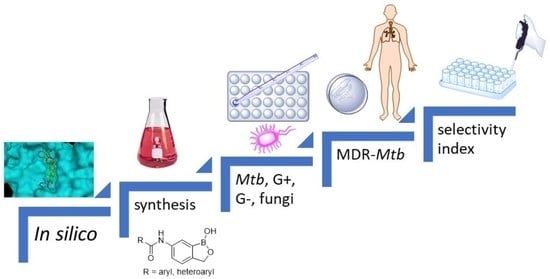
Design, Synthesis and Antimicrobial Evaluation of New N-(1-Hydroxy-1,3-dihydrobenzo[c][1,2]oxaborol-6-yl)(hetero)aryl-2-carboxamides as Potential Inhibitors of Mycobacterial Leucyl-tRNA Synthetase
Abstract
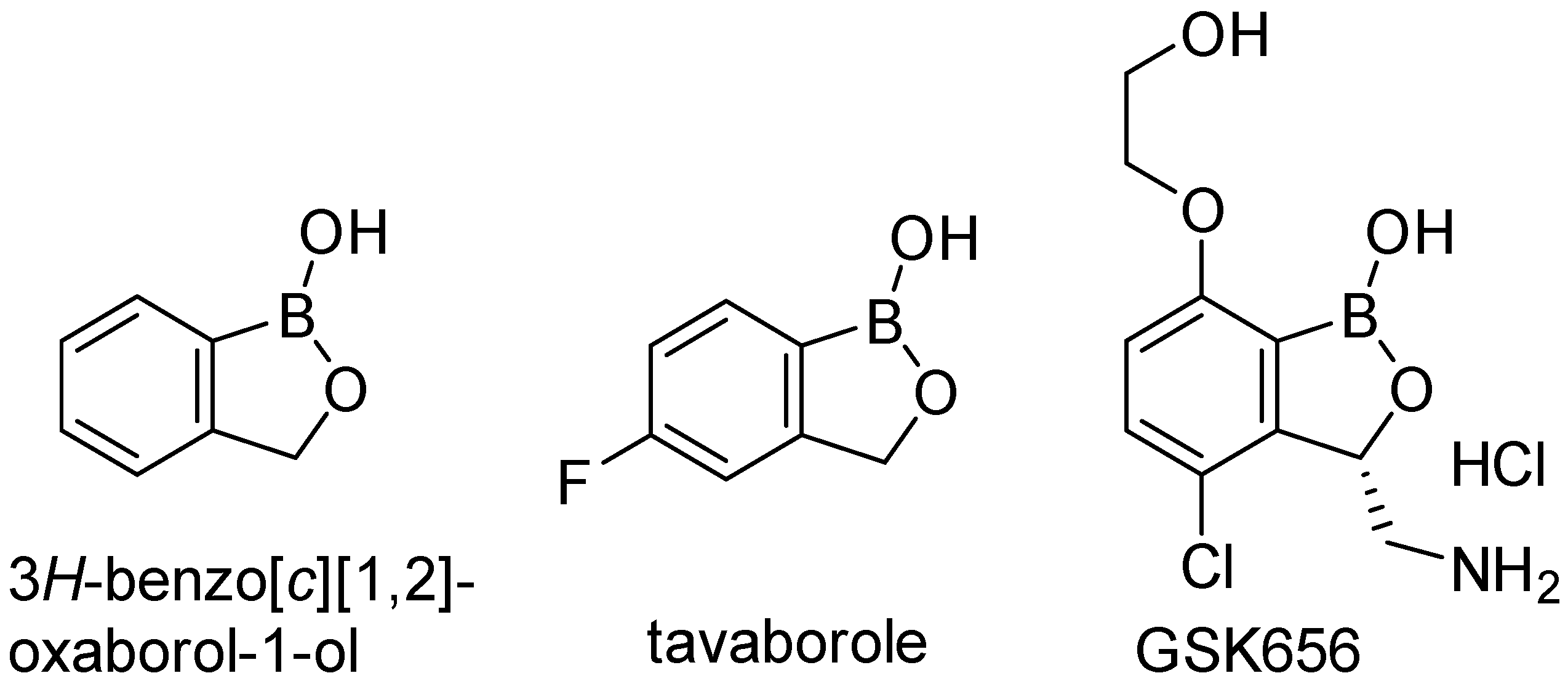
1. Introduction
2. Results and Discussion
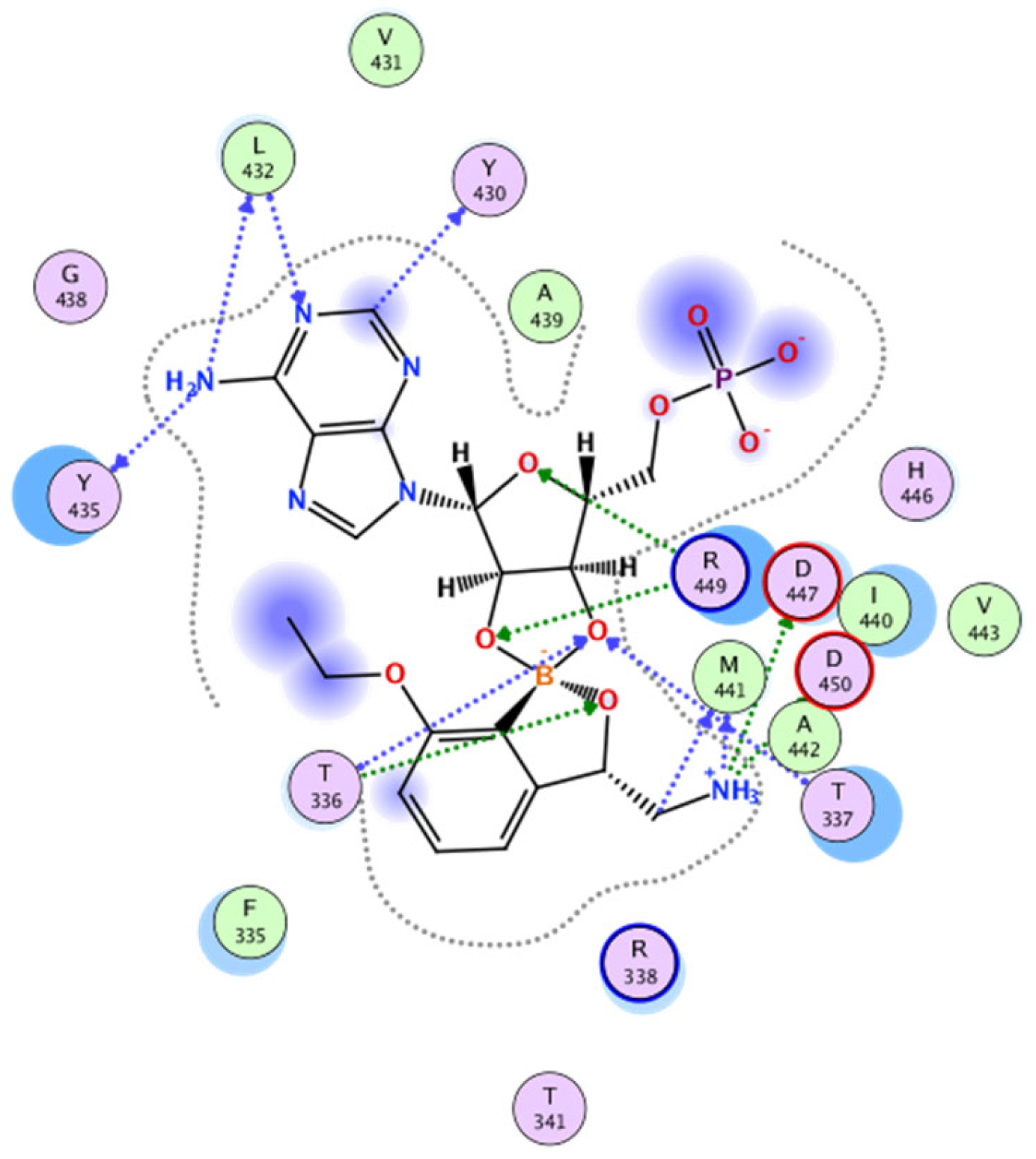
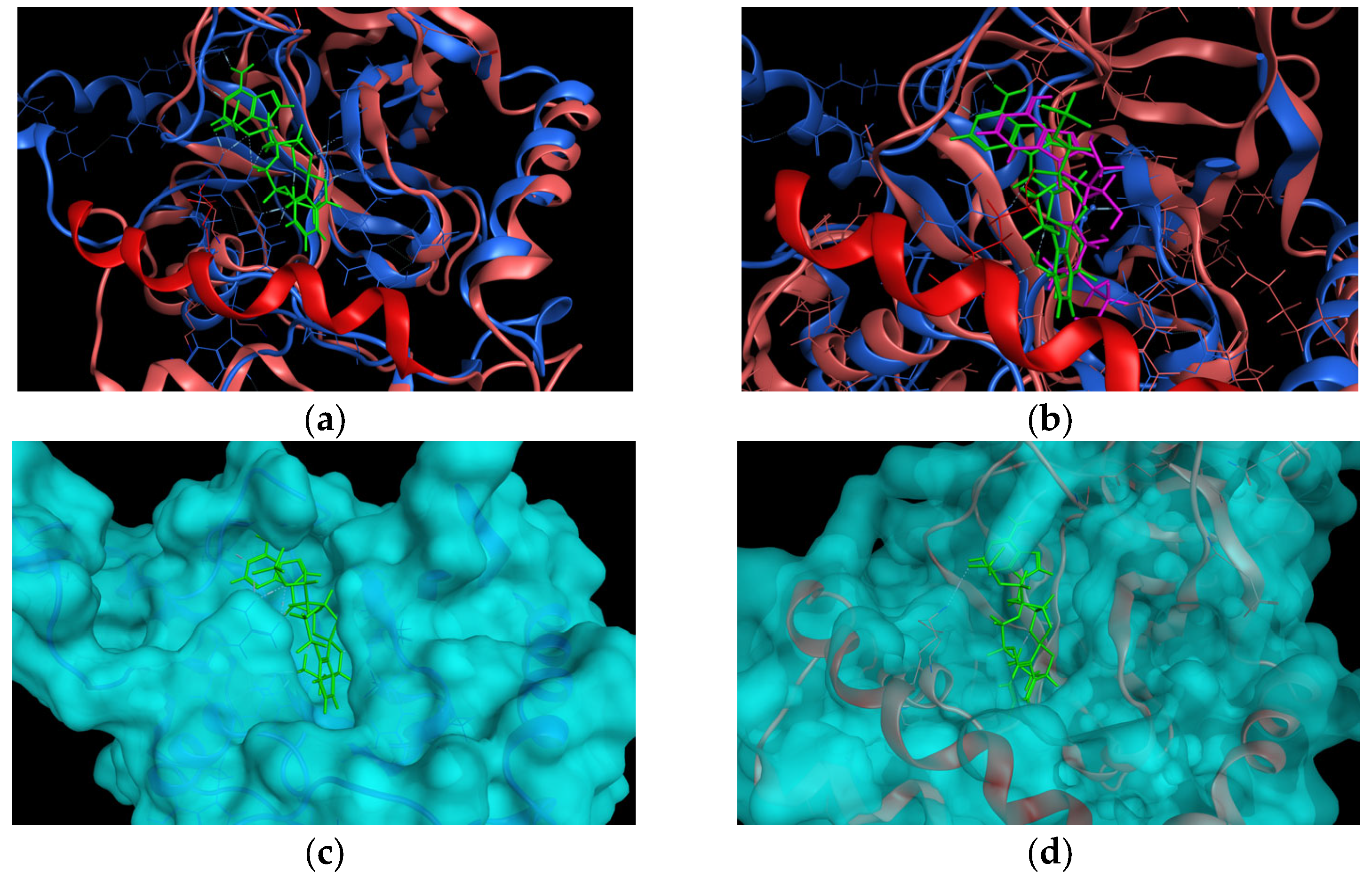
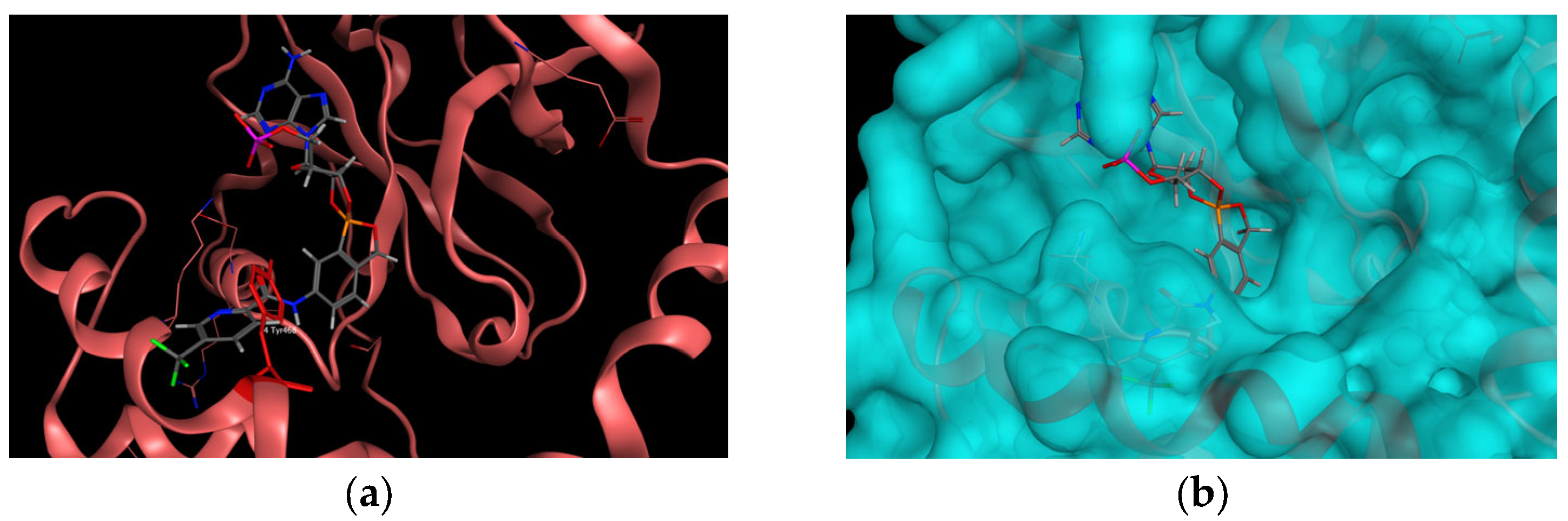
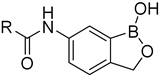
2.1. In Silico Studies
2.2. Synthesis of the Compounds
2.3. In Vitro Antimycobacterial Activity
2.3.1. Antimycobacterial Activity against Collection Strains of Mycobacteria
2.3.2. Antimycobacterial Activity against Multi-Drug-Resistant Clinical Isolates of M. tuberculosis
2.4. In Vitro Antibacterial and Antifungal Evaluation
2.5. Cytotoxicity Screening
3. Materials and Methods
3.1. In Silico Study
3.2. Synthesis of the Compounds
3.2.1. General Information
3.2.2. General Synthetic Procedure
3.2.3. Characterization and Purity of the Compounds 1–19
3.3. Biological Studies
3.3.1. Antimicrobial Screening
Antimycobacterial Screening against Mtb H37Rv and MDR Strains of Mtb
Susceptibility Profiles for the Used MDR Mtb Strains
3.3.2. Cytotoxicity Screening
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- WHO. Global Tuberculosis Report 2022; WHO: Geneve, Switzerland, 2022; p. 68. ISBN 978-92-4-006172-9. [Google Scholar]
- Coker, R.J. Review: Multidrug-resistant tuberculosis: Public health challenges. Trop. Med. Int. Health 2004, 9, 25–40. [Google Scholar] [CrossRef] [PubMed]
- Esposito, S.; D’Ambrosio, L.; Tadolini, M.; Schaaf, H.S.; Luna, J.C.; Centis, B.R.; Dara, M.; Matteelli, A.; Blasi, F.; Migliori, G.B. ERS/WHO Tuberculosis Consilium assistance with extensively drug-resistant tuberculosis management in a child: Case study of compassionate delamanid use. Eur. Respir. J. 2014, 44, 811–815. [Google Scholar] [CrossRef] [PubMed]
- Adamczyk-Wozniak, A.; Borys, K.M.; Sporzynski, A. Recent Developments in the Chemistry and Biological Applications of Benzoxaboroles. Chem. Rev. 2015, 115, 5224–5247. [Google Scholar] [CrossRef]
