Cardiac-Specific Expression of Cre Recombinase Leads to Age-Related Cardiac Dysfunction Associated with Tumor-like Growth of Atrial Cardiomyocyte and Ventricular Fibrosis and Ferroptosis
Abstract
1. Introduction
2. Results
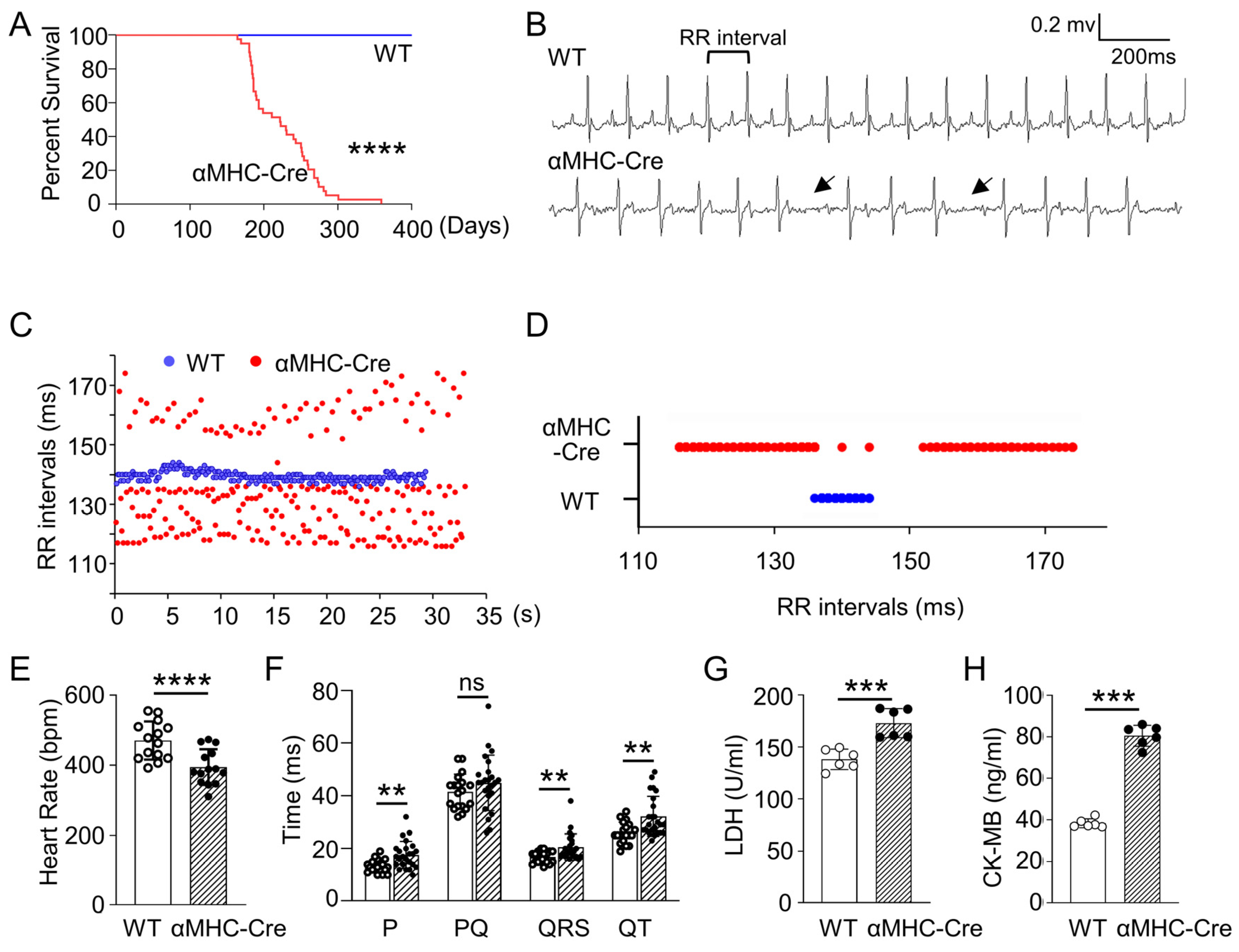
2.1. Age-Related Progressive Death Accompanied with Arrhythmia and Heart Injury in αMHC-Cre Transgenic Mice
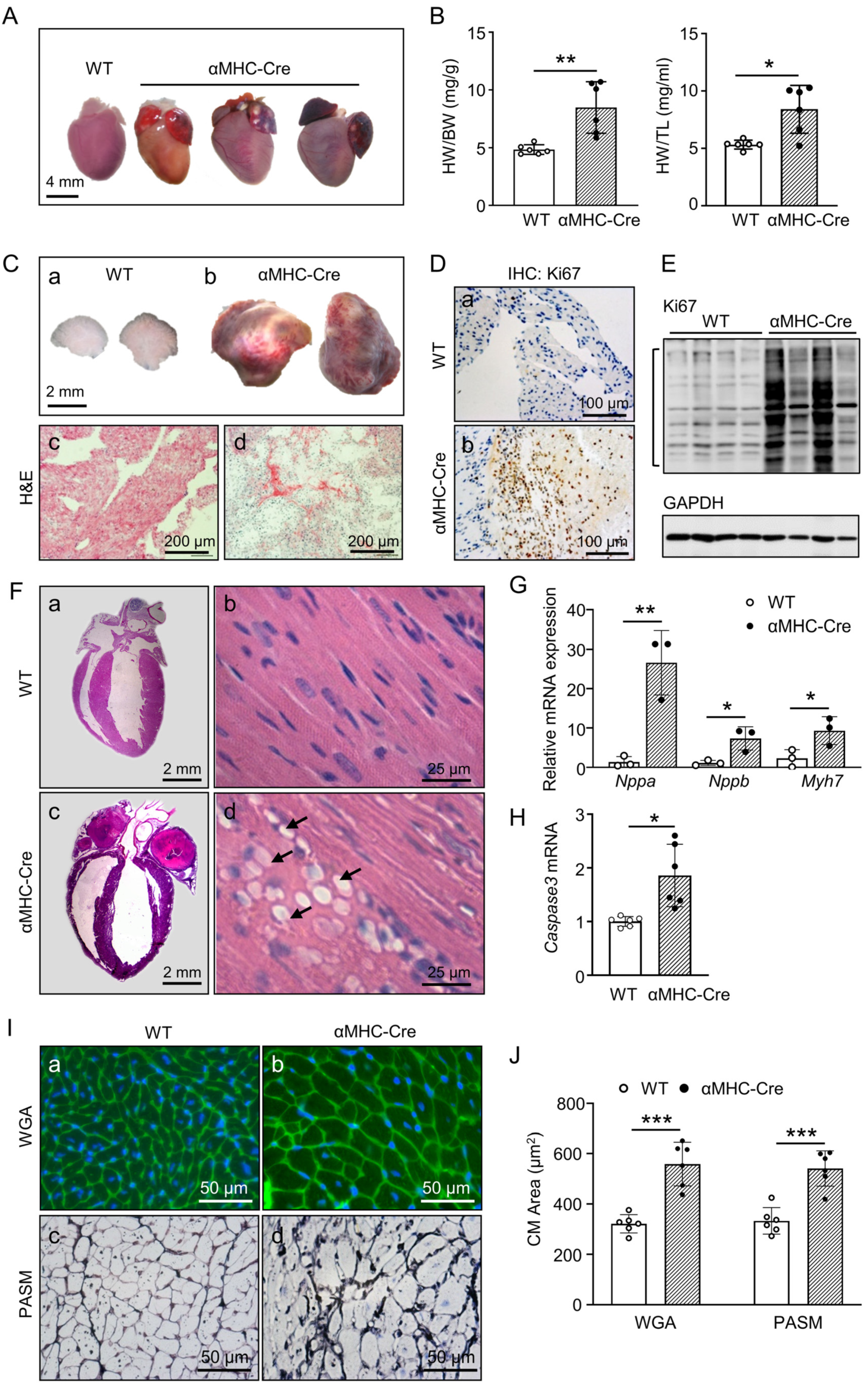
2.2. Atrial Tumor-like growth in αMHC-Cre Mice Led to Blockage of Blood Circulation and Heart Failure Bump
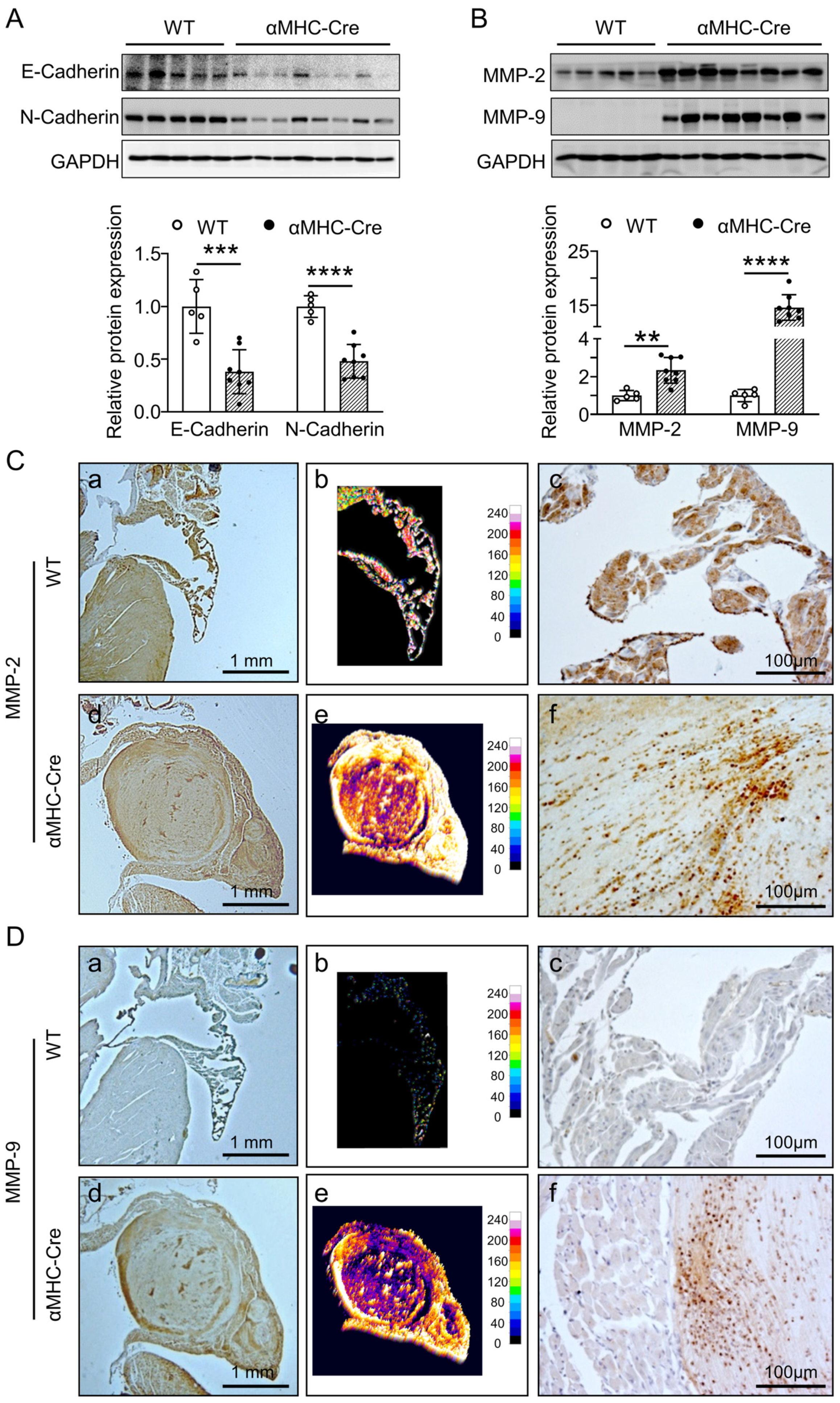
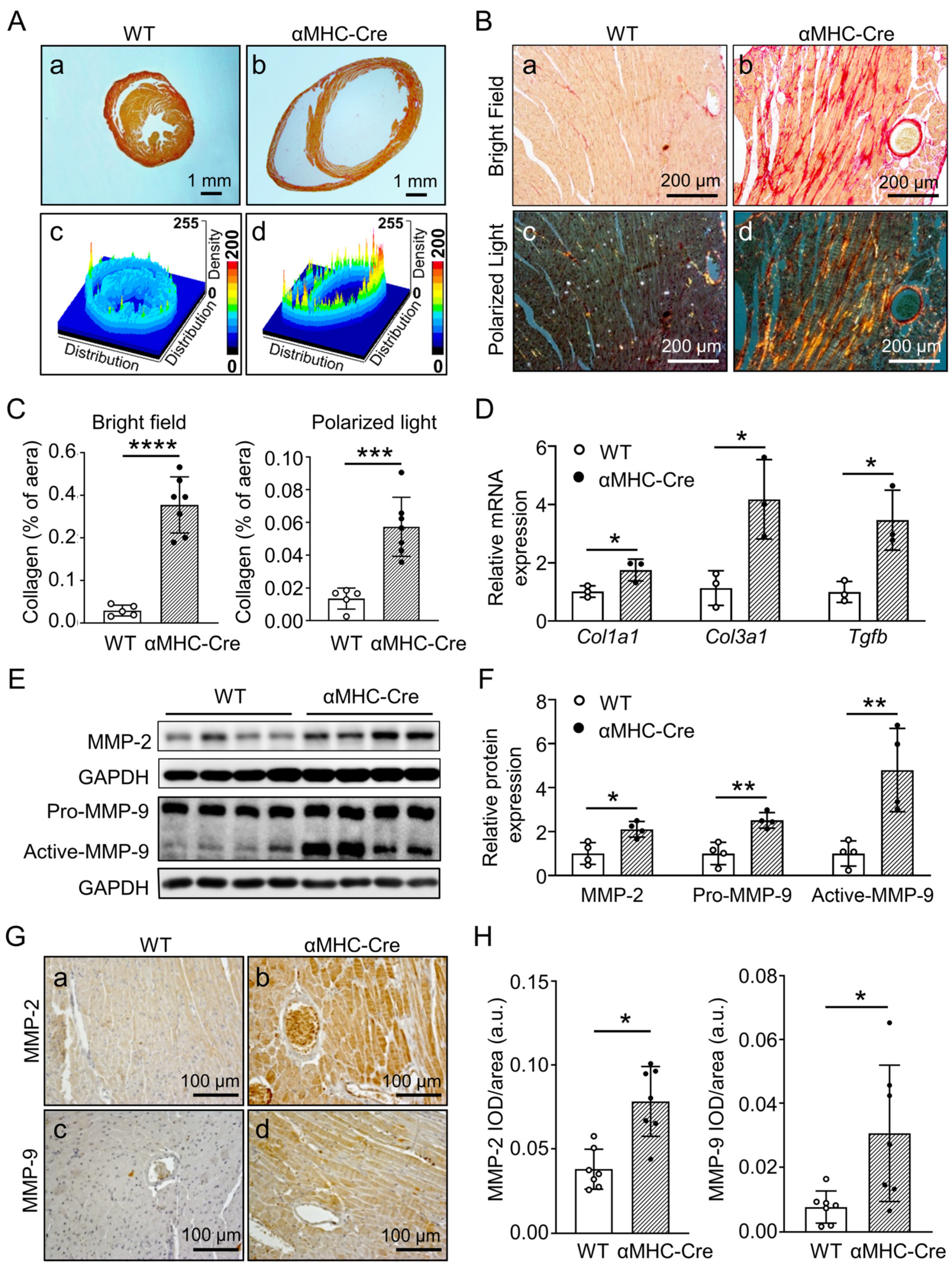
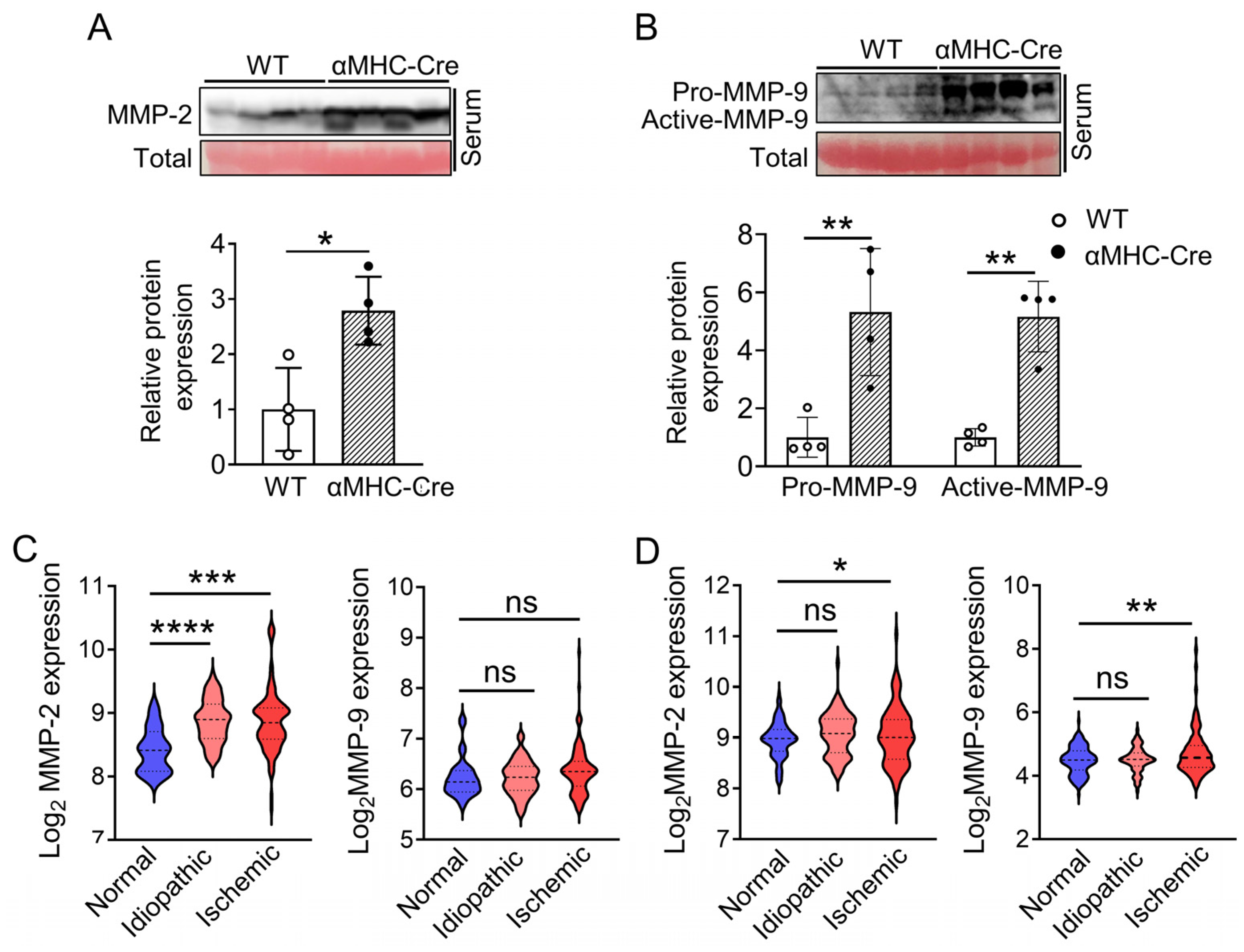
2.3. α. MHC-Cre Mice Developed Severe Cardiac Fibrosis with Enhanced MMP-2 and MMP-9 Level
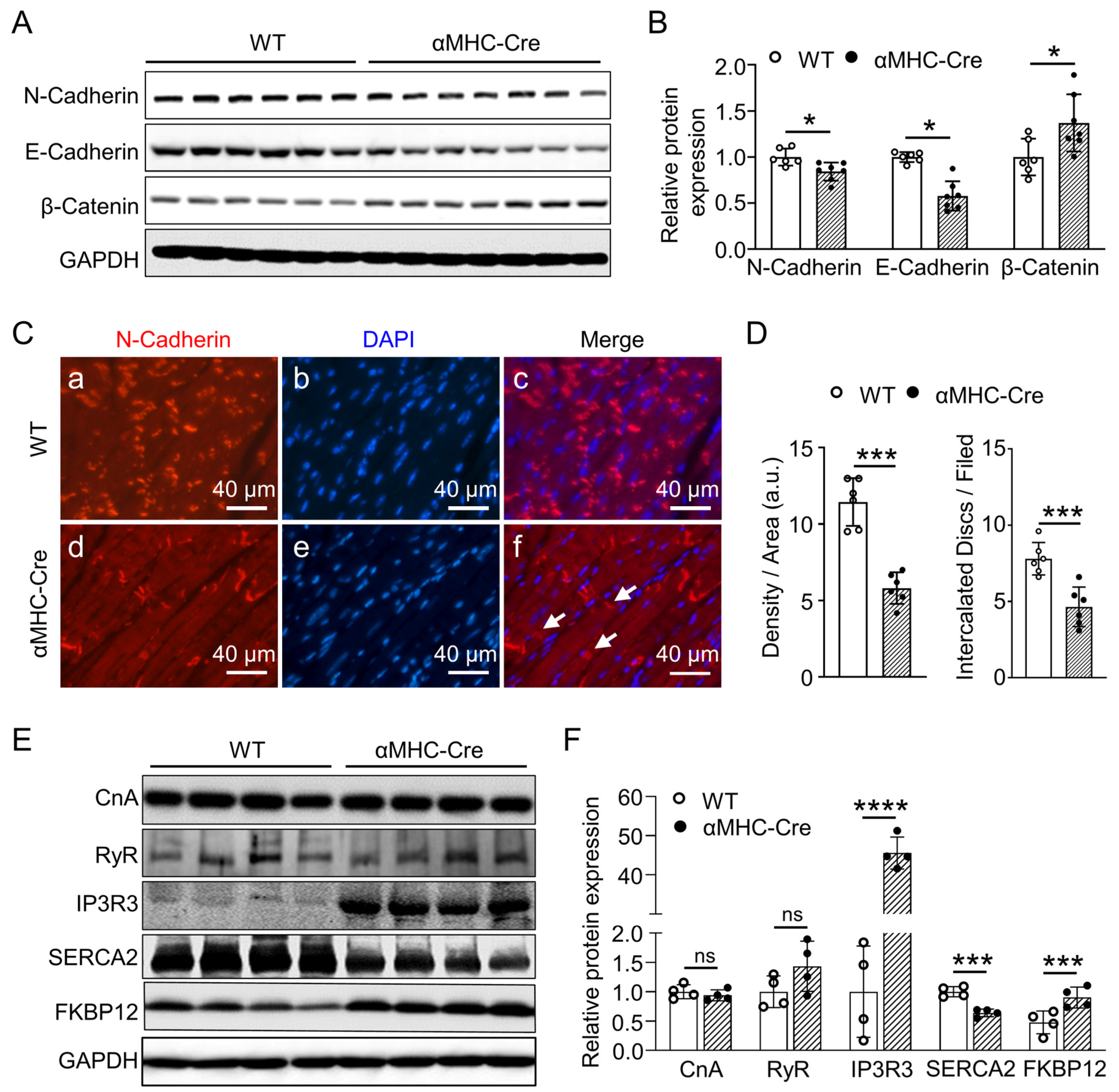
2.4. Alternation in Proteins Expression Associated with Intercalated Disc and Calcium Handling of αMHC-Cre Mice
2.5. Ferroptosis Signaling Pathway Is Involved in Heart Failure Caused by Cardiac-Specific Expression of Cre Recombinase
3. Discussion
4. Materials and Methods
4.1. α. MHC-Cre Transgenic Strains and Animal Care
4.2. α. MHC-Cre Transgenic Strains and Genotype Identification
4.3. Electrocardiography (ECG)
4.4. Measurement of Serum LDH and CK-MB
4.5. Histological Evaluations, Immunohistochemistry and Immunofluorescence
4.6. Quantitative Real-Time PCR
4.7. Western Blot
4.8. Analysis on Expression of MMP-2 and MMP-9 in Human Heart Failure
4.9. Statistical Analysis
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Kim, H.; Kim, M.; Im, S.K.; Fang, S. Mouse Cre-LoxP system: General principles to determine tissue-specific roles of target genes. Lab. Anim. Res. 2018, 34, 147–159. [Google Scholar] [CrossRef]
- Zhang, S.; Li, Y.; Huang, X.; Liu, K.; Wang, Q.D.; Chen, A.F.; Sun, K.; Lui, K.O.; Zhou, B. Seamless Genetic Recording of Transiently Activated Mesenchymal Gene Expression in Endothelial Cells During Cardiac Fibrosis. Circulation 2021, 144, 2004–2020. [Google Scholar] [CrossRef] [PubMed]
- Jakab, M.; Rostalski, T.; Lee, K.H.; Mogler, C.; Augustin, H.G. Tie2 Receptor in Tumor-Infiltrating Macrophages Is Dispensable for Tumor Angiogenesis and Tumor Relapse after Chemotherapy. Cancer Res. 2022, 82, 1353–1364. [Google Scholar] [CrossRef] [PubMed]
- Keuls, R.A.; Parchem, R.J. Single-Cell Multiomic Approaches Reveal Diverse Labeling of the Nervous System by Common Cre-Drivers. Front. Cell. Neurosci. 2021, 15, 648570. [Google Scholar] [CrossRef] [PubMed]
- Sauer, B.; Henderson, N. Targeted insertion of exogenous DNA into the eukaryotic genome by the Cre recombinase. New Biol. 1990, 2, 441–449. [Google Scholar] [PubMed]
- Agah, R.; Frenkel, P.A.; French, B.A.; Michael, L.H.; Overbeek, P.A.; Schneider, M.D. Gene recombination in postmitotic cells. Targeted expression of Cre recombinase provokes cardiac-restricted, site-specific rearrangement in adult ventricular muscle in vivo. J. Clin. Invest. 1997, 100, 169–179. [Google Scholar] [CrossRef] [PubMed]
- Nagy, A. Cre recombinase: The universal reagent for genome tailoring. Genesis 2000, 26, 99–109. [Google Scholar] [CrossRef]
- Minamino, T.; Gaussin, V.; DeMayo, F.J.; Schneider, M.D. Inducible gene targeting in postnatal myocardium by cardiac-specific expression of a hormone-activated Cre fusion protein. Circ. Res. 2001, 88, 587–592. [Google Scholar] [CrossRef]
- Sohal, D.S.; Nghiem, M.; Crackower, M.A.; Witt, S.A.; Kimball, T.R.; Tymitz, K.M.; Penninger, J.M.; Molkentin, J.D. Temporally regulated and tissue-specific gene manipulations in the adult and embryonic heart using a tamoxifen-inducible Cre protein. Circ Res. 2001, 89, 20–25. [Google Scholar] [CrossRef]
