Longevity, Centenarians and Modified Cellular Proteodynamics
Abstract
1. Introduction
2. The Question of DNA Maintenance
3. Transcription
3.1. mRNA: Quality (Transcription Fidelity) and Quantity
3.2. Noncoding RNAs: miRNA and lncRNA
3.3. Posttranscriptional RNA Modification
4. Translation: Effectiveness and Fidelity
5. Energy Requirement of Proteodynamics: Do Centenarians Stand Out from among Aged Individuals?
6. Effectiveness of Posttranslational Modifications
7. Proteomic Signatures in Centenarians
7.1. Proteostatic Systems and Autophagy in Longevity
7.1.1. Autophagy—The Ubiquitin-Proteasome System (UPS)
7.1.2. Autophagy—Lysosome Pathway
7.1.3. The Calpain–Calpastatin System
7.1.4. ADAMS and ADAMTS
8. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Borras, C.; Ingles, M.; Mas-Bargues, C.; Dromant, M.; Sanz-Ros, J.; Roman-Dominguez, A.; Gimeno-Mallench, L.; Gambini, J.; Vina, J. Centenarians: An excellent example of resilience for successful ageing. Mech. Ageing Dev. 2020, 186, 111199. [Google Scholar] [CrossRef] [PubMed]
- Frankowska, N.; Lisowska, K.; Witkowski, J.M. Proteolysis dysfunction in the process of aging and age-related diseases. Front. Aging 2022, 3, 927630. [Google Scholar] [CrossRef]
- Montesanto, A.; Dato, S.; Bellizzi, D.; Rose, G.; Passarino, G. Epidemiological, genetic and epigenetic aspects of the research on healthy ageing and longevity. Immun. Ageing 2012, 9, 6. [Google Scholar] [CrossRef] [PubMed]
- Ingles, M.; Mas-Bargues, C.; Berna-Erro, A.; Matheu, A.; Sanchis, P.; Avellana, J.A.; Borras, C.; Vina, J. Centenarians Overexpress Pluripotency-Related Genes. J. Gerontol. Ser. A 2019, 74, 1391–1395. [Google Scholar] [CrossRef] [PubMed]
- Sgarbi, G.; Matarrese, P.; Pinti, M.; Lanzarini, C.; Ascione, B.; Gibellini, L.; Dika, E.; Patrizi, A.; Tommasino, C.; Capri, M.; et al. Mitochondria hyperfusion and elevated autophagic activity are key mechanisms for cellular bioenergetic preservation in centenarians. Aging 2014, 6, 296–310. [Google Scholar] [CrossRef]
- Franceschi, C.; Bonafe, M.; Valensin, S.; Olivieri, F.; De Luca, M.; Ottaviani, E.; De Benedictis, G. Inflamm-aging. An evolutionary perspective on immunosenescence. Ann. N. Y. Acad. Sci. 2000, 908, 244–254. [Google Scholar] [CrossRef]
- Franceschi, C.; Bonafe, M. Centenarians as a model for healthy aging. Biochem. Soc. Trans. 2003, 31, 457–461. [Google Scholar] [CrossRef]
- Ostan, R.; Bucci, L.; Capri, M.; Salvioli, S.; Scurti, M.; Pini, E.; Monti, D.; Franceschi, C. Immunosenescence and immunogenetics of human longevity. Neuroimmunomodulation 2008, 15, 224–240. [Google Scholar] [CrossRef]
- Candore, G.; Caruso, C.; Colonna-Romano, G. Inflammation, genetic background and longevity. Biogerontology 2010, 11, 565–573. [Google Scholar] [CrossRef]
- Salvioli, S.; Monti, D.; Lanzarini, C.; Conte, M.; Pirazzini, C.; Bacalini, M.G.; Garagnani, P.; Giuliani, C.; Fontanesi, E.; Ostan, R.; et al. Immune system, cell senescence, aging and longevity--inflamm-aging reappraised. Curr. Pharm. Des. 2013, 19, 1675–1679. [Google Scholar]