- Adamczyk-Wozniak, A.; Cyranski, M.K.; Zubrowska, A.; Sporzynski, A. Benzoxaboroles-Old compounds with new applications. J. Organomet. Chem. 2009, 694, 3533–3541. [Google Scholar] [CrossRef]
- Snyder, H.R.; Reedy, A.J.; Lennarz, W.J. Synthesis of aromatic boronic acids. aldehydo boronic acids and a boronic acid analog of tyrosine1. J. Am. Chem. Soc. 1958, 80, 835–838. [Google Scholar] [CrossRef]
- Lennarz, W.J.; Snyder, H.R. Arylboronic Acids.4. Reactions of Boronophthalide. J. Am. Chem. Soc. 1960, 82, 2172–2175. [Google Scholar] [CrossRef]
- Baker, S.J.; Ding, C.Z.; Akama, T.; Zhang, Y.K.; Hernandez, V.; Xia, Y. Therapeutic potential of boron-containing compounds. Future Med. Chem. 2009, 1, 1275–1288. [Google Scholar] [CrossRef]
- Fernandes, G.F.S.; Denny, W.A.; Dos Santos, J.L. Boron in drug design: Recent advances in the development of new therapeutic agents. Eur. J. Med. Chem. 2019, 179, 791–804. [Google Scholar] [CrossRef]
- Dhawan, B.; Akhter, G.; Hamid, H.; Kesharwani, P.; Alam, M.S. Benzoxaboroles: New emerging and versatile scaffold with a plethora of pharmacological activities. J. Mol. Struct. 2022, 1252, 21. [Google Scholar] [CrossRef]
- Coghi, P.S.; Zhu, Y.H.; Xie, H.M.; Hosmane, N.S.; Zhang, Y.J. Organoboron Compounds: Effective Antibacterial and Antiparasitic Agents. Molecules 2021, 26, 26. [Google Scholar] [CrossRef]
- Rock, F.L.; Mao, W.M.; Yaremchuk, A.; Tukalo, M.; Crepin, T.; Zhou, H.C.; Zhang, Y.K.; Hernandez, V.; Akama, T.; Baker, S.J.; et al. An antifungal agent inhibits an aminoacyl-tRNA synthetase by trapping tRNA in the editing site. Science 2007, 316, 1759–1761. [Google Scholar] [CrossRef] [PubMed]
- Palencia, A.; Li, X.F.; Bu, W.; Choi, W.; Ding, C.Z.; Easom, E.E.; Feng, L.; Hernandez, V.; Houston, P.; Liu, L.; et al. Discovery of Novel Oral Protein Synthesis Inhibitors of Mycobacterium tuberculosis That Target Leucyl-tRNA Synthetase. Antimicrob. Agents Chemother. 2016, 60, 6271–6280. [Google Scholar] [CrossRef] [PubMed]