- Hoesl, E.; Stieber, J.; Herrmann, S.; Feil, S.; Tybl, E.; Hofmann, F.; Feil, R.; Ludwig, A. Tamoxifen-inducible gene deletion in the cardiac conduction system. J. Mol. Cell. Cardiol. 2008, 45, 62–69. [Google Scholar] [CrossRef]
- Maruyama, M.; Li, B.Y.; Chen, H.; Xu, X.; Song, L.S.; Guatimosim, S.; Zhu, W.; Yong, W.; Zhang, W.; Bu, G.; et al. FKBP12 is a critical regulator of the heart rhythm and the cardiac voltage-gated sodium current in mice. Circ Res. 2011, 108, 1042–1052. [Google Scholar] [CrossRef]
- Heine, H.L.; Leong, H.S.; Rossi, F.M.; McManus, B.M.; Podor, T.J. Strategies of conditional gene expression in myocardium: An overview. Methods Mol. Med. 2005, 112, 109–154. [Google Scholar] [CrossRef] [PubMed]
- Silver, D.P.; Livingston, D.M. Self-excising retroviral vectors encoding the Cre recombinase overcome Cre-mediated cellular toxicity. Mol. Cell. 2001, 8, 233–243. [Google Scholar] [CrossRef] [PubMed]
- Naiche, L.A.; Papaioannou, V.E. Cre activity causes widespread apoptosis and lethal anemia during embryonic development. Genesis 2007, 45, 768–775. [Google Scholar] [CrossRef] [PubMed]
- Schmidt, E.E.; Taylor, D.S.; Prigge, J.R.; Barnett, S.; Capecchi, M.R. Illegitimate Cre-dependent chromosome rearrangements in transgenic mouse spermatids. Proc. Natl. Acad. Sci. USA 2000, 97, 13702–13707. [Google Scholar] [CrossRef] [PubMed]
- Schmidt-Supprian, M.; Rajewsky, K. Vagaries of conditional gene targeting. Nat. Immunol. 2007, 8, 665–668. [Google Scholar] [CrossRef]
- Wang, X.; Lauth, A.; Wan, T.C.; Lough, J.W.; Auchampach, J.A. Myh6-driven Cre recombinase activates the DNA damage response and the cell cycle in the myocardium in the absence of loxP sites. Dis. Model. Mech. 2020, 13, dmm046375. [Google Scholar] [CrossRef]
- Loonstra, A.; Vooijs, M.; Beverloo, H.B.; Allak, B.A.; van Drunen, E.; Kanaar, R.; Berns, A.; Jonkers, J. Growth inhibition and DNA damage induced by Cre recombinase in mammalian cells. Proc. Natl. Acad. Sci. USA 2001, 98, 9209–9214. [Google Scholar] [CrossRef]
- Huh, W.J.; Mysorekar, I.U.; Mills, J.C. Inducible activation of Cre recombinase in adult mice causes gastric epithelial atrophy, metaplasia, and regenerative changes in the absence of “floxed” alleles. Am. J. Physiol. Gastrointest. Liver Physiol. 2010, 299, G368–G380. [Google Scholar] [CrossRef]
- Buerger, A.; Rozhitskaya, O.; Sherwood, M.C.; Dorfman, A.L.; Bisping, E.; Abel, E.D.; Pu, W.T.; Izumo, S.; Jay, P.Y. Dilated cardiomyopathy resulting from high-level myocardial expression of Cre-recombinase. J. Card. Fail. 2006, 12, 392–398. [Google Scholar] [CrossRef]
- Pugach, E.K.; Richmond, P.A.; Azofeifa, J.G.; Dowell, R.D.; Leinwand, L.A. Prolonged Cre expression driven by the α-myosin heavy chain promoter can be cardiotoxic. J. Mol. Cell. Cardiol. 2015, 86, 54–61. [Google Scholar] [CrossRef]
- Rehmani, T.; Salih, M.; Tuana, B.S. Cardiac-Specific Cre Induces Age-Dependent Dilated Cardiomyopathy (DCM) in Mice. Molecules 2019, 24, 1189. [Google Scholar] [CrossRef]
- Hougen, K.; Aronsen, J.M.; Stokke, M.K.; Enger, U.; Nygard, S.; Andersson, K.B.; Christensen, G.; Sejersted, O.M.; Sjaastad, I. Cre-loxP DNA recombination is possible with only minimal unspecific transcriptional changes and without cardiomyopathy in Tg(alphaMHC-MerCreMer) mice. American journal of physiology. Am. J. Physiol. Heart Circ. Physiol. 2010, 299, H1671–H1678. [Google Scholar] [CrossRef] [PubMed]
