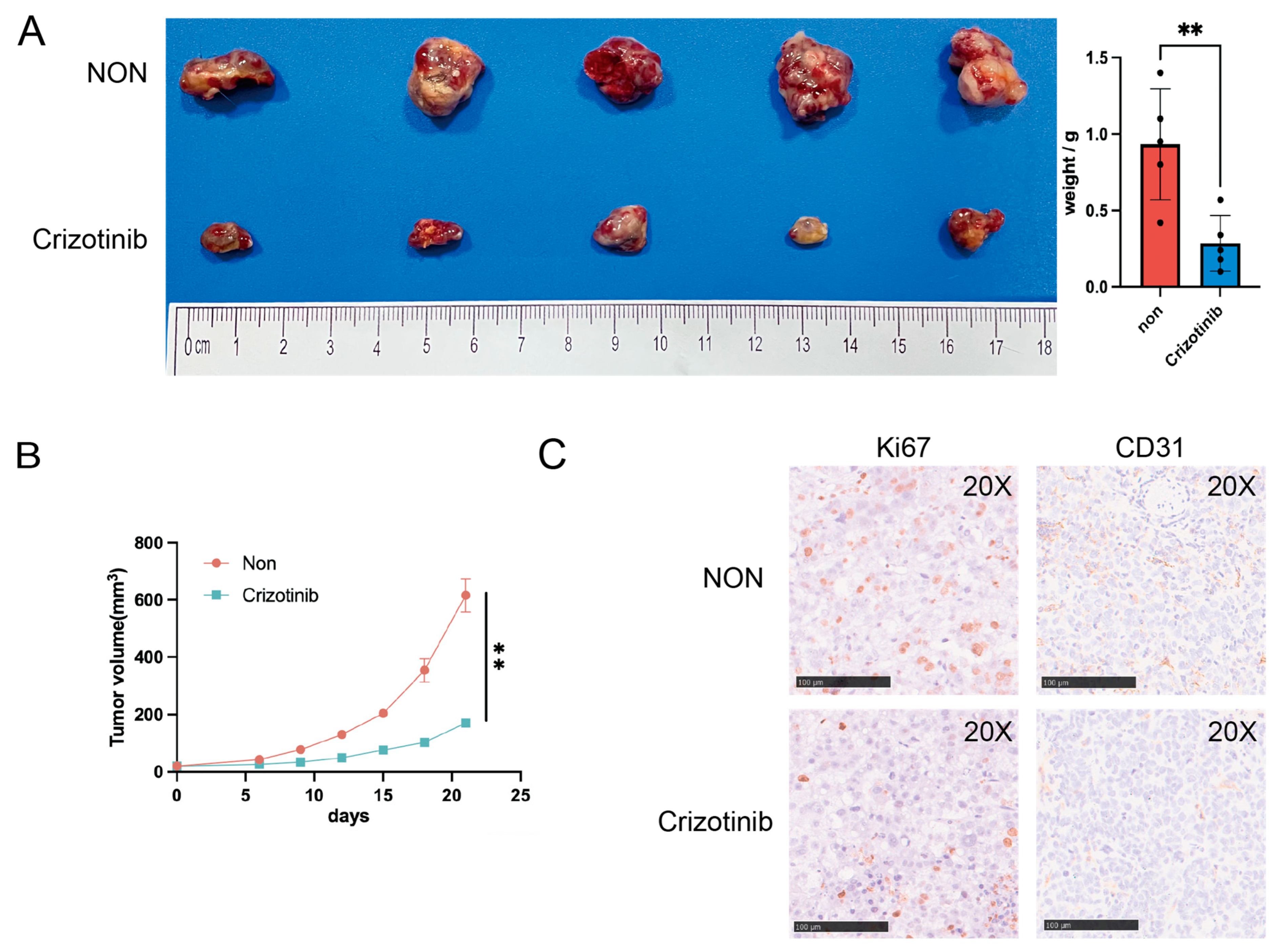
2.8. Investigation of Potential Drugs Based on the Model
3. Discussion
4. Materials and Methods
4.1. Sample and Data Collection
4.2. Establishment and Validation of a Prognostic Model
4.3. Construction and Evaluation of a Predictive Nomogram
4.4. TMB Calculation
4.5. Protein-Protein Interaction and Function Enrichment Analysis
4.6. Immune Status Analysis and Treatment Prediction
4.7. Prediction of Potential Therapeutic Agents in Patients with HCC
4.8. Cells and Animals
4.9. Migration Assay
4.10. Tube Formation Assays
4.11. Immunohistochemistry (IHC)
4.12. Flow Cytometry
4.13. Modeling HCC in Mice Using Subcutaneous Xenografts
4.14. cDNA Synthesis and Quantitative Real-Time PCR (qRT-PCR) Assay
4.15. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Craig, A.J.; von Felden, J.; Garcia-Lezana, T.; Sarcognato, S.; Villanueva, A. Tumour evolution in hepatocellular carcinoma. Nat. Rev. Gastroenterol. Hepatol. 2020, 17, 139–152. [Google Scholar] [CrossRef] [PubMed]
- Huang, D.Q.; El-Serag, H.B.; Loomba, R. Global epidemiology of NAFLD-related HCC: Trends, predictions, risk factors and prevention. Nat. Rev. Gastroenterol. Hepatol. 2021, 18, 223–238. [Google Scholar] [CrossRef] [PubMed]
- Singal, A.G.; Lampertico, P.; Nahon, P. Epidemiology and surveillance for hepatocellular carcinoma: New trends. J. Hepatol. 2020, 72, 250–261. [Google Scholar] [CrossRef] [PubMed]
- Huang, A.; Yang, X.R.; Chung, W.Y.; Dennison, A.R.; Zhou, J. Targeted therapy for hepatocellular carcinoma. Signal Transduct. Target. Ther. 2020, 5, 146. [Google Scholar] [CrossRef] [PubMed]
- Llovet, J.M.; Ricci, S.; Mazzaferro, V.; Hilgard, P.; Gane, E.; Blanc, J.F.; de Oliveira, A.C.; Santoro, A.; Raoul, J.L.; Forner, A.; et al. Sorafenib in advanced hepatocellular carcinoma. N. Engl. J. Med. 2008, 359, 378–390. [Google Scholar] [CrossRef]
- Llovet, J.M.; Kelley, R.K.; Villanueva, A.; Singal, A.G.; Pikarsky, E.; Roayaie, S.; Lencioni, R.; Koike, K.; Zucman-Rossi, J.; Finn, R.S. Hepatocellular carcinoma. Nat. Rev. Dis. Prim. 2021, 7, 6. [Google Scholar] [CrossRef] [PubMed]
- Emrich, S.J.; Barbazuk, W.B.; Li, L.; Schnable, P.S. Gene discovery and annotation using LCM-454 transcriptome sequencing. Genome Res. 2007, 17, 69–73. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Z.; Li, H.; Jiang, S.; Li, R.; Li, W.; Chen, H.; Bo, X. A survey and evaluation of Web-based tools/databases for variant analysis of TCGA data. Brief. Bioinform. 2019, 20, 1524–1541. [Google Scholar] [CrossRef]