- Minciullo, P.L.; Catalano, A.; Mandraffino, G.; Casciaro, M.; Crucitti, A.; Maltese, G.; Morabito, N.; Lasco, A.; Gangemi, S.; Basile, G. Inflammaging and Anti-Inflammaging: The Role of Cytokines in Extreme Longevity. Arch. Immunol. Ther. Exp. 2016, 64, 111–126. [Google Scholar] [CrossRef]
- Fulop, T.; Witkowski, J.M.; Olivieri, F.; Larbi, A. The integration of inflammaging in age-related diseases. Semin. Immunol. 2018, 40, 17–35. [Google Scholar] [CrossRef]
- Santos-Lozano, A.; Valenzuela, P.L.; Llavero, F.; Lista, S.; Carrera-Bastos, P.; Hampel, H.; Pareja-Galeano, H.; Galvez, B.G.; Lopez, J.A.; Vazquez, J.; et al. Successful aging: Insights from proteome analyses of healthy centenarians. Aging 2020, 12, 3502–3515. [Google Scholar] [CrossRef] [PubMed]
- Fulop, T.; Larbi, A.; Hirokawa, K.; Cohen, A.A.; Witkowski, J.M. Immunosenescence is both functional/adaptive and dysfunctional/maladaptive. Semin. Immunopathol. 2020, 42, 521–536. [Google Scholar] [CrossRef]
- Moskalev, A.; Stambler, I.; Caruso, C. Innate and Adaptive Immunity in Aging and Longevity: The Foundation of Resilience. Aging Dis. 2020, 11, 1363–1373. [Google Scholar] [CrossRef] [PubMed]
- Santoro, A.; Bientinesi, E.; Monti, D. Immunosenescence and inflammaging in the aging process: Age-related diseases or longevity? Ageing Res. Rev. 2021, 71, 101422. [Google Scholar] [CrossRef] [PubMed]
- Zhou, L.; Ge, M.; Zhang, Y.; Wu, X.; Leng, M.; Gan, C.; Mou, Y.; Zhou, J.; Valencia, C.A.; Hao, Q.; et al. Centenarians Alleviate Inflammaging by Changing the Ratio and Secretory Phenotypes of T Helper 17 and Regulatory T Cells. Front. Pharmacol. 2022, 13, 877709. [Google Scholar] [CrossRef]
- Vitale, G.; Barbieri, M.; Kamenetskaya, M.; Paolisso, G. GH/IGF-I/insulin system in centenarians. Mech. Ageing Dev. 2017, 165, 107–114. [Google Scholar] [CrossRef]
- Savage, N. Proteomics: High-protein research. Nature 2015, 527, S6–S7. [Google Scholar] [CrossRef]
- Witkowski, J.M.; Mikosik, A.; Bryl, E.; Fulop, T. Proteodynamics in aging human T cells-The need for its comprehensive study to understand the fine regulation of T lymphocyte functions. Exp. Gerontol. 2018, 107, 161–168. [Google Scholar] [CrossRef]
- Witkowski, J.M.; Bryl, E.; Fulop, T. Proteodynamics and aging of eukaryotic cells. Mech. Ageing Dev. 2021, 194, 111430. [Google Scholar] [CrossRef] [PubMed]
- Cenci, S.; van Anken, E.; Sitia, R. Proteostenosis and plasma cell pathophysiology. Curr. Opin. Cell Biol. 2011, 23, 216–222. [Google Scholar] [CrossRef] [PubMed]
- Cho, M.; Suh, Y. Genome maintenance and human longevity. Curr. Opin. Genet. Dev. 2014, 26, 105–115. [Google Scholar] [CrossRef] [PubMed]
- Vijg, J.; Perls, T.; Franceschi, C.; van Orsouw, N.J. BRCA1 gene sequence variation in centenarians. Ann. N. Y. Acad. Sci. 2001, 928, 85–96. [Google Scholar] [CrossRef]
- Orans, J.; McSweeney, E.A.; Iyer, R.R.; Hast, M.A.; Hellinga, H.W.; Modrich, P.; Beese, L.S. Structures of human exonuclease 1 DNA complexes suggest a unified mechanism for nuclease family. Cell 2011, 145, 212–223. [Google Scholar] [CrossRef] [PubMed]