- Hall, M.E.; Smith, G.; Hall, J.E.; Stec, D.E. Systolic dysfunction in cardiac-specific ligand-inducible MerCreMer transgenic mice. American journal of physiology. Am. J. Physiol. Heart Circ. Physiol. 2011, 301, H253–H260. [Google Scholar] [CrossRef] [PubMed]
- Lexow, J.; Poggioli, T.; Sarathchandra, P.; Santini, M.P.; Rosenthal, N. Cardiac fibrosis in mice expressing an inducible myocardial-specific Cre driver. Dis. Model. Mech. 2013, 6, 1470–1476. [Google Scholar] [CrossRef]
- Koitabashi, N.; Bedja, D.; Zaiman, A.L.; Pinto, Y.M.; Zhang, M.; Gabrielson, K.L.; Takimoto, E.; Kass, D.A. Avoidance of transient cardiomyopathy in cardiomyocyte-targeted tamoxifen-induced MerCreMer gene deletion models. Circ. Res. 2009, 105, 12–15. [Google Scholar] [CrossRef]
- Bersell, K.; Choudhury, S.; Mollova, M.; Polizzotti, B.D.; Ganapathy, B.; Walsh, S.; Wadugu, B.; Arab, S.; Kühn, B. Moderate and high amounts of tamoxifen in αMHC-MerCreMer mice induce a DNA damage response, leading to heart failure and death. Dis. Model. Mech. 2013, 6, 1459–1469. [Google Scholar] [CrossRef]
- Stockwell, B.R.; Friedmann Angeli, J.P.; Bayir, H.; Bush, A.I.; Conrad, M.; Dixon, S.J.; Fulda, S.; Gascón, S.; Hatzios, S.K.; Kagan, V.E.; et al. Ferroptosis: A Regulated Cell Death Nexus Linking Metabolism, Redox Biology, and Disease. Cell 2017, 171, 273–285. [Google Scholar] [CrossRef] [PubMed]
- Dixon, S.J.; Lemberg, K.M.; Lamprecht, M.R.; Skouta, R.; Zaitsev, E.M.; Gleason, C.E.; Patel, D.N.; Bauer, A.J.; Cantley, A.M.; Yang, W.S.; et al. Ferroptosis: An iron-dependent form of nonapoptotic cell death. Cell 2012, 149, 1060–1072. [Google Scholar] [CrossRef]
- Fang, X.; Wang, H.; Han, D.; Xie, E.; Yang, X.; Wei, J.; Gu, S.; Gao, F.; Zhu, N.; Yin, X.; et al. Ferroptosis as a target for protection against cardiomyopathy. Proc. Natl. Acad. Sci. USA 2019, 116, 2672–2680. [Google Scholar] [CrossRef]
- Bai, T.; Li, M.; Liu, Y.; Qiao, Z.; Wang, Z. Inhibition of ferroptosis alleviates atherosclerosis through attenuating lipid peroxidation and endothelial dysfunction in mouse aortic endothelial cell. Free Radic. Biol. Med. 2020, 160, 92–102. [Google Scholar] [CrossRef] [PubMed]
- Fang, X.; Cai, Z.; Wang, H.; Han, D.; Cheng, Q.; Zhang, P.; Gao, F.; Yu, Y.; Song, Z.; Wu, Q.; et al. Loss of Cardiac Ferritin H Facilitates Cardiomyopathy via Slc7a11-Mediated Ferroptosis. Circ. Res. 2020, 127, 486–501. [Google Scholar] [CrossRef]
- Liu, B.; Zhao, C.; Li, H.; Chen, X.; Ding, Y.; Xu, S. Puerarin protects against heart failure induced by pressure overload through mitigation of ferroptosis. Biochem. Biophys. Res. Commun. 2018, 497, 233–240. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.; Fan, H.; Wang, S.; Tang, G.; Zhai, C.; Shen, L. Ferroptosis: A Novel Therapeutic Target for Ischemia-Reperfusion Injury. Front. Cell Dev. Biol. 2021, 9, 688605. [Google Scholar] [CrossRef] [PubMed]
- Sales Gil, R.; Vagnarelli, P. Ki-67: More Hidden behind a Classic Proliferation Marker. Trends Biochem. Sci. 2018, 43, 747–748. [Google Scholar] [CrossRef] [PubMed]
- Jokinen, M.P.; Lieuallen, W.G.; Boyle, M.C.; Johnson, C.L.; Malarkey, D.E.; Nyska, A. Morphologic aspects of rodent cardiotoxicity in a retrospective evaluation of National Toxicology Program studies. Toxicol. Pathol. 2011, 39, 850–860. [Google Scholar] [CrossRef]
- Zhang, Y.; Weinberg, R.A. Epithelial-to-mesenchymal transition in cancer: Complexity and opportunities. Front. Med. 2018, 12, 361–373. [Google Scholar] [CrossRef]