- Hellmann, M.D.; Ciuleanu, T.E.; Pluzanski, A.; Lee, J.S.; Otterson, G.A.; Audigier-Valette, C.; Minenza, E.; Linardou, H.; Burgers, S.; Salman, P.; et al. Nivolumab plus Ipilimumab in Lung Cancer with a High Tumor Mutational Burden. N. Engl. J. Med. 2018, 378, 2093–2104. [Google Scholar] [CrossRef]
- Jardim, D.L.; Goodman, A.; de Melo Gagliato, D.; Kurzrock, R. The Challenges of Tumor Mutational Burden as an Immunotherapy Biomarker. Cancer Cell 2021, 39, 154–173. [Google Scholar] [CrossRef]
- Rebouissou, S.; Franconi, A.; Calderaro, J.; Letouze, E.; Imbeaud, S.; Pilati, C.; Nault, J.C.; Couchy, G.; Laurent, A.; Balabaud, C.; et al. Genotype-phenotype correlation of CTNNB1 mutations reveals different ss-catenin activity associated with liver tumor progression. Hepatology 2016, 64, 2047–2061. [Google Scholar] [CrossRef] [PubMed]
- He, X.; Xu, C. Immune checkpoint signaling and cancer immunotherapy. Cell Res. 2020, 30, 660–669. [Google Scholar] [CrossRef] [PubMed]
- Charoentong, P.; Finotello, F.; Angelova, M.; Mayer, C.; Efremova, M.; Rieder, D.; Hackl, H.; Trajanoski, Z. Pan-cancer Immunogenomic Analyses Reveal Genotype-Immunophenotype Relationships and Predictors of Response to Checkpoint Blockade. Cell Rep. 2017, 18, 248–262. [Google Scholar] [CrossRef] [PubMed]
- Sato, K.; Tanaka, S.; Mitsunori, Y.; Mogushi, K.; Yasen, M.; Aihara, A.; Ban, D.; Ochiai, T.; Irie, T.; Kudo, A.; et al. Contrast-enhanced intraoperative ultrasonography for vascular imaging of hepatocellular carcinoma: Clinical and biological significance. Hepatology 2013, 57, 1436–1447. [Google Scholar] [CrossRef]
- Zhu, A.X.; Duda, D.G.; Sahani, D.V.; Jain, R.K. HCC and angiogenesis: Possible targets and future directions. Nat. Rev. Clin. Oncol. 2011, 8, 292–301. [Google Scholar] [CrossRef]
- Zhang, M.S.; Cui, J.D.; Lee, D.; Yuen, V.W.; Chiu, D.K.; Goh, C.C.; Cheu, J.W.; Tse, A.P.; Bao, M.H.; Wong, B.P.Y.; et al. Hypoxia-induced macropinocytosis represents a metabolic route for liver cancer. Nat. Commun. 2022, 13, 954. [Google Scholar] [CrossRef]
- Wu, Q.; Zhou, W.; Yin, S.; Zhou, Y.; Chen, T.; Qian, J.; Su, R.; Hong, L.; Lu, H.; Zhang, F.; et al. Blocking Triggering Receptor Expressed on Myeloid Cells-1-Positive Tumor-Associated Macrophages Induced by Hypoxia Reverses Immunosuppression and Anti-Programmed Cell Death Ligand 1 Resistance in Liver Cancer. Hepatology 2019, 70, 198–214. [Google Scholar] [CrossRef]
