Structure and Microbiological Activity of 1H-benzo[d]imidazole Derivatives
Abstract
:1. Introduction
2. Results and Discussion
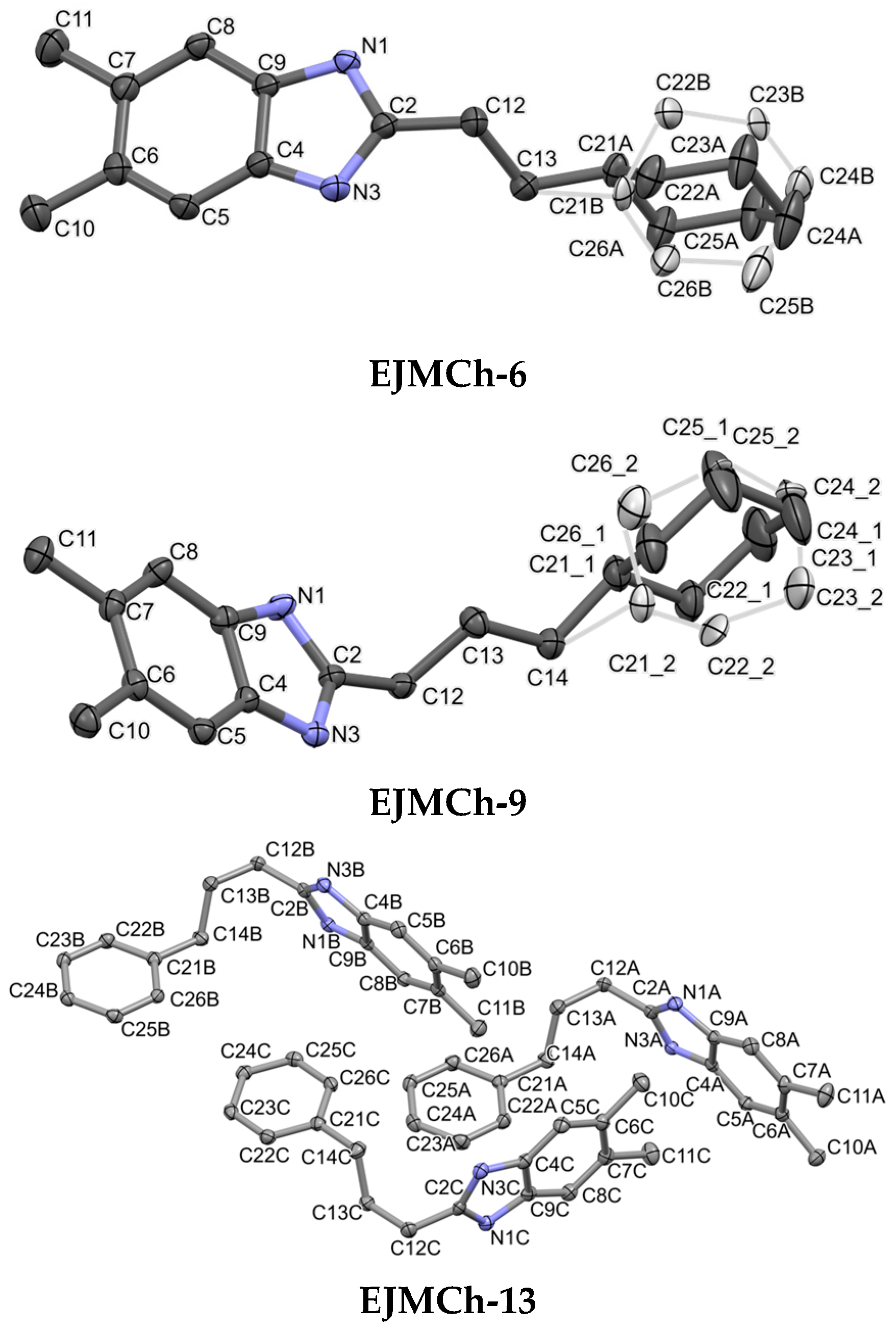
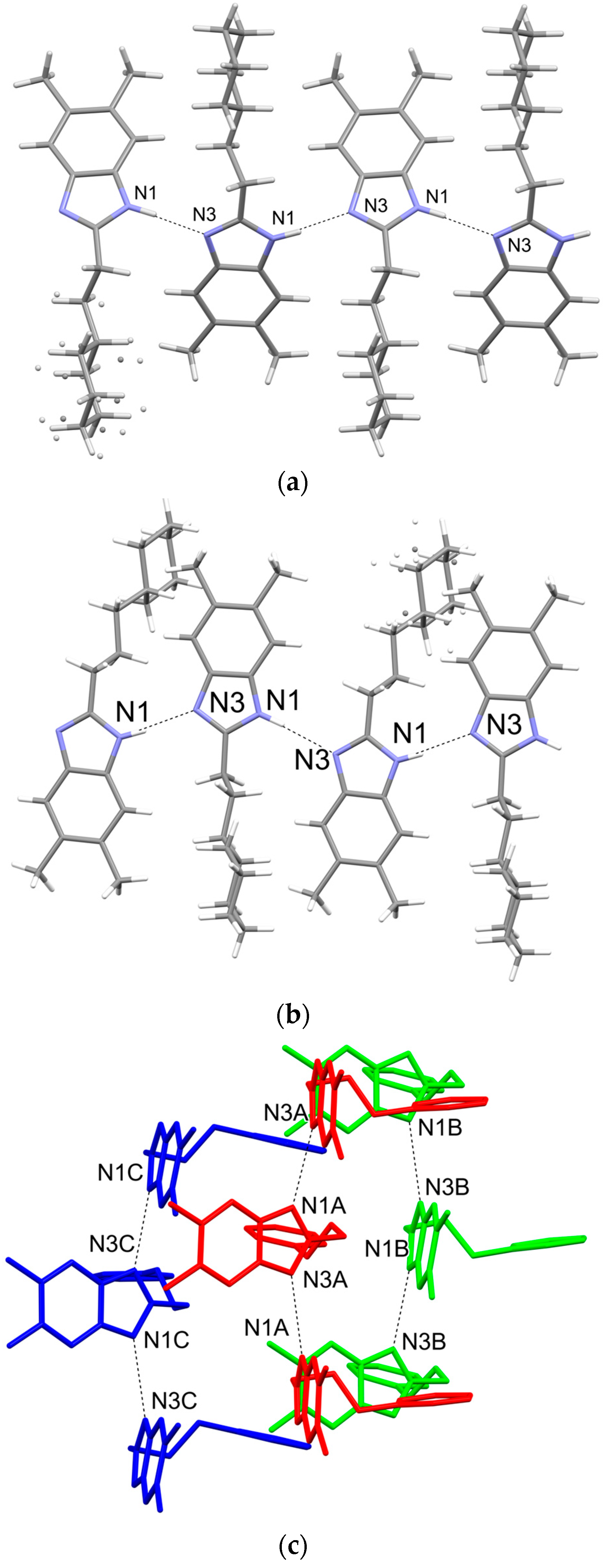
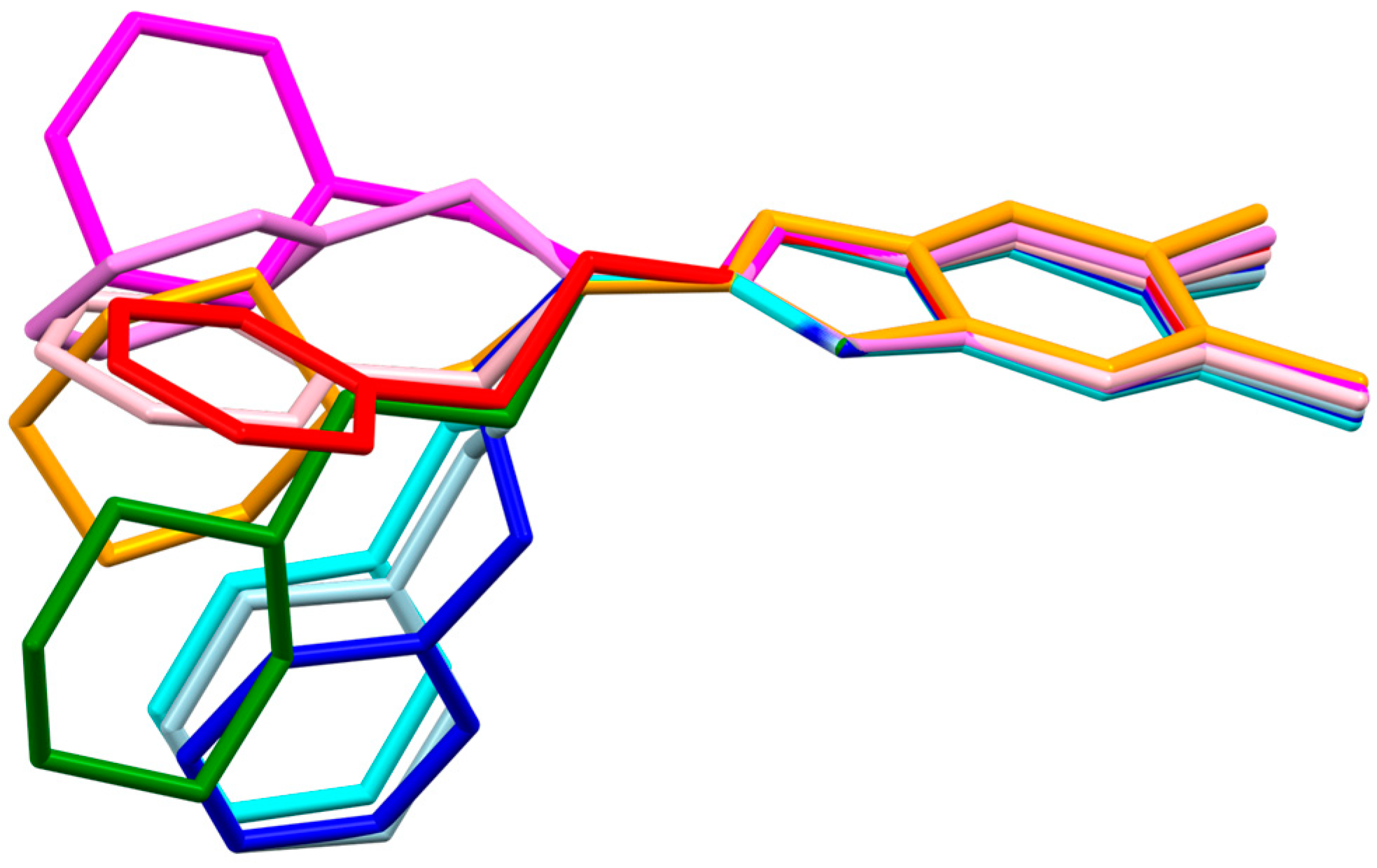
2.1. Structure Analysis