- Loh, C.Y.; Chai, J.Y.; Tang, T.F.; Wong, W.F.; Sethi, G.; Shanmugam, M.K.; Chong, P.P.; Looi, C.Y. The E-Cadherin and N-Cadherin Switch in Epithelial-to-Mesenchymal Transition: Signaling, Therapeutic Implications, and Challenges. Cells 2019, 8, 1118. [Google Scholar] [CrossRef]
- Rhee, J.S.; Coussens, L.M. RECKing MMP function: Implications for cancer development. Trends Cell Biol. 2002, 12, 209–211. [Google Scholar] [CrossRef]
- Talman, V.; Ruskoaho, H. Cardiac fibrosis in myocardial infarction-from repair and remodeling to regeneration. Cell Tissue Res. 2016, 365, 563–581. [Google Scholar] [CrossRef] [PubMed]
- DeLeon-Pennell, K.Y.; Meschiari, C.A.; Jung, M.; Lindsey, M.L. Matrix Metalloproteinases in Myocardial Infarction and Heart Failure. Prog. Mol. Biol. Transl. Sci. 2017, 147, 75–100. [Google Scholar] [CrossRef] [PubMed]
- Lindsey, M.L. Assigning matrix metalloproteinase roles in ischaemic cardiac remodelling. Nat. Rev. Cardiol. 2018, 15, 471–479. [Google Scholar] [CrossRef]
- Giannandrea, M.; Parks, W.C. Diverse functions of matrix metalloproteinases during fibrosis. Dis. Model. Mech. 2014, 7, 193–203. [Google Scholar] [CrossRef]
- Radosinska, J.; Barancik, M.; Vrbjar, N. Heart failure and role of circulating MMP-2 and MMP-9. Panminerva Med. 2017, 59, 241–253. [Google Scholar] [CrossRef] [PubMed]
- Kostetskii, I.; Li, J.; Xiong, Y.; Zhou, R.; Ferrari, V.A.; Patel, V.V.; Molkentin, J.D.; Radice, G.L. Induced deletion of the N-cadherin gene in the heart leads to dissolution of the intercalated disc structure. Circ. Res. 2005, 96, 346–354. [Google Scholar] [CrossRef]
- Ferreira-Cornwell, M.C.; Luo, Y.; Narula, N.; Lenox, J.M.; Lieberman, M.; Radice, G.L. Remodeling the intercalated disc leads to cardiomyopathy in mice misexpressing cadherins in the heart. J. Cell Sci. 2002, 115, 1623–1634. [Google Scholar] [CrossRef]
- Ito, Y.; Yoshida, M.; Masuda, H.; Maeda, D.; Kudo-Asabe, Y.; Umakoshi, M.; Nanjo, H.; Goto, A. Disorganization of intercalated discs in dilated cardiomyopathy. Sci. Rep. 2021, 11, 11852. [Google Scholar] [CrossRef] [PubMed]
- Eisner, D.A.; Caldwell, J.L.; Kistamás, K.; Trafford, A.W. Calcium and Excitation-Contraction Coupling in the Heart. Circ. Res. 2017, 121, 181–195. [Google Scholar] [CrossRef]
- Gambardella, J.; Trimarco, B.; Iaccarino, G.; Santulli, G. New Insights in Cardiac Calcium Handling and Excitation-Contraction Coupling. Adv. Exp. Med. Biol. 2018, 1067, 373–385. [Google Scholar] [CrossRef]
- Bögeholz, N.; Muszynski, A.; Pott, C. The physiology of cardiac calcium handling. Wien. Med. Wochenschr. 2012, 162, 278–282. [Google Scholar] [CrossRef]
- Xu, X.; Balk, S.P.; Isaacs, W.B.; Ma, J. Calcium signaling: An underlying link between cardiac disease and carcinogenesis. Cell Biosci. 2018, 8, 39. [Google Scholar] [CrossRef]
- Rajewsky, K.; Gu, H.; Kühn, R.; Betz, U.A.; Müller, W.; Roes, J.; Schwenk, F. Conditional gene targeting. J. Clin. Investig. 1996, 98, 600–603. [Google Scholar] [CrossRef] [PubMed]
- Kos, C.H. Cre/loxP system for generating tissue-specific knockout mouse models. Nutr. Rev. 2004, 62, 243–246. [Google Scholar] [CrossRef]
- Higashi, A.Y.; Ikawa, T.; Muramatsu, M.; Economides, A.N.; Niwa, A.; Okuda, T.; Murphy, A.J.; Rojas, J.; Heike, T.; Nakahata, T.; et al. Direct hematological toxicity and illegitimate chromosomal recombination caused by the systemic activation of CreERT2. J. Immunol. 2009, 182, 5633–5640. [Google Scholar] [CrossRef] [PubMed]