- Li, X.F.; Hernandez, V.; Rock, F.L.; Choi, W.; Mak, Y.S.L.; Mohan, M.; Mao, W.M.; Zhou, Y.; Easom, E.E.; Plattner, J.J.; et al. Discovery of a Potent and Specific M-tuberculosis Leucyl-tRNA Synthetase Inhibitor: (S)-3-(Aminomethyl)-4-chloro-7-(2hydroxyethoxy)benzo c 1,2 oxaborol-1(3 H)-ol (GSK656). J. Med. Chem. 2017, 60, 8011–8026. [Google Scholar] [CrossRef]
- Edwards, B.D.; Field, S.K. The Struggle to End a Millennia-Long Pandemic: Novel Candidate and Repurposed Drugs for the Treatment of Tuberculosis. Drugs 2022, 82, 1695–1715. [Google Scholar] [CrossRef] [PubMed]
- Hu, Q.H.; Liu, R.J.; Fang, Z.P.; Zhang, J.; Ding, Y.Y.; Tan, M.; Wang, M.; Pan, W.; Zhou, H.C.; Wang, E.D. Discovery of a potent benzoxaborole-based anti-pneumococcal agent targeting leucyl-tRNA synthetase. Sci. Rep. 2013, 3, 2475. [Google Scholar] [CrossRef]
- Seiradake, E.; Mao, W.; Hernandez, V.; Baker, S.J.; Plattner, J.J.; Alley, M.R.K.; Cusack, S. Crystal Structures of the Human and Fungal Cytosolic Leucyl-tRNA Synthetase Editing Domains: A Structural Basis for the Rational Design of Antifungal Benzoxaboroles. J. Mol. Biol. 2009, 390, 196–207. [Google Scholar] [CrossRef]
- Korkegian, A.; O’Malley, T.; Xia, Y.; Zhou, Y.S.; Carter, D.S.; Sunde, B.; Flint, L.; Thompson, D.; Ioerger, T.R.; Sacchettini, J.; et al. The 7-phenyl benzoxaborole series is active against Mycobacterium tuberculosis. Tuberculosis 2018, 108, 96–98. [Google Scholar] [CrossRef]
- Alam, M.A.; Arora, K.; Gurrapu, S.; Jonnalagadda, S.K.; Nelson, G.L.; Kiprof, P.; Jonnalagadda, S.C.; Mereddy, V.R. Synthesis and evaluation of functionalized benzoboroxoles as potential anti-tuberculosis agents. Tetrahedron 2016, 72, 3795–3801. [Google Scholar] [CrossRef]
- Patel, N.; O’Malley, T.; Zhang, Y.K.; Xia, Y.; Sunde, B.; Flint, L.; Korkegian, A.; Ioerger, T.R.; Sacchettini, J.; Alley, M.R.K.; et al. A Novel 6-Benzyl Ether Benzoxaborole Is Active against Mycobacterium tuberculosis In Vitro. Antimicrob. Agents Chemother. 2017, 61, 3. [Google Scholar] [CrossRef]
- Gumbo, M.; Beteck, R.M.; Mandizvo, T.; Seldon, R.; Warner, D.F.; Hoppe, H.C.; Isaacs, M.; Laming, D.; Tam, C.C.; Cheng, L.W.; et al. Cinnamoyl-Oxaborole Amides: Synthesis and Their in Vitro Biological Activity. Molecules 2018, 23, 13. [Google Scholar] [CrossRef]