Sample Quality Verification via Solid-State NMR
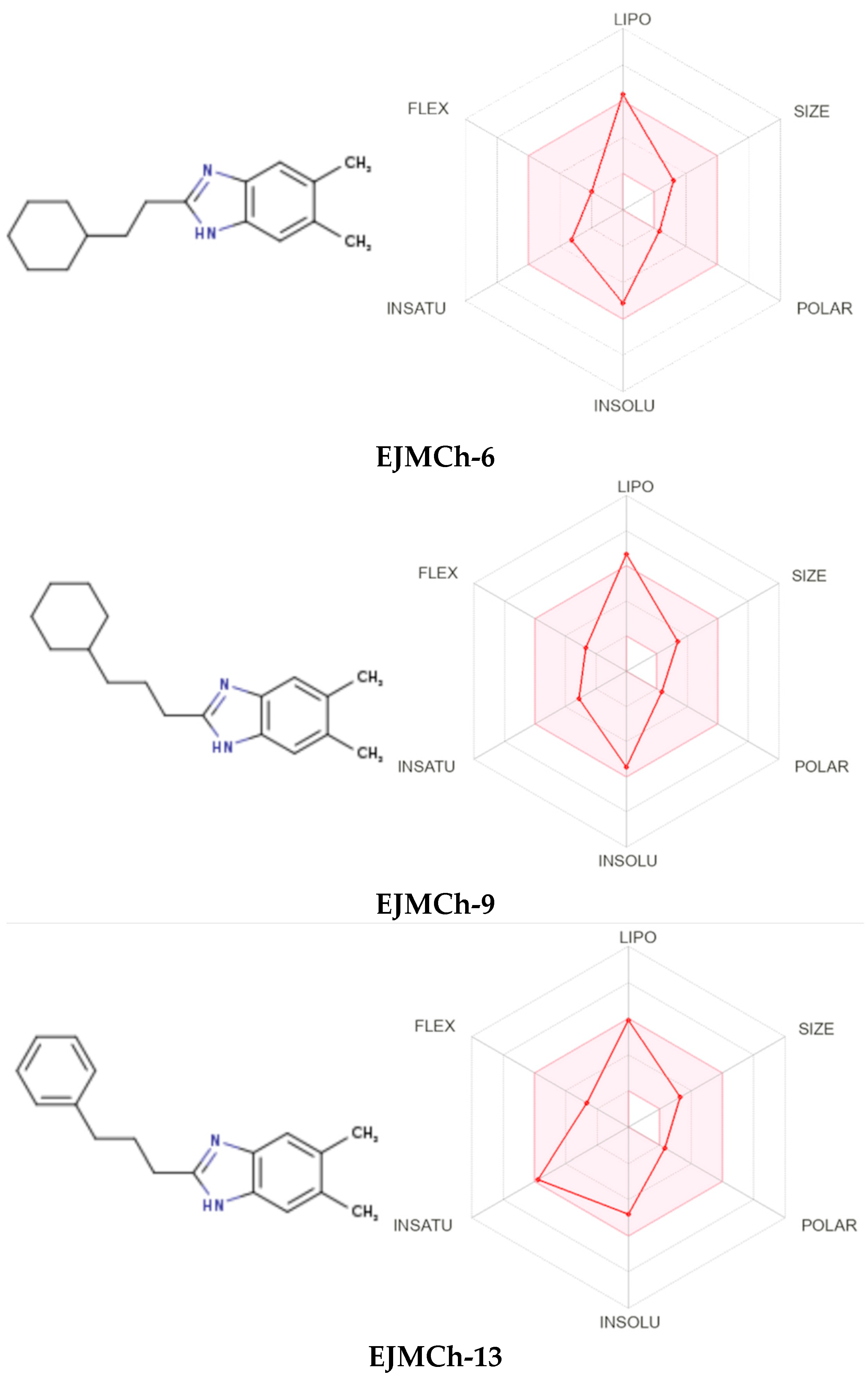
2.2. ADME Analysis
2.3. Antimicrobial Activity
3. Materials and Methods
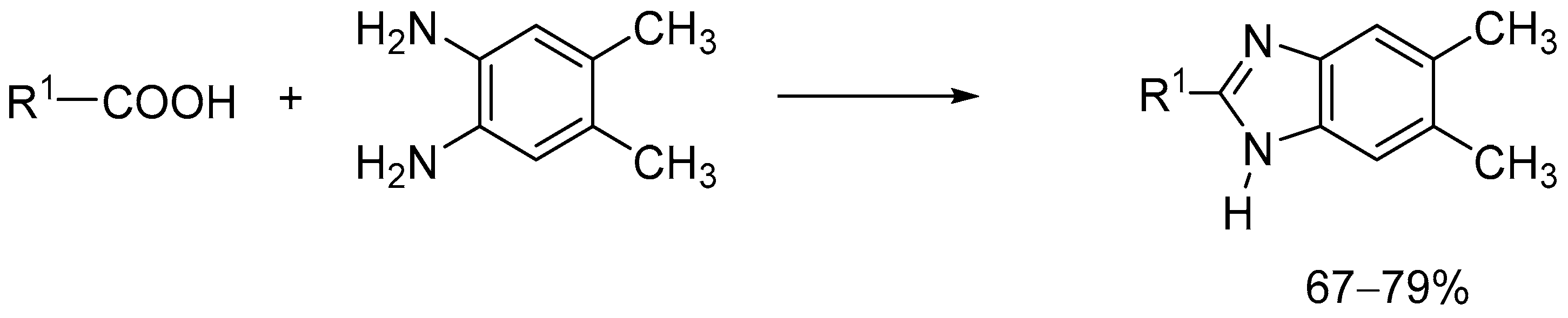
3.1. Chemistry
3.2. X-ray Study
3.3. NMR Study
3.4. ADME
3.5. In Vitro Antibacterial Activity Assay
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Centers for Disease Control and Prevention. Antibiotic Resistance Threats in the United States. 2019. Available online: https://www.cdc.gov/drugresistance/pdf/threats-report/2019-ar-threats-report-508.pdf (accessed on 20 December 2022).
- President’s Council of Advisors on Science and Technology. Report to the President on Combating Antibiotic Resistance. Available online: https://obamawhitehouse.archives.gov/sites/default/files/microsites/ostp/PCAST/pcast_carb_report_sept2014.pdf (accessed on 20 December 2022).