- Hameyer, D.; Loonstra, A.; Eshkind, L.; Schmitt, S.; Antunes, C.; Groen, A.; Bindels, E.; Jonkers, J.; Krimpenfort, P.; Meuwissen, R.; et al. Toxicity of ligand-dependent Cre recombinases and generation of a conditional Cre deleter mouse allowing mosaic recombination in peripheral tissues. Physiol. Genom. 2007, 31, 32–41. [Google Scholar] [CrossRef] [PubMed]
- Forni, P.E.; Scuoppo, C.; Imayoshi, I.; Taulli, R.; Dastrù, W.; Sala, V.; Betz, U.A.; Muzzi, P.; Martinuzzi, D.; Vercelli, A.E.; et al. High levels of Cre expression in neuronal progenitors cause defects in brain development leading to microencephaly and hydrocephaly. J. Neurosci. 2006, 26, 9593–9602. [Google Scholar] [CrossRef]
- Lee, J.Y.; Ristow, M.; Lin, X.; White, M.F.; Magnuson, M.A.; Hennighausen, L. RIP-Cre revisited, evidence for impairments of pancreatic beta-cell function. J. Biol. Chem. 2006, 281, 2649–2653. [Google Scholar] [CrossRef]
- Radice, G.L. N-cadherin-mediated adhesion and signaling from development to disease: Lessons from mice. Prog. Mol. Biol. Transl. Sci. 2013, 116, 263–289. [Google Scholar] [CrossRef] [PubMed]
- Piven, O.O.; Kostetskii, I.E.; Macewicz, L.L.; Kolomiets, Y.M.; Radice, G.L.; Lukash, L.L. Requirement for N-cadherin-catenin complex in heart development. Exp. Biol. Med. 2011, 236, 816–822. [Google Scholar] [CrossRef]
- Prasad, V.; Lorenz, J.N.; Lasko, V.M.; Nieman, M.L.; Huang, W.; Wang, Y.; Wieczorek, D.W.; Shull, G.E. SERCA2 Haploinsufficiency in a Mouse Model of Darier Disease Causes a Selective Predisposition to Heart Failure. Biom. Res. Int. 2015, 2015, 251598. [Google Scholar] [CrossRef]
- Sayed, A.I. Gender Differences in Coronary Artery Disease, Clinical Characteristics, and Angiographic Features in the Jazan Region, Saudi Arabia. Cureus 2022, 14, e30239. [Google Scholar] [CrossRef]
- Skelding, K.A.; Boga, G.; Sartorius, J.; Wood, G.C.; Berger, P.B.; Mascarenhas, V.H.; Good, C.W.; Scott, T.D.; Blankenship, J.C. Frequency of coronary angiography and revascularization among men and women with myocardial infarction and their relationship to mortality at one year: An analysis of the Geisinger myocardial infarction cohort. J. Interv. Cardiol. 2013, 26, 14–21. [Google Scholar] [CrossRef]
- Minhas, A.; Cubero Salazar, I.; Kazzi, B.; Hays, A.G.; Choi, A.D.; Arbab-Zadeh, A.; Michos, E.D. Sex-Specific Plaque Signature: Uniqueness of Atherosclerosis in Women. Curr. Cardiol. Rep. 2021, 23, 84. [Google Scholar] [CrossRef]
- Xiao, Y.; Zeng, Z.; Guan, X.; Wang, L.; Wang, C.; Shi, H.; Shou, W.; Deng, K.; Xin, H. FKBP12.6 protects heart from AngII-induced hypertrophy through inhibiting Ca2+/calmodulin-mediated signalling pathways in vivo and in vitro. J. Cell. Mol. Med. 2018, 22, 3638–3651. [Google Scholar] [CrossRef] [PubMed]
- Lehnart, S.E.; Terrenoire, C.; Reiken, S.; Wehrens, X.H.; Song, L.; Tillman, E.J.; Mancarella, S.; Coromilas, J.; Lederer, W.J.; Kass, R.S.; et al. Stabilization of cardiac ryanodine receptor prevents intracellular calcium leak and arrhythmias. Proc. Natl. Acad. Sci. USA 2006, 103, 7906–7910. [Google Scholar] [CrossRef]
- Huang, F.; Shan, J.; Reiken, S.; Wehrens, X.H.; Marks, A.R. Analysis of calstabin2 (FKBP12.6)-ryanodine receptor interactions: Rescue of heart failure by calstabin2 in mice. Proc. Natl. Acad. Sci. USA 2006, 103, 3456–3461. [Google Scholar] [CrossRef]
- Yun, H.H.; Jung, S.Y.; Park, B.W.; Ko, J.S.; Yoo, K.; Yeo, J.; Kim, H.L.; Park, H.J.; Youn, H.J.; Lee, J.H. An Adult Mouse Model of Dilated Cardiomyopathy Caused by Inducible Cardiac-Specific Bis Deletion. Int. J. Mol. Sci. 2021, 22, 1343. [Google Scholar] [CrossRef]