- Han, J.; Ryu, S.; Moskowitz, D.M.; Rothenberg, D.; Leahy, D.J.; Atzmon, G.; Barzilai, N.; Suh, Y. Discovery of novel non-synonymous SNP variants in 988 candidate genes from 6 centenarians by target capture and next-generation sequencing. Mech. Ageing Dev. 2013, 134, 478–485. [Google Scholar] [CrossRef] [PubMed]
- Nebel, A.; Flachsbart, F.; Till, A.; Caliebe, A.; Blanche, H.; Arlt, A.; Hasler, R.; Jacobs, G.; Kleindorp, R.; Franke, A.; et al. A functional EXO1 promoter variant is associated with prolonged life expectancy in centenarians. Mech. Ageing Dev. 2009, 130, 691–699. [Google Scholar] [CrossRef]
- Polosak, J.; Roszkowska-Gancarz, M.; Kurylowicz, A.; Owczarz, M.; Dobosz, P.; Mossakowska, M.; Szybinska, A.; Puzianowska-Kuznicka, M. Decreased expression and the Lys751Gln polymorphism of the XPD gene are associated with extreme longevity. Biogerontology 2010, 11, 287–297. [Google Scholar] [CrossRef]
- Piaceri, I.; Bagnoli, S.; Tedde, A.; Sorbi, S.; Nacmias, B. Ataxia-telangiectasia mutated (ATM) genetic variant in Italian centenarians. Neurol. Sci. 2013, 34, 573–575. [Google Scholar] [CrossRef]
- Chen, T.; Dong, B.; Lu, Z.; Tian, B.; Zhang, J.; Zhou, J.; Wu, H.; Zhang, Y.; Wu, J.; Lin, P.; et al. A functional single nucleotide polymorphism in promoter of ATM is associated with longevity. Mech. Ageing Dev. 2010, 131, 636–640. [Google Scholar] [CrossRef]
- Hadar, A.; Voinsky, I.; Parkhomenko, O.; Puzianowska-Kuznicka, M.; Kuznicki, J.; Gozes, I.; Gurwitz, D. Higher ATM expression in lymphoblastoid cell lines from centenarian compared with younger women. Drug Dev. Res. 2022, 83, 1419–1424. [Google Scholar] [CrossRef]
- Storci, G.; De Carolis, S.; Papi, A.; Bacalini, M.G.; Gensous, N.; Marasco, E.; Tesei, A.; Fabbri, F.; Arienti, C.; Zanoni, M.; et al. Genomic stability, anti-inflammatory phenotype, and up-regulation of the RNAseH2 in cells from centenarians. Cell Death Differ. 2019, 26, 1845–1858. [Google Scholar] [CrossRef] [PubMed]
- Lee, C.K.; Klopp, R.G.; Weindruch, R.; Prolla, T.A. Gene expression profile of aging and its retardation by caloric restriction. Science 1999, 285, 1390–1393. [Google Scholar] [CrossRef] [PubMed]
- Borras, C.; Abdelaziz, K.M.; Gambini, J.; Serna, E.; Ingles, M.; de la Fuente, M.; Garcia, I.; Matheu, A.; Sanchis, P.; Belenguer, A.; et al. Human exceptional longevity: Transcriptome from centenarians is distinct from septuagenarians and reveals a role of Bcl-xL in successful aging. Aging 2016, 8, 3185–3208. [Google Scholar] [CrossRef]
- Capri, M.; Salvioli, S.; Sevini, F.; Valensin, S.; Celani, L.; Monti, D.; Pawelec, G.; De Benedictis, G.; Gonos, E.S.; Franceschi, C. The genetics of human longevity. Ann. N. Y. Acad. Sci. 2006, 1067, 252–263. [Google Scholar] [CrossRef]
- Field, A.E.; Adams, P.D. Targeting chromatin aging-The epigenetic impact of longevity-associated interventions. Exp. Gerontol. 2017, 94, 29–33. [Google Scholar] [CrossRef]
- Balistreri, C.R.; Candore, G.; Accardi, G.; Bova, M.; Buffa, S.; Bulati, M.; Forte, G.I.; Listi, F.; Martorana, A.; Palmeri, M.; et al. Genetics of longevity. data from the studies on Sicilian centenarians. Immun. Ageing 2012, 9, 8. [Google Scholar] [CrossRef]