- Adamczyk-Wozniak, A.; Komarovska-Porokhnyavets, O.; Misterkiewicz, B.; Novikov, V.P.; Sporzynski, A. Biological activity of selected boronic acids and their derivatives. Appl. Organomet. Chem. 2012, 26, 390–393. [Google Scholar] [CrossRef]
- Si, Y.Y.; Basak, S.; Li, Y.; Merino, J.; Iuliano, J.N.; Walker, S.G.; Tonge, P.J. Antibacterial Activity and Mode of Action of a Sulfonamide-Based Class of Oxaborole Leucyl-tRNA-Synthetase Inhibitors. ACS Infect. Dis. 2019, 5, 1231–1238. [Google Scholar] [CrossRef] [PubMed]
- Ganapathy, U.S.; del Rio, R.G.; Cacho-Izquierdo, M.; Ortega, F.; Lelievre, J.; Barros-Aguirre, D.; Lindman, M.; Dartois, V.; Gengenbacher, M.; Dick, T. A Leucyl-tRNA Synthetase Inhibitor with Broad-Spectrum Antimycobacterial Activity. Antimicrob. Agents Chemother. 2021, 65, 13. [Google Scholar] [CrossRef] [PubMed]
- Ganapathy, U.S.; Gengenbacher, M.; Dick, T. Epetraborole Is Active against Mycobacterium abscessus. Antimicrob. Agents Chemother. 2021, 65, 5. [Google Scholar] [CrossRef]
- Kim, T.; Hanh, B.T.B.; Heo, B.; Quang, N.; Park, Y.; Shin, J.; Jeon, S.; Park, J.W.; Samby, K.; Jang, J. A Screening of the MMV Pandemic Response Box Reveals Epetraborole as A New Potent Inhibitor against Mycobacterium abscessus. Int. J. Mol. Sci. 2021, 22, 5936. [Google Scholar] [CrossRef]
- Zhang, P.P.; Ma, S.T. Recent development of leucyl-tRNA synthetase inhibitors as antimicrobial agents. MedChemComm 2019, 10, 1329–1341. [Google Scholar] [CrossRef]
- Lincecum, T.L.; Tukalo, M.; Yaremchuk, A.; Mursinna, R.S.; Williams, A.M.; Sproat, B.S.; Van Den Eynde, W.; Link, A.; Van Calenbergh, S.; Grotli, M.; et al. Structural and mechanistic basis of pre- and posttransfer editing by leucyl-tRNA synthetase. Mol. Cell 2003, 11, 951–963. [Google Scholar] [CrossRef]
- Palencia, A.; Crepin, T.; Vu, M.T.; Lincecum, T.L.; Martinis, S.A.; Cusack, S. Structural dynamics of the aminoacylation and proofreading functional cycle of bacterial leucyl-tRNA synthetase. Nat. Struct. Mol. Biol. 2012, 19, 677–684. [Google Scholar] [CrossRef]