- Khan, K.A.; Kerbel, R.S. Improving immunotherapy outcomes with anti-angiogenic treatments and vice versa. Nat. Rev. Clin. Oncol. 2018, 15, 310–324. [Google Scholar] [CrossRef]
- Tian, L.; Goldstein, A.; Wang, H.; Ching Lo, H.; Sun Kim, I.; Welte, T.; Sheng, K.; Dobrolecki, L.E.; Zhang, X.; Putluri, N.; et al. Mutual regulation of tumour vessel normalization and immunostimulatory reprogramming. Nature 2017, 544, 250–254. [Google Scholar] [CrossRef]
- Zhao, Y.; Zhang, Y.N.; Wang, K.T.; Chen, L. Lenvatinib for hepatocellular carcinoma: From preclinical mechanisms to anti-cancer therapy. Biochim. Biophys. Acta Rev. Cancer 2020, 1874, 188391. [Google Scholar] [CrossRef]
- Yi, C.; Chen, L.; Lin, Z.; Liu, L.; Shao, W.; Zhang, R.; Lin, J.; Zhang, J.; Zhu, W.; Jia, H.; et al. Lenvatinib Targets FGF Receptor 4 to Enhance Antitumor Immune Response of Anti-Programmed Cell Death-1 in HCC. Hepatology 2021, 74, 2544–2560. [Google Scholar] [CrossRef] [PubMed]
- Finn, R.S.; Ikeda, M.; Zhu, A.X.; Sung, M.W.; Baron, A.D.; Kudo, M.; Okusaka, T.; Kobayashi, M.; Kumada, H.; Kaneko, S.; et al. Phase Ib Study of Lenvatinib Plus Pembrolizumab in Patients with Unresectable Hepatocellular Carcinoma. J. Clin. Oncol. 2020, 38, 2960–2970. [Google Scholar] [CrossRef]
- Torrens, L.; Montironi, C.; Puigvehi, M.; Mesropian, A.; Leslie, J.; Haber, P.K.; Maeda, M.; Balaseviciute, U.; Willoughby, C.E.; Abril-Fornaguera, J.; et al. Immunomodulatory Effects of Lenvatinib Plus Anti-Programmed Cell Death Protein 1 in Mice and Rationale for Patient Enrichment in Hepatocellular Carcinoma. Hepatology 2021, 74, 2652–2669. [Google Scholar] [CrossRef] [PubMed]
- Deng, H.; Kan, A.; Lyu, N.; Mu, L.; Han, Y.; Liu, L.; Zhang, Y.; Duan, Y.; Liao, S.; Li, S.; et al. Dual Vascular Endothelial Growth Factor Receptor and Fibroblast Growth Factor Receptor Inhibition Elicits Antitumor Immunity and Enhances Programmed Cell Death-1 Checkpoint Blockade in Hepatocellular Carcinoma. Liver Cancer 2020, 9, 338–357. [Google Scholar] [CrossRef] [PubMed]
- Okkenhaug, K.; Graupera, M.; Vanhaesebroeck, B. Targeting PI3K in Cancer: Impact on Tumor Cells, Their Protective Stroma, Angiogenesis, and Immunotherapy. Cancer Discov. 2016, 6, 1090–1105. [Google Scholar] [CrossRef]