- Heyn, H.; Li, N.; Ferreira, H.J.; Moran, S.; Pisano, D.G.; Gomez, A.; Diez, J.; Sanchez-Mut, J.V.; Setien, F.; Carmona, F.J.; et al. Distinct DNA methylomes of newborns and centenarians. Proc. Natl. Acad. Sci. USA 2012, 109, 10522–10527. [Google Scholar] [CrossRef]
- Winnefeld, M.; Lyko, F. The aging epigenome: DNA methylation from the cradle to the grave. Genome Biol. 2012, 13, 165. [Google Scholar] [CrossRef] [PubMed]
- Gentilini, D.; Mari, D.; Castaldi, D.; Remondini, D.; Ogliari, G.; Ostan, R.; Bucci, L.; Sirchia, S.M.; Tabano, S.; Cavagnini, F.; et al. Role of epigenetics in human aging and longevity: Genome-wide DNA methylation profile in centenarians and centenarians’ offspring. Age 2013, 35, 1961–1973. [Google Scholar] [CrossRef] [PubMed]
- Brooks-Wilson, A.R. Genetics of healthy aging and longevity. Hum. Genet. 2013, 132, 1323–1338. [Google Scholar] [CrossRef] [PubMed]
- Xiao, F.H.; He, Y.H.; Li, Q.G.; Wu, H.; Luo, L.H.; Kong, Q.P. A genome-wide scan reveals important roles of DNA methylation in human longevity by regulating age-related disease genes. PLoS ONE 2015, 10, e0120388. [Google Scholar] [CrossRef] [PubMed]
- Horvath, S.; Pirazzini, C.; Bacalini, M.G.; Gentilini, D.; Di Blasio, A.M.; Delledonne, M.; Mari, D.; Arosio, B.; Monti, D.; Passarino, G.; et al. Decreased epigenetic age of PBMCs from Italian semi-supercentenarians and their offspring. Aging 2015, 7, 1159–1170. [Google Scholar] [CrossRef] [PubMed]
- Horvath, S. DNA methylation age of human tissues and cell types. Genome Biol. 2013, 14, R115. [Google Scholar] [CrossRef]
- Zhao, M.; Qin, J.; Yin, H.; Tan, Y.; Liao, W.; Liu, Q.; Luo, S.; He, M.; Liang, G.; Shi, Y.; et al. Distinct epigenomes in CD4(+) T cells of newborns, middle-ages and centenarians. Sci. Rep. 2016, 6, 38411. [Google Scholar] [CrossRef]
- Armstrong, N.J.; Mather, K.A.; Thalamuthu, A.; Wright, M.J.; Trollor, J.N.; Ames, D.; Brodaty, H.; Schofield, P.R.; Sachdev, P.S.; Kwok, J.B. Aging, exceptional longevity and comparisons of the Hannum and Horvath epigenetic clocks. Epigenomics 2017, 9, 689–700. [Google Scholar] [CrossRef]
- Gutman, D.; Rivkin, E.; Fadida, A.; Sharvit, L.; Hermush, V.; Rubin, E.; Kirshner, D.; Sabin, I.; Dwolatzky, T.; Atzmon, G. Exceptionally Long-Lived Individuals (ELLI) Demonstrate Slower Aging Rate Calculated by DNA Methylation Clocks as Possible Modulators for Healthy Longevity. Int. J. Mol. Sci. 2020, 21, 615. [Google Scholar] [CrossRef] [PubMed]
- Daunay, A.; Hardy, L.M.; Bouyacoub, Y.; Sahbatou, M.; Touvier, M.; Blanche, H.; Deleuze, J.F.; How-Kit, A. Centenarians consistently present a younger epigenetic age than their chronological age with four epigenetic clocks based on a small number of CpG sites. Aging 2022, 14, 7718–7733. [Google Scholar] [CrossRef] [PubMed]
- Puca, A.A.; Spinelli, C.; Accardi, G.; Villa, F.; Caruso, C. Centenarians as a model to discover genetic and epigenetic signatures of healthy ageing. Mech. Ageing Dev. 2018, 174, 95–102. [Google Scholar] [CrossRef]