- Liu, R.J.; Long, T.; Li, H.; Zhao, J.H.; Li, J.; Wang, M.Z.; Palencia, A.; Lin, J.Z.; Cusack, S.; Wang, E.D. Molecular basis of the multifaceted functions of human leucyl-tRNA synthetase in protein synthesis and beyond. Nucleic Acids Res. 2020, 48, 4946–4959. [Google Scholar] [CrossRef]
- Cummings, W.M.; Cox, C.H.; Snyder, H.R. Arylboronic acids. Medium-size ring-containing boronic ester groups. J. Org. Chem. 1969, 34, 1669–1674. [Google Scholar] [CrossRef]
- Franzblau, S.G.; Witzig, R.S.; McLaughlin, J.C.; Torres, P.; Madico, G.; Hernandez, A.; Degnan, M.T.; Cook, M.B.; Quenzer, V.K.; Ferguson, R.M.; et al. Rapid, low-technology MIC determination with clinical Mycobacterium tuberculosis isolates by using the microplate Alamar Blue assay. J. Clin. Microbiol. 1998, 36, 362–366. [Google Scholar] [CrossRef] [PubMed]
- Sundarsingh, T.J.A.; Ranjitha, J.; Rajan, A.; Shankar, V. Features of the biochemistry of Mycobacterium smegmatis, as a possible model for Mycobacterium tuberculosis. J. Infect. Public Health 2020, 13, 1255–1264. [Google Scholar] [CrossRef]
- Gupta, R.S.; Lo, B.; Son, J. Phylogenomics and Comparative Genomic Studies Robustly Support Division of the Genus Mycobacterium into an Emended Genus Mycobacterium and Four Novel Genera. Front. Microbiol. 2018, 9, 67. [Google Scholar] [CrossRef] [PubMed]
- Namouchi, A.; Cimino, M.; Favre-Rochex, S.; Charles, P.; Gicquel, B. Phenotypic and genomic comparison of Mycobacterium aurum and surrogate model species to Mycobacterium tuberculosis: Implications for drug discovery. BMC Genom. 2017, 18, 530. [Google Scholar] [CrossRef]
- Chaturvedi, V.; Dwivedi, N.; Tripathi, R.P.; Sinha, S. Evaluation of Mycobacterium smegmatis as a possible surrogate screen for selecting molecules active against multi-drug resistant Mycobacterium tuberculosis. J. Gen. Appl. Microbiol. 2007, 53, 333–337. [Google Scholar] [CrossRef]
- Heinrichs, M.T.; May, R.J.; Heider, F.; Reimers, T.; Sy, S.K.B.; Peloquin, C.A.; Derendorf, H. Mycobacterium tuberculosis Strains H37ra and H37rv have Equivalent Minimum Inhibitory Concentrations to Most Antituberculosis Drugs. Int. J. Mycobacteriol. 2018, 7, 156–161. [Google Scholar] [CrossRef]