- Kratsios, P.; Huth, M.; Temmerman, L.; Salimova, E.; Al Banchaabouchi, M.; Sgoifo, A.; Manghi, M.; Suzuki, K.; Rosenthal, N.; Mourkioti, F. Antioxidant amelioration of dilated cardiomyopathy caused by conditional deletion of NEMO/IKKgamma in cardiomyocytes. Circ. Res. 2010, 106, 133–144. [Google Scholar] [CrossRef]
- Kurosaka, S.; Leu, N.A.; Pavlov, I.; Han, X.; Ribeiro, P.A.; Xu, T.; Bunte, R.; Saha, S.; Wang, J.; Cornachione, A.; et al. Arginylation regulates myofibrils to maintain heart function and prevent dilated cardiomyopathy. J. Mol. Cell. Cardiol. 2012, 53, 333–341. [Google Scholar] [CrossRef] [PubMed]
- Zhao, J.; Yin, M.; Deng, H.; Jin, F.Q.; Xu, S.; Lu, Y.; Mastrangelo, M.A.; Luo, H.; Jin, Z.G. Cardiac Gab1 deletion leads to dilated cardiomyopathy associated with mitochondrial damage and cardiomyocyte apoptosis. Cell Death Differ. 2016, 23, 695–706. [Google Scholar] [CrossRef] [PubMed]
- Zheng, M.; Cheng, H.; Li, X.; Zhang, J.; Cui, L.; Ouyang, K.; Han, L.; Zhao, T.; Gu, Y.; Dalton, N.D.; et al. Cardiac-specific ablation of Cypher leads to a severe form of dilated cardiomyopathy with premature death. Hum. Mol. Genet. 2009, 18, 701–713. [Google Scholar] [CrossRef]
- Xiong, D.; He, H.; James, J.; Tokunaga, C.; Powers, C.; Huang, Y.; Osinska, H.; Towbin, J.A.; Purevjav, E.; Balschi, J.A.; et al. Cardiac-specific VLCAD deficiency induces dilated cardiomyopathy and cold intolerance. Am. J. Physiol. Heart Circ. Physiol. 2014, 306, H326–H338. [Google Scholar] [CrossRef]
- Jia, Y.; Chang, H.C.; Schipma, M.J.; Liu, J.; Shete, V.; Liu, N.; Sato, T.; Thorp, E.B.; Barger, P.M.; Zhu, Y.J.; et al. Cardiomyocyte-Specific Ablation of Med1 Subunit of the Mediator Complex Causes Lethal Dilated Cardiomyopathy in Mice. PLoS ONE 2016, 11, e0160755. [Google Scholar] [CrossRef]
- Saito, T.; Uchiumi, T.; Yagi, M.; Amamoto, R.; Setoyama, D.; Matsushima, Y.; Kang, D. Cardiomyocyte-specific loss of mitochondrial p32/C1qbp causes cardiomyopathy and activates stress responses. Cardiovasc. Res. 2017, 113, 1173–1185. [Google Scholar] [CrossRef] [PubMed]
- Balatskyi, V.V.; Macewicz, L.L.; Gan, A.M.; Goncharov, S.V.; Pawelec, P.; Portnichenko, G.V.; Lapikova-Bryginska, T.Y.; Navrulin, V.O.; Dosenko, V.E.; Olichwier, A.; et al. Cardiospecific deletion of αE-catenin leads to heart failure and lethality in mice. Pflugers. Arch. Eur. J. Physiol. 2018, 470, 1485–1499. [Google Scholar] [CrossRef]
- Rowe, G.C.; Asimaki, A.; Graham, E.L.; Martin, K.D.; Margulies, K.B.; Das, S.; Saffitz, J.; Arany, Z. Development of dilated cardiomyopathy and impaired calcium homeostasis with cardiac-specific deletion of ESRRβ. Am. J. Physiol. Heart Circ. Physiol. 2017, 312, H662–H671. [Google Scholar] [CrossRef]
- Ding, J.H.; Xu, X.; Yang, D.; Chu, P.H.; Dalton, N.D.; Ye, Z.; Yeakley, J.M.; Cheng, H.; Xiao, R.P.; Ross, J.; et al. Dilated cardiomyopathy caused by tissue-specific ablation of SC35 in the heart. EMBO J. 2004, 23, 885–896. [Google Scholar] [CrossRef]
- Harding, P.; Yang, X.P.; Yang, J.; Shesely, E.; He, Q.; LaPointe, M.C. Gene expression profiling of dilated cardiomyopathy in older male EP4 knockout mice. Am. J. Physiol. Heart Circ. Physiol. 2010, 298, H623–H632. [Google Scholar] [CrossRef] [PubMed]