- Ingles, M.; Belenguer-Varea, A.; Serna, E.; Mas-Bargues, C.; Tarazona-Santabalbina, F.J.; Borras, C.; Vina, J. Functional Transcriptomic Analysis of Centenarians’ Offspring Reveals a Specific Genetic Footprint That May Explain That They Are Less Frail Than Age-Matched Noncentenarians’ Offspring. J. Gerontol. Ser. A 2022, 77, 1931–1938. [Google Scholar] [CrossRef]
- Olivieri, F.; Spazzafumo, L.; Santini, G.; Lazzarini, R.; Albertini, M.C.; Rippo, M.R.; Galeazzi, R.; Abbatecola, A.M.; Marcheselli, F.; Monti, D.; et al. Age-related differences in the expression of circulating microRNAs: miR-21 as a new circulating marker of inflammaging. Mech. Ageing Dev. 2012, 133, 675–685. [Google Scholar] [CrossRef] [PubMed]
- Jiang, J.; Cheng, L.; Yan, L.; Ge, M.; Yang, L.; Ying, H.; Kong, Q. Decoding the role of long noncoding RNAs in the healthy aging of centenarians. Brief. Bioinform. 2021, 22, bbaa439. [Google Scholar] [CrossRef] [PubMed]
- Jiapaer, Z.; Su, D.; Hua, L.; Lehmann, H.I.; Gokulnath, P.; Vulugundam, G.; Song, S.; Zhang, L.; Gong, Y.; Li, G. Regulation and roles of RNA modifications in aging-related diseases. Aging Cell 2022, 21, e13657. [Google Scholar] [CrossRef] [PubMed]
- Roundtree, I.A.; Evans, M.E.; Pan, T.; He, C. Dynamic RNA Modifications in Gene Expression Regulation. Cell 2017, 169, 1187–1200. [Google Scholar] [CrossRef]
- Sun, J.; Cheng, B.; Su, Y.; Li, M.; Ma, S.; Zhang, Y.; Zhang, A.; Cai, S.; Bao, Q.; Wang, S.; et al. The Potential Role of m6A RNA Methylation in the Aging Process and Aging-Associated Diseases. Front. Genet. 2022, 13, 869950. [Google Scholar] [CrossRef]
- Casella, G.; Tsitsipatis, D.; Abdelmohsen, K.; Gorospe, M. mRNA methylation in cell senescence. Wiley Interdiscip. Rev. RNA 2019, 10, e1547. [Google Scholar] [CrossRef]
- Cai, X.; Hu, Y.; Tang, H.; Hu, H.; Pang, L.; Xing, J.; Liu, Z.; Luo, Y.; Jiang, B.; Liu, T.; et al. RNA methyltransferase NSUN2 promotes stress-induced HUVEC senescence. Oncotarget 2016, 7, 19099–19110. [Google Scholar] [CrossRef]
- De Jesus, D.F.; Zhang, Z.; Kahraman, S.; Brown, N.K.; Chen, M.; Hu, J.; Gupta, M.K.; He, C.; Kulkarni, R.N. m(6)A mRNA Methylation Regulates Human beta-Cell Biology in Physiological States and in Type 2 Diabetes. Nat. Metab. 2019, 1, 765–774. [Google Scholar] [CrossRef]
- Zhao, T.; Li, X.; Sun, D.; Zhang, Z. Oxidative stress: One potential factor for arsenite-induced increase of N(6)-methyladenosine in human keratinocytes. Environ. Toxicol. Pharmacol. 2019, 69, 95–103. [Google Scholar] [CrossRef]
- Wu, J.; Frazier, K.; Zhang, J.; Gan, Z.; Wang, T.; Zhong, X. Emerging role of m(6) A RNA methylation in nutritional physiology and metabolism. Obes. Rev. 2020, 21, e12942. [Google Scholar] [CrossRef]
- Li, J.; Han, Y.; Zhang, H.; Qian, Z.; Jia, W.; Gao, Y.; Zheng, H.; Li, B. The m6A demethylase FTO promotes the growth of lung cancer cells by regulating the m6A level of USP7 mRNA. Biochem. Biophys. Res. Commun. 2019, 512, 479–485. [Google Scholar] [CrossRef] [PubMed]
- Shafik, A.M.; Zhang, F.; Guo, Z.; Dai, Q.; Pajdzik, K.; Li, Y.; Kang, Y.; Yao, B.; Wu, H.; He, C.; et al. N6-methyladenosine dynamics in neurodevelopment and aging, and its potential role in Alzheimer’s disease. Genome Biol. 2021, 22, 17. [Google Scholar] [CrossRef] [PubMed]