- Ambrozkiewicz, W.; Kucerova-Chlupacova, M.; Jand’ourek, O.; Konecna, K.; Paterova, P.; Barta, P.; Vinsova, J.; Dolezal, M.; Zitko, J. 5-Alkylamino-N-phenylpyrazine-2-carboxamides: Design, Preparation, and Antimycobacterial Evaluation. Molecules 2020, 25, 21. [Google Scholar] [CrossRef]
- Nawrot, D.; Suchankova, E.; Jandourek, O.; Konecna, K.; Barta, P.; Dolezal, M.; Zitko, J. N-pyridinylbenzamides: An isosteric approach towards new antimycobacterial compounds. Chem. Biol. Drug Des. 2021, 97, 686–700. [Google Scholar] [CrossRef]
- Konecna, K.; Diepoltova, A.; Holmanova, P.; Jand’ourek, O.; Vejsova, M.; Voxova, B.; Barta, P.; Maixnerova, J.; Trejtnar, F.; Kucerova-Chlupacova, M. Comprehensive insight into anti-staphylococcal and anti-enterococcal action of brominated and chlorinated pyrazine-based chalcones. Front. Microbiol. 2022, 13, 14. [Google Scholar] [CrossRef]
- Baker, S.J.; Zhang, Y.K.; Akama, T.; Lau, A.; Zhou, H.; Hernandez, V.; Mao, W.M.; Alley, M.R.K.; Sanders, V.; Plattner, J.J. Discovery of a new boron-containing antifungal agent, 5-fluoro-1,3-dihydro-1-hydroxy-2,1-benzoxaborole (AN2690), for the potential treatment of onychomycosis. J. Med. Chem. 2006, 49, 4447–4450. [Google Scholar] [CrossRef]
- European Committee for Antimicrobial Susceptibility Testing (EUCAST) of the European Society for Clinical Microbiology and Infectious Diseases (ESCMID). Eucast Discussion Document E. Dis 5.1: Determination of minimum inhibitory concentrations (MICs) of antibacterial agents by broth dilution. Clin. Microbiol. Infect. 2003, 9, 9–15. [CrossRef]
- Arendrup, M.C.; Meletiadis, J.; Mouton, J.W.; Lagrou, K.; Hama, P.; Guinea, J.; Afst-Eucast. Eucast Definitive Document E. Def 7.3.1. Method for the Determination of Broth Dilution Minimum Inhibitory Concentrations of Antifungal Agents for Yeasts. 2017. Available online: https://www.eucast.org/fileadmin/src/media/PDFs/EUCAST_files/AFST/Files/EUCAST_E_Def_7_3_1_Yeast_testing__definitive.pdf (accessed on 20 December 2022).