- Sriram, A.; Kalanxhi, E.; Kapoor, G.; Craig, J.; Balasubramanian, R.; Brar, S.; Criscuolo, N.; Hamilton, A.; Klein, E.; Tseng, K.; et al. State of the World’s Antibiotics, 2021. Washington, DC: Center for Disease Dynamics, Economics, and Policy. 2021. Available online: https://onehealthtrust.org/wp-content/uploads/2021/02/The-State-of-the-Worlds-Antibiotics-in-2021.pdf (accessed on 20 December 2022).
- European Centre for Disease Prevention and Control. Antimicrobial Resistance in the EU/EEA (EARS-Net). Annual Epidemiological Report for 2019. Available online: https://www.ecdc.europa.eu/sites/default/files/documents/AER-EARS-Net%202019-final.pdf (accessed on 20 December 2022).
- O’Neill, J. Tackling Drug-Resistant Infections Globally: Final Report and Recommendations. Review on Antimicrobial Resistance, May 2016. Available online: https://amr-review.org/sites/default/files/160518_Final%20paper_with%20cover.pdf (accessed on 20 December 2022).
- World Health Organization: Antimicrobial Resistance: Global Report on Surveillance. 2022. Available online: https://www.who.int/publications/i/item/9789240062702 (accessed on 20 December 2022).
- Andersson, D.I.; Hughes, D. Evolution of antibiotic resistance at non-lethal drug concentrations. Drug Resist. Update. 2012, 15, 162–172. [Google Scholar] [CrossRef] [PubMed]
- Hammarskjöld, F.; Mernelius, S.; Andersson, R.E.; Berg, S.; Hanberger, H.; Löfgren, S.; Malmvall, B.-E.; Petzold, M.; Matussek, A. Possible transmission of Candida albicans on an intensive care unit: Genotype and temporal cluster analyses. J. Hosp. Infect. 2013, 85, 60–65. [Google Scholar] [CrossRef] [PubMed]
- Pfaller, M.A.; Diekema, D.J. Epidemiology of invasive candidiasis: A persistent public health problem. Clin. Microbiol. Rev. 2007, 20, 133–163. [Google Scholar] [CrossRef] [PubMed]
- Siscar-Lewin, S.; Hube, B.; Brunke, S. Emergence and evolution of virulence in human pathogenic fungi. Trends Microbiol. 2022, 30, 693–704. [Google Scholar] [CrossRef]
- Pathare, B.; Bansode, T. Review- biological active benzimidazole derivatives. Results Chem. 2021, 3, 100200. [Google Scholar] [CrossRef]
- Dokla, E.M.E.; Abutaleb, N.S.; Milik, S.N.; Li, D.; El-Baz, K.; Shalaby, M.-A.W.; Al-Karaki, R.; Nasr, M.; Klein, C.D.; Abouzid, K.A.M.; et al. Development of benzimidazole-based derivatives as antimicrobial agents and their syn-ergistic effect with colistin against gram-negative bacteria. Eur. J. Med. Chem. 2020, 186, 111850. [Google Scholar] [CrossRef]
- Huang, D.; Qiu, F.; Zhang, Z.; Shi, L.; Cao, C.; Ke, S. Synthesis and antifungal activity of substituted 2-Aryl benzimidaz-oles derivatives. J. Het. Chem. 2019, 56, 2494–2498. [Google Scholar] [CrossRef]
- Gobis, K.; Foks, H.; Serocki, M.; Augustynowicz-Kopeć, E.; Napiórkowska. Synthesis and evaluation of in vitro antimycobacterial activity of novel 1H-benzo[d]imidazole derivatives and analogues. Eur. J. Med. Chem. 2015, 89, 13–20. [Google Scholar] [CrossRef]
- Global Tuberculosis Report 2022; World Health Organization: Geneva, Switzerland, 2022; Available online: https://www.who.int/publications/i/item/9789240061729 (accessed on 20 December 2022).