- Zhao, F.; Xu, Y.; Gao, S.; Qin, L.; Austria, Q.; Siedlak, S.L.; Pajdzik, K.; Dai, Q.; He, C.; Wang, W.; et al. METTL3-dependent RNA m(6)A dysregulation contributes to neurodegeneration in Alzheimer’s disease through aberrant cell cycle events. Mol. Neurodegener. 2021, 16, 70. [Google Scholar] [CrossRef] [PubMed]
- Thomas, S.; Mukherjee, A.B. A longitudinal study of human age-related ribosomal RNA gene activity as detected by silver-stained NORs. Mech. Ageing Dev. 1996, 92, 101–109. [Google Scholar] [CrossRef]
- Reis-Rodrigues, P.; Czerwieniec, G.; Peters, T.W.; Evani, U.S.; Alavez, S.; Gaman, E.A.; Vantipalli, M.; Mooney, S.D.; Gibson, B.W.; Lithgow, G.J.; et al. Proteomic analysis of age-dependent changes in protein solubility identifies genes that modulate lifespan. Aging Cell 2012, 11, 120–127. [Google Scholar] [CrossRef]
- Jung, M.; Jin, S.G.; Zhang, X.; Xiong, W.; Gogoshin, G.; Rodin, A.S.; Pfeifer, G.P. Longitudinal epigenetic and gene expression profiles analyzed by three-component analysis reveal down-regulation of genes involved in protein translation in human aging. Nucleic Acids Res. 2015, 43, e100. [Google Scholar] [CrossRef]
- MacInnes, A.W. The role of the ribosome in the regulation of longevity and lifespan extension. Wiley Interdiscip. Rev. RNA 2016, 7, 198–212. [Google Scholar] [CrossRef]
- Hadar, A.; Milanesi, E.; Walczak, M.; Puzianowska-Kuznicka, M.; Kuznicki, J.; Squassina, A.; Niola, P.; Chillotti, C.; Attems, J.; Gozes, I.; et al. SIRT1, miR-132 and miR-212 link human longevity to Alzheimer’s Disease. Sci. Rep. 2018, 8, 8465. [Google Scholar] [CrossRef]
- Blagosklonny, M.V. Why human lifespan is rapidly increasing: Solving “longevity riddle” with “revealed-slow-aging” hypothesis. Aging 2010, 2, 177–182. [Google Scholar] [CrossRef]
- Robert, L.; Fulop, T. Longevity and its regulation: Centenarians and beyond. Interdiscip. Top. Gerontol. 2014, 39, 198–211. [Google Scholar] [CrossRef]
- Lopez-Otin, C.; Blasco, M.A.; Partridge, L.; Serrano, M.; Kroemer, G. The hallmarks of aging. Cell 2013, 153, 1194–1217. [Google Scholar] [CrossRef] [PubMed]
- Mota-Martorell, N.; Jove, M.; Pamplona, R. mTOR Complex 1 Content and Regulation Is Adapted to Animal Longevity. Int. J. Mol. Sci. 2022, 23, 8747. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Z.; Sun, B.; Yu, D.; Bian, M. Roles of tRNA metabolism in aging and lifespan. Cell Death Dis. 2021, 12, 548. [Google Scholar] [CrossRef] [PubMed]
- Tang, H.; Fan, X.; Xing, J.; Liu, Z.; Jiang, B.; Dou, Y.; Gorospe, M.; Wang, W. NSun2 delays replicative senescence by repressing p27 (KIP1) translation and elevating CDK1 translation. Aging 2015, 7, 1143–1158. [Google Scholar] [CrossRef] [PubMed]
- Lewinska, A.; Adamczyk-Grochala, J.; Kwasniewicz, E.; Deregowska, A.; Semik, E.; Zabek, T.; Wnuk, M. Reduced levels of methyltransferase DNMT2 sensitize human fibroblasts to oxidative stress and DNA damage that is accompanied by changes in proliferation-related miRNA expression. Redox Biol. 2018, 14, 20–34. [Google Scholar] [CrossRef]