- Myers, V.D.; Tomar, D.; Madesh, M.; Wang, J.; Song, J.; Zhang, X.Q.; Gupta, M.K.; Tahrir, F.G.; Gordon, J.; McClung, J.M.; et al. Haplo-insufficiency of Bcl2-associated athanogene 3 in mice results in progressive left ventricular dysfunction, β-adrenergic insensitivity, and increased apoptosis. J. Cell. Physiol. 2018, 233, 6319–6326. [Google Scholar] [CrossRef] [PubMed]
- Blaich, A.; Pahlavan, S.; Tian, Q.; Oberhofer, M.; Poomvanicha, M.; Lenhardt, P.; Domes, K.; Wegener, J.W.; Moosmang, S.; Ruppenthal, S.; et al. Mutation of the calmodulin binding motif IQ of the L-type Ca(v)1.2 Ca2+ channel to EQ induces dilated cardiomyopathy and death. J. Biol. Chem. 2012, 287, 22616–22625. [Google Scholar] [CrossRef] [PubMed]
- Cho, E.; Kang, H.; Kang, D.K.; Lee, Y. Myocardial-specific ablation of Jumonji and AT-rich interaction domain-containing 2 (Jarid2) leads to dilated cardiomyopathy in mice. J. Biol. Chem. 2019, 294, 4981–4996. [Google Scholar] [CrossRef]
- Jia, Y.; Liu, N.; Viswakarma, N.; Sun, R.; Schipma, M.J.; Shang, M.; Thorp, E.B.; Kanwar, Y.S.; Thimmapaya, B.; Reddy, J.K. PIMT/NCOA6IP Deletion in the Mouse Heart Causes Delayed Cardiomyopathy Attributable to Perturbation in Energy Metabolism. Int. J. Mol. Sci. 2018, 19, 1485. [Google Scholar] [CrossRef]
- Parlakian, A.; Charvet, C.; Escoubet, B.; Mericskay, M.; Molkentin, J.D.; Gary-Bobo, G.; De Windt, L.J.; Ludosky, M.A.; Paulin, D.; Daegelen, D.; et al. Temporally controlled onset of dilated cardiomyopathy through disruption of the SRF gene in adult heart. Circulation 2005, 112, 2930–2939. [Google Scholar] [CrossRef]
- Gozalo, A.S.; Zerfas, P.M.; Elkins, W.R.; Gieseck, R.L. Retrospective Study of Intercalated Disk Defects Associated with Dilated Cardiomyopathy, Atrial Thrombosis, and Heart Failure in BALB/c Mice Deficient in IL4 Receptor α. Comp. Med. 2020, 70, 266–276. [Google Scholar] [CrossRef]
- Kong, P.; Christia, P.; Frangogiannis, N.G. The pathogenesis of cardiac fibrosis. Cell. Mol. Life Sci. 2014, 71, 549–574. [Google Scholar] [CrossRef]
- Hijova, E. Matrix metalloproteinases: Their biological functions and clinical implications. Brati. Lek. Listy. 2005, 106, 127–132. [Google Scholar]
- Li, Y.Y.; Feldman, A.M. Matrix metalloproteinases in the progression of heart failure: Potential therapeutic implications. Drugs 2001, 61, 1239–1252. [Google Scholar] [CrossRef]
- Sheikh, F.; Ross, R.S.; Chen, J. Cell-cell connection to cardiac disease. Trends Cardiovasc. Med. 2009, 19, 182–190. [Google Scholar] [CrossRef]
- Siri-Angkul, N.; Dadfar, B.; Jaleel, R.; Naushad, J.; Parambathazhath, J.; Doye, A.A.; Xie, L.H.; Gwathmey, J.K. Calcium and Heart Failure: How Did We Get Here and Where Are We Going. Int. J. Mol. Sci. 2021, 22, 7392. [Google Scholar] [CrossRef]
- Ikeda, Y.; Hoshijima, M.; Chien, K.R. Toward biologically targeted therapy of calcium cycling defects in heart failure. Physiology 2008, 23, 6–16. [Google Scholar] [CrossRef]
- Marks, A.R. Calcium cycling proteins and heart failure: Mechanisms and therapeutics. J. Clin. Invest. 2013, 123, 46–52. [Google Scholar] [CrossRef]
- Dridi, H.; Kushnir, A.; Zalk, R.; Yuan, Q.; Melville, Z.; Marks, A.R. Intracellular calcium leak in heart failure and atrial fibrillation: A unifying mechanism and therapeutic target. Nat. Rev. Cardiol. 2020, 17, 732–747. [Google Scholar] [CrossRef]
- Lei, G.; Mao, C.; Yan, Y.; Zhuang, L.; Gan, B. Ferroptosis, radiotherapy, and combination therapeutic strategies. Protein Cell 2021, 12, 836–857. [Google Scholar] [CrossRef]