- Arendrup, M.C.; Meletiadis, J.; Mouton, J.W.; Lagrou, K.; Hamal, P.; Guinea, J.; Afst-Eucast. Eucast Definitive Document E.Def 9.3.1. Method for the Determination of Broth Dilution Minimum Inhibitory Concentrations of Antifungal Agents for Conidia Forming Moulds. 2017. Available online: https://www.eucast.org/fileadmin/src/media/PDFs/EUCAST_files/AFST/Files/EUCAST_E_Def_9_3_1_Mould_testing__definitive.pdf (accessed on 20 December 2022).
- Ramappa, V.; Aithal, G.P. Hepatotoxicity Related to Anti-tuberculosis Drugs: Mechanisms and Management. J. Clin. Exp. Hepatol. 2013, 3, 37–49. [Google Scholar] [CrossRef] [PubMed]

 | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| LogP | MIC | IC50 | |||||||
| Comp. | R | Mtb H37Ra (µM) | Mtb H37Rv (µM) | M. kansasii (µM) | M. avium (µM) | M. aurum (µM) | M. smeg. (µM) | HepG2 (µM) | |
| 1 | 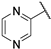 | 0.42 | 122.53 | 49.01 | 30.62 | 122.53 | 15.33 | 30.62 | >1000 |
| 2 |  | 0.92 | 58.07 | 46.46 | 58.07 | 58.07 | 14.53 | 29.03 | >500 |
| 3 |  | 0.58 | >1851.4 | 92.91 | 58.07 | >1851.4 | 14.53 | 14.53 | >500 |
| 4 | 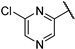 | 1.16 | 107.95 | 86.36 | 26.98 | 26.98 | 13.51 | 26.98 | >1000 |
| 5 |  | 1.16 | 26.98 | 21.59 | 107.95 | 53.98 | 13.51 | 53.98 | 82.44 |
| 6 |  | 0.67 | >1851.47 | >370.29 | >1851.47 | >1851.47 | 14.48 | 462.87 | >100 |
| 7 | 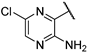 | 1.40 | >1851.47 | >328.41 | >1851.47 | 410.51 | >1851.47 | 410.51 | >100 |
| 8 | 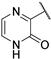 | −0.61 | 230.59 | 92.24 | 230.59 | 57.65 | 28.82 | 28.82 | >1000 |
| 9 | 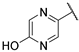 | 1.05 | 115.30 | 184.48 | 115.30 | 230.59 | 230.59 | 57.65 | >1000 |
| 10 | 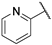 | 1.20 | 61.50 | 49.20 | 30.74 | 61.50 | 15.39 | 61.50 | >1000 |
| 11 | 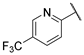 | 2.19 | 97.03 | 9.72 | 12.14 | 97.03 | 24.25 | 24.25 | >100 |
| 12 |  | 1.84 | 194.07 | 155.26 | 194.07 | 388.14 | 12.14 | 24.25 | >250 |
| 13 | 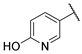 | 1.19 | 462.88 | >370.30 | 115.72 | 462.88 | 115.72 | 115.72 | >1000 |
| 14 |  | 2.52 | 434.77 | 43.48 | 217.39 | 434.77 | 13.60 | 217.39 | >1000 |
| 15 |  | 2.27 | 53.23 | 42.59 | 106.46 | 425.85 | 53.23 | 851.71 | 210.5 |
| 16 | 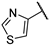 | 1.06 | 120.16 | 96.13 | 30.03 | 240.32 | 15.03 | 30.03 | >1000 |
| 17 | 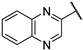 | 2.01 | >819.40 | 20.49 | 25.60 | 102.43 | 12.82 | 12.82 | >250 |
| 18 | 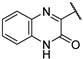 | 0.98 | >1851.47 | >311.43 | >1851.47 | 48.66 | 24.32 | 24.32 | 243.7 |
| 19 | 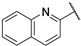 | 2.59 | 205.52 | 82.21 | 205.52 | 25.68 | 25.68 | 25.68 | 466.7 |
| INH | - | 3.65 | 2.84 | 45.57 | 7291.87 | 28.51 | 227.87 | na | |
| RIF | - | 0.00759 | na | 0.06 | 0.08 | 0.47 | 15.19 | na | |
| CIP | - | 1.51 | na | 0.75 | 1.51 | 0.05 | 0.19 | na | |