- Yew, W.-W. Management of multidrug-resistant tuberculosis and extensively drug-resistant tuberculosis: Current status and future prospects. Kekkaku 2011, 86, 9–16. [Google Scholar]
- Parsons, S.; Flack, H.D.; Wagner, T. Use of intensity quotients and differences in absolute structure refinement. Acta Cryst. 2013, B69, 249–259. [Google Scholar] [CrossRef]
- Macrae, C.F.; Sovago, I.; Cottrell, S.J.; Galek, P.T.A.; McCabe, P.; Pidcock, E.; Platings, M.; Shields, G.P.; Stevens, J.S.; Towler, M.; et al. Mercury 4.0: From visualization to analysis, design and prediction. J. Appl. Cryst. 2020, 53, 226–235. [Google Scholar] [CrossRef] [PubMed]
- Bernstein, J.; Davis, R.E.; Shimoni, L.; Chang, N.-L. Patterns in Hydrogen Bonding Functionality and Graph Set Analysis in Crystals. Angew. Chem. Int. Ed. Engl. 1995, 34, 1555–1573. [Google Scholar] [CrossRef]
- Groom, C.R.; Bruno, I.J.; Lightfoot, M.P.; Ward, S.C. The Cambridge Structural Database. Acta Cryst. 2016, B72, 171–179. [Google Scholar] [CrossRef] [PubMed]
- Główka, M.L.; Kałużyńska, S.; Krause, M.; Gobis, K.; Foks, H.; Szczesio, M.; Olczak, A. The structures of benzimidazole de-rivatives and their potential as tuberculostatics. Acta Cryst. 2018, C74, 1684–1691. [Google Scholar]
- Lipinski, C.A.; Lombardo, F.; Dominy, B.W.; Feeney, P.J. Experimental and computational approaches to estimate solubility and permeability in drug discovery and development settings. Adv. Drug Deliv. Rev. 2001, 46, 3–26. [Google Scholar] [CrossRef]
- Ghose, A.K.; Viswanadhan, V.N.; Wendolski, J.J. A knowledge-based approach in designing combinatorial or medicinal chemistry libraries for drug discovery. 1. A qualitative and quantitative characterization of known drug databases. J. Comb. Chem. 1999, 1, 55–68. [Google Scholar] [CrossRef]
- Egan, W.J.; Merz, K.M.; Baldwin, J.J. Prediction of drug absorption using multivariate statistics. J. Med. Chem. 2000, 3, 3867–3877. [Google Scholar] [CrossRef]
- Veber, D.F.; Johnson, S.R.; Cheng, H.Y.; Smith, B.R.; Ward, K.W.; Kopple, K.D. Molecular properties that influence the oral bioavailability of drug candidates. J. Med. Chem. 2002, 45, 2615–2623. [Google Scholar] [CrossRef]
- Muegge, I.; Heald, S.L.; Brittelli, D. Simple selection criteria f or drug-like chemical matter. J. Med. Chem. 2001, 44, 1841–1946. [Google Scholar] [CrossRef]
- Huynh, J.; Donovan, J.; Hoan Phu, N.; Dang Trung Nghia, H.; Thuy Thuong Thuong, N.; Thwaites, G.E. Tuberculous men-ingitis: Progress and remaining questions. Lancet Neurol. 2022, 21, 450–464. [Google Scholar] [CrossRef]
- Tahlan, S.; Kumar, S.; Narasimhan, B. Antimicrobial potential of 1H-benzo[d]imidazole scaffold: A review. BMC Chem. 2019, 13, 18–40. [Google Scholar] [CrossRef] [PubMed]
- Sheldrick, G.M. SHELXT—Integrated space-group and crystal-structure determination. Acta Cryst. 2015, C71, 3–8. [Google Scholar] [CrossRef] [PubMed]