- Schosserer, M.; Minois, N.; Angerer, T.B.; Amring, M.; Dellago, H.; Harreither, E.; Calle-Perez, A.; Pircher, A.; Gerstl, M.P.; Pfeifenberger, S.; et al. Methylation of ribosomal RNA by NSUN5 is a conserved mechanism modulating organismal lifespan. Nat. Commun. 2015, 6, 6158. [Google Scholar] [CrossRef]
- Li, H.; Ren, M.; Li, Q. (1)H NMR-Based Metabolomics Reveals the Intrinsic Interaction of Age, Plasma Signature Metabolites, and Nutrient Intake in the Longevity Population in Guangxi, China. Nutrients 2022, 14, 2539. [Google Scholar] [CrossRef]
- Han, K.; Ma, J.; Dou, J.; Hao, D.; Zhu, W.; Yu, X.; Zheng, W.; Song, Y.; Shi, F.; Li, Q. A Clinical Trial of the Effects of a Dietary Pattern on Health Metrics and Fecal Metabolites in Volunteers With Risk of Cardiovascular Disease. Front. Nutr. 2022, 9, 853365. [Google Scholar] [CrossRef]
- Orgel, L.E. The maintenance of the accuracy of protein synthesis and its relevance to ageing. Proc. Natl. Acad. Sci. USA 1963, 49, 517–521. [Google Scholar] [CrossRef] [PubMed]
- Ke, Z.; Seluanov, A.; Gorbunova, V. Accurate translation is important for longevity. Aging 2018, 10, 297–298. [Google Scholar] [CrossRef] [PubMed]
- Azpurua, J.; Ke, Z.; Chen, I.X.; Zhang, Q.; Ermolenko, D.N.; Zhang, Z.D.; Gorbunova, V.; Seluanov, A. Naked mole-rat has increased translational fidelity compared with the mouse, as well as a unique 28S ribosomal RNA cleavage. Proc. Natl. Acad. Sci. USA 2013, 110, 17350–17355. [Google Scholar] [CrossRef] [PubMed]
- Ke, Z.; Mallik, P.; Johnson, A.B.; Luna, F.; Nevo, E.; Zhang, Z.D.; Gladyshev, V.N.; Seluanov, A.; Gorbunova, V. Translation fidelity coevolves with longevity. Aging Cell 2017, 16, 988–993. [Google Scholar] [CrossRef] [PubMed]
- Varanda, A.S.; Santos, M.; Soares, A.R.; Vitorino, R.; Oliveira, P.; Oliveira, C.; Santos, M.A.S. Human cells adapt to translational errors by modulating protein synthesis rate and protein turnover. RNA Biol. 2020, 17, 135–149. [Google Scholar] [CrossRef]
- Francisco, S.; Ferreira, M.; Moura, G.; Soares, A.R.; Santos, M.A.S. Does proteostasis get lost in translation? Implications for protein aggregation across the lifespan. Ageing Res. Rev. 2020, 62, 101119. [Google Scholar] [CrossRef] [PubMed]
- Richman, T.R.; Ermer, J.A.; Siira, S.J.; Kuznetsova, I.; Brosnan, C.A.; Rossetti, G.; Baker, J.; Perks, K.L.; Cserne Szappanos, H.; Viola, H.M.; et al. Mitochondrial mistranslation modulated by metabolic stress causes cardiovascular disease and reduced lifespan. Aging Cell 2021, 20, e13408. [Google Scholar] [CrossRef] [PubMed]
- Bender, A.; Krishnan, K.J.; Morris, C.M.; Taylor, G.A.; Reeve, A.K.; Perry, R.H.; Jaros, E.; Hersheson, J.S.; Betts, J.; Klopstock, T.; et al. High levels of mitochondrial DNA deletions in substantia nigra neurons in aging and Parkinson disease. Nat. Genet. 2006, 38, 515–517. [Google Scholar] [CrossRef]
- Tranah, G.J. Mitochondrial-nuclear epistasis: Implications for human aging and longevity. Ageing Res. Rev. 2011, 10, 238–252. [Google Scholar] [CrossRef]
- Short, K.R.; Bigelow, M.L.; Kahl, J.; Singh, R.; Coenen-Schimke, J.; Raghavakaimal, S.; Nair, K.S. Decline in skeletal muscle mitochondrial function with aging in humans. Proc. Natl. Acad. Sci. USA 2005, 102, 5618–5623. [Google Scholar] [CrossRef]