- Murillo, M.M.; Zelenay, S.; Nye, E.; Castellano, E.; Lassailly, F.; Stamp, G.; Downward, J. RAS interaction with PI3K p110alpha is required for tumor-induced angiogenesis. J. Clin. Investig. 2014, 124, 3601–3611. [Google Scholar] [CrossRef] [PubMed]
- Soler, A.; Serra, H.; Pearce, W.; Angulo, A.; Guillermet-Guibert, J.; Friedman, L.S.; Vinals, F.; Gerhardt, H.; Casanovas, O.; Graupera, M.; et al. Inhibition of the p110alpha isoform of PI 3-kinase stimulates nonfunctional tumor angiogenesis. J. Exp. Med. 2013, 210, 1937–1945. [Google Scholar] [CrossRef]
- Polverino, F.; Naso, F.D.; Asteriti, I.A.; Palmerini, V.; Singh, D.; Valente, D.; Bird, A.W.; Rosa, A.; Mapelli, M.; Guarguaglini, G. The Aurora-A/TPX2 Axis Directs Spindle Orientation in Adherent Human Cells by Regulating NuMA and Microtubule Stability. Curr. Biol. 2021, 31, 658–667.e655. [Google Scholar] [CrossRef]
- Huang, D.H.; Jian, J.; Li, S.; Zhang, Y.; Liu, L.Z. TPX2 silencing exerts antitumor effects on hepatocellular carcinoma by regulating the PI3K/AKT signaling pathway. Int. J. Mol. Med. 2019, 44, 2113–2122. [Google Scholar] [CrossRef]
- Huang, Y.; Wang, H.; Lian, Y.; Wu, X.; Zhou, L.; Wang, J.; Deng, M.; Huang, Y. Upregulation of kinesin family member 4A enhanced cell proliferation via activation of Akt signaling and predicted a poor prognosis in hepatocellular carcinoma. Cell Death Dis. 2018, 9, 141. [Google Scholar] [CrossRef]
- Lee, Y.M.; Lee, S.; Lee, E.; Shin, H.; Hahn, H.; Choi, W.; Kim, W. Human kinesin superfamily member 4 is dominantly localized in the nuclear matrix and is associated with chromosomes during mitosis. Biochem. J. 2001, 360, 549–556. [Google Scholar] [CrossRef] [PubMed]
- Yuen, H.F.; Chan, K.K.; Grills, C.; Murray, J.T.; Platt-Higgins, A.; Eldin, O.S.; O’Byrne, K.; Janne, P.; Fennell, D.A.; Johnston, P.G.; et al. Ran is a potential therapeutic target for cancer cells with molecular changes associated with activation of the PI3K/Akt/mTORC1 and Ras/MEK/ERK pathways. Clin. Cancer Res. 2012, 18, 380–391. [Google Scholar] [CrossRef] [PubMed]
- Tan, Y.; Zhang, Y.; Zhou, Q.; Jia, D.; Sun, Q. Design and structural characterization of autoinhibition-compromised full-length Ran. Signal Transduct. Target. Ther. 2021, 6, 44. [Google Scholar] [CrossRef] [PubMed]
- Near, R.I.; Zhang, Y.; Makkinje, A.; Vanden Borre, P.; Lerner, A. AND-34/BCAR3 differs from other NSP homologs in induction of anti-estrogen resistance, cyclin D1 promoter activation and altered breast cancer cell morphology. J. Cell. Physiol. 2007, 212, 655–665. [Google Scholar] [CrossRef] [PubMed]