- Sheldrick, G.M. A short history of SHELX. Acta Cryst. 2008, A64, 112. [Google Scholar] [CrossRef] [PubMed]
- Hübschle, C.B.; Sheldrick, G.M.; Dittrich, B. ShelXle: A Qt graphical user interface for SHELXL. J. Appl. Cryst. 2011, 44, 1281–1284. [Google Scholar] [CrossRef] [PubMed]
- Westrip, S.P. publCIF: Software for editing, validating and formatting crystallographic information files. J. Apply. Cryst. 2010, 43, 920–925. [Google Scholar] [CrossRef]
- Kratzert, D.; Krossing, I. Recent improvements in DSR. J. Appl. Cryst. 2018, 51, 928–934. [Google Scholar] [CrossRef]
- Daina, A.; Michielin, O.; Zoete, V. SwissADME: A free web tool to evaluate pharmacokinetics, drug-likeness and medicinal chemistry friendliness of small molecules. Sci. Rep. 2017, 7, 42717. [Google Scholar] [CrossRef]
- Daina, A.; Michielin, O.; Zoete, V. iLOGP: A simple, robust, and efficient description of n-octanol/water partition coefficient for drug design using the GB/SA approach. J. Chem. Inf. Model. 2014, 54, 3284–3301. [Google Scholar] [CrossRef]
- Daina, A.; Zoete, V. A BOILED-Egg To Predict Gastrointestinal Absorption and Brain Penetration of Small Molecules. ChemMedChem 2016, 11, 1117–1121. [Google Scholar] [CrossRef]
- Zesławska, E.; Korona-Głowniak, I.; Szczesio, M.; Olczak, A.; Zylewska, A.; Tejchman, W.; Malm, A. Structural analysis and antimicrobial activity of 2[1H]-pyrimidinethione/selenone derivatives. J. Mol. Struct. 2017, 1142, 261–266. [Google Scholar] [CrossRef]
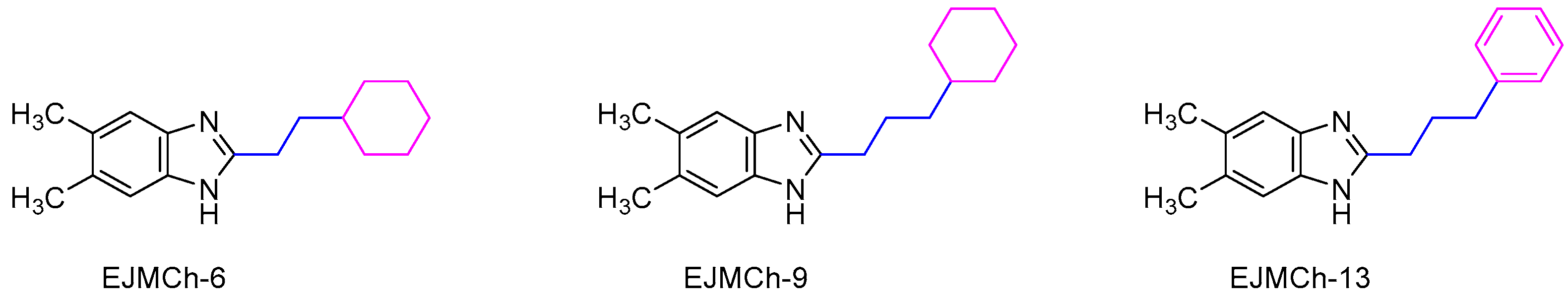


| EJMCh-6 | EJMCh-9 | EJMCh-13 | |
|---|---|---|---|
| Crystal data | |||
| Chemical formula | C17H24N2 | C18H26N2 | C18H20N2 |
| Mr | 256.38 | 270.41 | 264.36 |
| Crystal system, space group | Monoclinic, Cc | Monoclinic, P21/c | Monoclinic, Cc |
| a, b, c (Å) | 15.2276 (3), 12.7173 (3), 9.7501 (2) | 14.2078 (4), 9.9210 (3), 11.5761 (4) | 28.8210 (9), 15.4123 (5), 10.1758 (3) |
| β (°) | 126.1959 (5) | 104.7986 (9) | 98.2484 (13) |
| V (Å3) | 1523.74 (6) | 1577.59 (9) | 4473.3 (2) |
| Z | 4 | 4 | 12 |
| Data collection | |||
| No. of measured, independent and observed [I > 2σ(I)] reflections | 15,809, 2939, 2936 | 3012, 3012, 2963 | 31367, 7814, 7731 |
| Rint | 0.020 | 0.024 | 0.022 |
| (sin θ/λ)max (Å−1) | 0.617 | 0.618 | 0.618 |
| Refinement | |||
| R[F2 > 2σ(F2)], wR(F2), S | 0.026, 0.072, 1.06 | 0.034, 0.092, 1.05 | 0.033, 0.090, 1.05 |
| No. of reflections | 2939 | 3012 | 7814 |
| No. of parameters | 234 | 240 | 547 |
| No. of restraints | 2 | 30 | 2 |
| Δmax, Δmin (eÅ−3) | 0.19, −0.12 | 0.22, −0.14 | 0.28, −0.20 |
| Absolute structure | Flack x determined using 1439 quotients [(I+)-(I−)]/[(I+)+(I−)] [17]. | – | Flack x determined using 3275 quotients [(I+)-(I−)]/[(I+)+(I−)] [17]. |
| Absolute structure parameter | 0.07 (5) | – | 0.09 (8) |