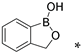 | na | 55.99 * | na | na | na | na | na | ||
 | na | 11.04 * | na | na | na | na | na | ||
| Comp. | MIC Mtb H37Rv (µM) | MIC Mtb IZAK (µM) | MIC Mtb MATI (µM) |
|---|---|---|---|
| 4 | 86.36 | 86.36 | 86.36 |
| 5 | 21.59 | 43.18 | 43.18 |
| 10 | 49.20 | 98.41 | 98.41 |
| 17 | 20.49 | 40.97 | 40.97 |
| INH | 2.84 | 91.15 | >91.15 |
| CPX | 0.6 | 0.6 | 0.6 |
| EMB | 1.91 | 7.64 | 7.64 |
| Compound | HepG2 IC50 (µM) | Mtb H37Rv MIC (µM) | SI |
|---|---|---|---|
| 1 | >1000 | 49.01 | >20.40 |
| 2 | >500 | 46.46 | >10.76 |
| 5 | 82.44 | 21.59 | 3.82 |
| 10 | >1000 | 49.20 | >20.33 |
| 11 | >100 | 9.72 | >10.29 |
| 14 | >1000 | 43.48 | >23.00 |
| 15 | 210.5 | 42.59 | 4.94 |
| 17 | >250 | 20.49 | >12.20 |
| Laboratory ID | Drug | Concentration (µM) | Susceptibility |
|---|---|---|---|
| IZAK | STM | 6.88 | resistant |
| INH | 29.17 | resistant | |
| RIF | >9.72 | resistant | |
| EMB | 2.45 | sensitive | |
| PZA | >129.96 | resistant | |
| MATI | STM | >27.51 | resistant |
| INH | >58.33 | resistant | |
| RIF | >9.72 | resistant | |
| EMB | 2.45 | sensitive | |
| PZA | >1039.70 | resistant |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Šlechta, P.; Needle, A.A.; Jand’ourek, O.; Paterová, P.; Konečná, K.; Bárta, P.; Kuneš, J.; Kubíček, V.; Doležal, M.; Kučerová-Chlupáčová, M. Design, Synthesis and Antimicrobial Evaluation of New N-(1-Hydroxy-1,3-dihydrobenzo[c][1,2]oxaborol-6-yl)(hetero)aryl-2-carboxamides as Potential Inhibitors of Mycobacterial Leucyl-tRNA Synthetase. Int. J. Mol. Sci. 2023, 24, 2951. https://doi.org/10.3390/ijms24032951
Šlechta P, Needle AA, Jand’ourek O, Paterová P, Konečná K, Bárta P, Kuneš J, Kubíček V, Doležal M, Kučerová-Chlupáčová M. Design, Synthesis and Antimicrobial Evaluation of New N-(1-Hydroxy-1,3-dihydrobenzo[c][1,2]oxaborol-6-yl)(hetero)aryl-2-carboxamides as Potential Inhibitors of Mycobacterial Leucyl-tRNA Synthetase. International Journal of Molecular Sciences. 2023; 24(3):2951. https://doi.org/10.3390/ijms24032951
Chicago/Turabian StyleŠlechta, Petr, Adam Anthony Needle, Ondřej Jand’ourek, Pavla Paterová, Klára Konečná, Pavel Bárta, Jiří Kuneš, Vladimír Kubíček, Martin Doležal, and Marta Kučerová-Chlupáčová. 2023. "Design, Synthesis and Antimicrobial Evaluation of New N-(1-Hydroxy-1,3-dihydrobenzo[c][1,2]oxaborol-6-yl)(hetero)aryl-2-carboxamides as Potential Inhibitors of Mycobacterial Leucyl-tRNA Synthetase" International Journal of Molecular Sciences 24, no. 3: 2951. https://doi.org/10.3390/ijms24032951
APA StyleŠlechta, P., Needle, A. A., Jand’ourek, O., Paterová, P., Konečná, K., Bárta, P., Kuneš, J., Kubíček, V., Doležal, M., & Kučerová-Chlupáčová, M. (2023). Design, Synthesis and Antimicrobial Evaluation of New N-(1-Hydroxy-1,3-dihydrobenzo[c][1,2]oxaborol-6-yl)(hetero)aryl-2-carboxamides as Potential Inhibitors of Mycobacterial Leucyl-tRNA Synthetase. International Journal of Molecular Sciences, 24(3), 2951. https://doi.org/10.3390/ijms24032951