- Ni, J.; Wang, J.; Fu, Y.; Yan, C.; Zhu, M.; Jiang, Y.; Chen, J.; Ding, Y.; Fan, X.; Li, G.; et al. Functional genetic variants in centrosome-related genes CEP72 and YWHAG confer susceptibility to gastric cancer. Arch. Toxicol. 2020, 94, 2861–2872. [Google Scholar] [CrossRef]
- Vanhaesebroeck, B.; Perry, M.W.D.; Brown, J.R.; Andre, F.; Okkenhaug, K. PI3K inhibitors are finally coming of age. Nat. Rev. Drug Discov. 2021, 20, 741–769. [Google Scholar] [CrossRef]
- Sun, H.X.; Xu, Y.; Yang, X.R.; Wang, W.M.; Bai, H.; Shi, R.Y.; Nayar, S.K.; Devbhandari, R.P.; He, Y.Z.; Zhu, Q.F.; et al. Hypoxia inducible factor 2 alpha inhibits hepatocellular carcinoma growth through the transcription factor dimerization partner 3/E2F transcription factor 1-dependent apoptotic pathway. Hepatology 2013, 57, 1088–1097. [Google Scholar] [CrossRef]
- Herraez, E.; Lozano, E.; Macias, R.I.; Vaquero, J.; Bujanda, L.; Banales, J.M.; Marin, J.J.; Briz, O. Expression of SLC22A1 variants may affect the response of hepatocellular carcinoma and cholangiocarcinoma to sorafenib. Hepatology 2013, 58, 1065–1073. [Google Scholar] [CrossRef]
- Weng, S.; Zhou, L.; Deng, Q.; Wang, J.; Yu, Y.; Zhu, J.; Yuan, Y. Niclosamide induced cell apoptosis via upregulation of ATF3 and activation of PERK in Hepatocellular carcinoma cells. BMC Gastroenterol. 2016, 16, 25. [Google Scholar] [CrossRef]
- Li, Y.; Li, B.; Xu, Y.; Qian, L.; Xu, T.; Meng, G.; Li, H.; Wang, Y.; Zhang, L.; Jiang, X.; et al. GOT2 Silencing Promotes Reprogramming of Glutamine Metabolism and Sensitizes Hepatocellular Carcinoma to Glutaminase Inhibitors. Cancer Res. 2022, 82, 3223–3235. [Google Scholar] [CrossRef]
- Stelzer, G.; Rosen, N.; Plaschkes, I.; Zimmerman, S.; Twik, M.; Fishilevich, S.; Stein, T.I.; Nudel, R.; Lieder, I.; Mazor, Y.; et al. The GeneCards Suite: From Gene Data Mining to Disease Genome Sequence Analyses. Curr. Protoc. Bioinform. 2016, 54, 1.30.1–1.30.33. [Google Scholar] [CrossRef] [PubMed]
- Mayakonda, A.; Lin, D.C.; Assenov, Y.; Plass, C.; Koeffler, H.P. Maftools: Efficient and comprehensive analysis of somatic variants in cancer. Genome Res. 2018, 28, 1747–1756. [Google Scholar] [CrossRef]
- Zhou, Y.; Zhou, B.; Pache, L.; Chang, M.; Khodabakhshi, A.H.; Tanaseichuk, O.; Benner, C.; Chanda, S.K. Metascape provides a biologist-oriented resource for the analysis of systems-level datasets. Nat. Commun. 2019, 10, 1523. [Google Scholar] [CrossRef] [PubMed]