| Chemical | EJMCh-6 | EJMCh-9 | EJMCh-13 | Standard Drug | |||||
|---|---|---|---|---|---|---|---|---|---|
| Microorganism | mg/L | Vancomycin | |||||||
| Gram-positive bacteria | MIC | MBC | MIC | MBC | MIC | MBC | MIC | MBC | |
| S. aureus ATCC 25923 | >1000 | Nd | 31.25 | 1000 | 15.6 | 125 | 0.98 | 0.98 | |
| S. aureus ATCC BAA-1707 * | >1000 | Nd | 31.25 | 1000 | 15.6 | 1000 | 0.98 | 0.98 | |
| S. epidermidis ATCC 12228 | >1000 | Nd | 7.8 | 1000 | 15.6 | 15.6 | 0.98 | 0.98 | |
| M. luteus ATCC 10240 | >1000 | Nd | 7.8 | 1000 | 7.8 | 62.5 | 0.12 | 0.12 | |
| E. faecalis ATCC 29212 | >1000 | Nd | 15.6 | 1000 | 15.6 | 1000 | 1.95 | 3.9 | |
| B. cereus ATCC 10876 | >1000 | Nd | 15.6 | 1000 | 15.6 | 15.6 | 0.98 | 15.6 | |
| Gram-negative bacteria | Ciprofloxacin | ||||||||
| S. typhimurium ATCC 14028 | >1000 | Nd | >1000 | >1000 | >1000 | Nd | 0.061 | 0.06 | |
| E. coli ATCC 25922 | >1000 | Nd | >1000 | >1000 | >1000 | Nd | 0.015 | 0.08 | |
| P. mirabilis ATCC 12453 | >1000 | Nd | 250 | 1000 | >1000 | Nd | 0.030 | 0.03 | |
| K. pneumoniae ATCC 13883 | >1000 | Nd | >1000 | >1000 | >1000 | Nd | 0.122 | 0.24 | |
| P. aeruginosa ATCC 9027 | >1000 | Nd | >1000 | >1000 | >1000 | Nd | 0.488 | 0.98 | |
| Yeasts | MIC | MFC | MIC | MFC | MIC | MFC | Nystatin | ||
| C. albicans ATCC 102231 | >1000 | Nd | >1000 | >1000 | >1000 | >1000 | 0.48 | 0.48 | |
| C. parapsilosis ATCC 22019 | >1000 | Nd | >1000 | >1000 | >1000 | >1000 | 0.24 | 0.48 | |
| C. glabrata ATCC 90030 | >1000 | Nd | >1000 | >1000 | 500 | >1000 | 0.24 | 0.48 | |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Olczak, A.; Pawlak, T.; Kałużyńska, S.; Gobis, K.; Korona-Głowniak, I.; Suśniak, K.; Zaborowski, M.; Szczesio, M. Structure and Microbiological Activity of 1H-benzo[d]imidazole Derivatives. Int. J. Mol. Sci. 2023, 24, 3319. https://doi.org/10.3390/ijms24043319
Olczak A, Pawlak T, Kałużyńska S, Gobis K, Korona-Głowniak I, Suśniak K, Zaborowski M, Szczesio M. Structure and Microbiological Activity of 1H-benzo[d]imidazole Derivatives. International Journal of Molecular Sciences. 2023; 24(4):3319. https://doi.org/10.3390/ijms24043319
Chicago/Turabian StyleOlczak, Andrzej, Tomasz Pawlak, Sylwia Kałużyńska, Katarzyna Gobis, Izabela Korona-Głowniak, Katarzyna Suśniak, Marcin Zaborowski, and Małgorzata Szczesio. 2023. "Structure and Microbiological Activity of 1H-benzo[d]imidazole Derivatives" International Journal of Molecular Sciences 24, no. 4: 3319. https://doi.org/10.3390/ijms24043319
APA StyleOlczak, A., Pawlak, T., Kałużyńska, S., Gobis, K., Korona-Głowniak, I., Suśniak, K., Zaborowski, M., & Szczesio, M. (2023). Structure and Microbiological Activity of 1H-benzo[d]imidazole Derivatives. International Journal of Molecular Sciences, 24(4), 3319. https://doi.org/10.3390/ijms24043319