- Friedmann Angeli, J.P.; Schneider, M.; Proneth, B.; Tyurina, Y.Y.; Tyurin, V.A.; Hammond, V.J.; Herbach, N.; Aichler, M.; Walch, A.; Eggenhofer, E.; et al. Inactivation of the ferroptosis regulator Gpx4 triggers acute renal failure in mice. Nat. Cell Biol. 2014, 16, 1180–1191. [Google Scholar] [CrossRef]
- Yang, W.S.; SriRamaratnam, R.; Welsch, M.E.; Shimada, K.; Skouta, R.; Viswanathan, V.S.; Cheah, J.H.; Clemons, P.A.; Shamji, A.F.; Clish, C.B.; et al. Regulation of ferroptotic cancer cell death by GPX4. Cell 2014, 156, 317–331. [Google Scholar] [CrossRef]
- Koppula, P.; Zhuang, L.; Gan, B. Cystine transporter SLC7A11/xCT in cancer: Ferroptosis, nutrient dependency, and cancer therapy. Protein Cell 2021, 12, 599–620. [Google Scholar] [CrossRef]
- Cheng, H.; Lederer, W.J. Calcium sparks. Physiol. Rev. 2008, 88, 1491–1545. [Google Scholar] [CrossRef]
- Zhou, X.; Li, Z.; Wang, Z.; Chen, E.; Jones, O.; Tan, T.; Takeshima, H.; Bryant, J.; Ma, J.; Xu, X.H.; et al. Syncytium calcium signaling and macrophage function in the heart. Cell Biosci. 2018, 8, 24. [Google Scholar] [CrossRef]
- Cameron, A.M.; Steiner, J.P.; Roskams, A.J.; Ali, S.M.; Ronnett, G.V.; Snyder, S.H. Calcineurin associated with the inositol 1,4,5-trisphosphate receptor-FKBP12 complex modulates Ca2+ flux. Cell 1995, 83, 463–472. [Google Scholar] [CrossRef]
- Hannenhalli, S.; Putt, M.E.; Gilmore, J.M.; Wang, J.; Parmacek, M.S.; Epstein, J.A.; Morrisey, E.E.; Margulies, K.B.; Cappola, T.P. Transcriptional genomics associates FOX transcription factors with human heart failure. Circulation 2006, 114, 1269–1276. [Google Scholar] [CrossRef]
- Liu, Y.; Morley, M.; Brandimarto, J.; Hannenhalli, S.; Hu, Y.; Ashley, E.A.; Tang, W.H.; Moravec, C.S.; Margulies, K.B.; Cappola, T.P.; et al. RNA-Seq identifies novel myocardial gene expression signatures of heart failure. Genomics 2015, 105, 83–89. [Google Scholar] [CrossRef]

Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Li, Z.; Duan, Q.; Cui, Y.; Jones, O.D.; Shao, D.; Zhang, J.; Gao, Y.; Cao, X.; Wang, S.; Li, J.; et al. Cardiac-Specific Expression of Cre Recombinase Leads to Age-Related Cardiac Dysfunction Associated with Tumor-like Growth of Atrial Cardiomyocyte and Ventricular Fibrosis and Ferroptosis. Int. J. Mol. Sci. 2023, 24, 3094. https://doi.org/10.3390/ijms24043094
Li Z, Duan Q, Cui Y, Jones OD, Shao D, Zhang J, Gao Y, Cao X, Wang S, Li J, et al. Cardiac-Specific Expression of Cre Recombinase Leads to Age-Related Cardiac Dysfunction Associated with Tumor-like Growth of Atrial Cardiomyocyte and Ventricular Fibrosis and Ferroptosis. International Journal of Molecular Sciences. 2023; 24(4):3094. https://doi.org/10.3390/ijms24043094
Chicago/Turabian StyleLi, Zhongguang, Qinchun Duan, Ying Cui, Odell D. Jones, Danyang Shao, Jianfei Zhang, Yuru Gao, Xixi Cao, Shulin Wang, Jiali Li, and et al. 2023. "Cardiac-Specific Expression of Cre Recombinase Leads to Age-Related Cardiac Dysfunction Associated with Tumor-like Growth of Atrial Cardiomyocyte and Ventricular Fibrosis and Ferroptosis" International Journal of Molecular Sciences 24, no. 4: 3094. https://doi.org/10.3390/ijms24043094
APA StyleLi, Z., Duan, Q., Cui, Y., Jones, O. D., Shao, D., Zhang, J., Gao, Y., Cao, X., Wang, S., Li, J., Lei, X., Zhang, W., Wang, L., Zhou, X., Xu, M., Liu, Y., Ma, J., & Xu, X. (2023). Cardiac-Specific Expression of Cre Recombinase Leads to Age-Related Cardiac Dysfunction Associated with Tumor-like Growth of Atrial Cardiomyocyte and Ventricular Fibrosis and Ferroptosis. International Journal of Molecular Sciences, 24(4), 3094. https://doi.org/10.3390/ijms24043094