- Yu, G.; Wang, L.G.; Han, Y.; He, Q.Y. clusterProfiler: An R package for comparing biological themes among gene clusters. OMICS 2012, 16, 284–287. [Google Scholar] [CrossRef] [PubMed]

| Symbol | Forward Primer | Reverse Primer |
|---|---|---|
| TPX2 | ATGGAACTGGAGGGCTTTTTC | TGTTGTCAACTGGTTTCAAAGGT |
| KIF4A | TACTGCGGTGGAGCAAGAAG | CATCTGCGCTTGACGGAGAG |
| RAB11A | CAACAAGAAGCATCCAGGTTGA | GCACCTACAGCTCCACGATAAT |
| UBE2S | ACAAGGAGGTGACGACACTGA | CCACGTTCGGGTGGAAGAT |
| GPLD1 | ATGTCTGCTTTCAGGTTGTGG | ACGCATCCTGGTGTTCTAGTAA |
| EPAS1 | CGGAGGTGTTCTATGAGCTGG | AGCTTGTGTGTTCGCAGGAA |
| GOT2 | AAGAGGGACACCAATAGCAAAAA | GCAGAACGTAAGGCTTTCCAT |
| KDR | GGCCCAATAATCAGAGTGGCA | CCAGTGTCATTTCCGATCACTTT |
| RAN | GGTGGTACTGGAAAAACGACC | CCCAAGGTGGCTACATACTTCT |
| BCAR3 | CAGAAACATGCCGGTGAATCA | GTGGGGATTTGGAGTGGGG |
| ATF3 | CCTCTGCGCTGGAATCAGTC | TTCTTTCTCGTCGCCTCTTTTT |
| KLKB1 | TCCTTGTTTGCTACAGTTTCCTG | TCTGGCAGTATTGGGCATTTG |
| YWHAG | AGCCACTGTCGAATGAGGAAC | CTGCTCAATGCTACTGATGACC |
| SLC22A1 | ACGGTGGCGATCATGTACC | CCCATTCTTTTGAGCGATGTGG |
| GAPDH(reference gene) | GGAGCGAGATCCCTCCAAAAT | GGCTGTTGTCATACTTCTCATGG |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ci, H.; Wang, X.; Shen, K.; Du, W.; Zhou, J.; Fu, Y.; Dong, Q.; Jia, H. An Angiogenic Gene Signature for Prediction of the Prognosis and Therapeutic Responses of Hepatocellular Carcinoma. Int. J. Mol. Sci. 2023, 24, 3324. https://doi.org/10.3390/ijms24043324
Ci H, Wang X, Shen K, Du W, Zhou J, Fu Y, Dong Q, Jia H. An Angiogenic Gene Signature for Prediction of the Prognosis and Therapeutic Responses of Hepatocellular Carcinoma. International Journal of Molecular Sciences. 2023; 24(4):3324. https://doi.org/10.3390/ijms24043324
Chicago/Turabian StyleCi, Hongfei, Xufeng Wang, Keyu Shen, Wei Du, Jiaming Zhou, Yan Fu, Qiongzhu Dong, and Huliang Jia. 2023. "An Angiogenic Gene Signature for Prediction of the Prognosis and Therapeutic Responses of Hepatocellular Carcinoma" International Journal of Molecular Sciences 24, no. 4: 3324. https://doi.org/10.3390/ijms24043324
APA StyleCi, H., Wang, X., Shen, K., Du, W., Zhou, J., Fu, Y., Dong, Q., & Jia, H. (2023). An Angiogenic Gene Signature for Prediction of the Prognosis and Therapeutic Responses of Hepatocellular Carcinoma. International Journal of Molecular Sciences, 24(4), 3324. https://doi.org/10.3390/ijms24043324