- Layec, G.; Trinity, J.D.; Hart, C.R.; Le Fur, Y.; Zhao, J.; Reese, V.; Jeong, E.K.; Richardson, R.S. Impaired Muscle Efficiency but Preserved Peripheral Hemodynamics and Mitochondrial Function With Advancing Age: Evidence From Exercise in the Young, Old, and Oldest-Old. J. Gerontol. Ser. A 2018, 73, 1303–1312. [Google Scholar] [CrossRef]
- De Benedictis, G.; Rose, G.; Carrieri, G.; De Luca, M.; Falcone, E.; Passarino, G.; Bonafe, M.; Monti, D.; Baggio, G.; Bertolini, S.; et al. Mitochondrial DNA inherited variants are associated with successful aging and longevity in humans. FASEB J. 1999, 13, 1532–1536. [Google Scholar] [CrossRef]
- Niemi, A.K.; Hervonen, A.; Hurme, M.; Karhunen, P.J.; Jylha, M.; Majamaa, K. Mitochondrial DNA polymorphisms associated with longevity in a Finnish population. Hum. Genet. 2003, 112, 29–33. [Google Scholar] [CrossRef] [PubMed]
- Dato, S.; Passarino, G.; Rose, G.; Altomare, K.; Bellizzi, D.; Mari, V.; Feraco, E.; Franceschi, C.; De Benedictis, G. Association of the mitochondrial DNA haplogroup J with longevity is population specific. Eur. J. Hum. Genet. 2004, 12, 1080–1082. [Google Scholar] [CrossRef] [PubMed]
- Ross, O.A.; McCormack, R.; Curran, M.D.; Duguid, R.A.; Barnett, Y.A.; Rea, I.M.; Middleton, D. Mitochondrial DNA polymorphism: Its role in longevity of the Irish population. Exp. Gerontol. 2001, 36, 1161–1178. [Google Scholar] [CrossRef]
- de Benedictis, G.; Carrieri, G.; Varcasia, O.; Bonafe, M.; Franceschi, C. Inherited variability of the mitochondrial genome and successful aging in humans. Ann. N. Y. Acad. Sci. 2000, 908, 208–218. [Google Scholar] [CrossRef]
- Ristow, M.; Zarse, K. How increased oxidative stress promotes longevity and metabolic health: The concept of mitochondrial hormesis (mitohormesis). Exp. Gerontol. 2010, 45, 410–418. [Google Scholar] [CrossRef] [PubMed]
- Barcena, C.; Mayoral, P.; Quiros, P.M. Mitohormesis, an Antiaging Paradigm. Int. Rev. Cell Mol. Biol. 2018, 340, 35–77. [Google Scholar] [CrossRef] [PubMed]
- Wei, Y.; Zhang, Y.J.; Cai, Y.; Xu, M.H. The role of mitochondria in mTOR-regulated longevity. Biol. Rev. Camb. Philos. Soc. 2015, 90, 167–181. [Google Scholar] [CrossRef]
- Miura, Y.; Hashii, N.; Tsumoto, H.; Takakura, D.; Ohta, Y.; Abe, Y.; Arai, Y.; Kawasaki, N.; Hirose, N.; Endo, T.; et al. Change in N-Glycosylation of Plasma Proteins in Japanese Semisupercentenarians. PLoS ONE 2015, 10, e0142645. [Google Scholar] [CrossRef]
- Dall’Olio, F.; Vanhooren, V.; Chen, C.C.; Slagboom, P.E.; Wuhrer, M.; Franceschi, C. N-glycomic biomarkers of biological aging and longevity: A link with inflammaging. Ageing Res. Rev. 2013, 12, 685–698. [Google Scholar] [CrossRef]
- Vanhooren, V.; Desmyter, L.; Liu, X.E.; Cardelli, M.; Franceschi, C.; Federico, A.; Libert, C.; Laroy, W.; Dewaele, S.; Contreras, R.; et al. N-glycomic changes in serum proteins during human aging. Rejuvenation Res. 2007, 10, 521–531a. [Google Scholar] [CrossRef]
- Ohtsubo, K.; Marth, J.D. Glycosylation in cellular mechanisms of health and disease. Cell 2006, 126, 855–867. [Google Scholar] [CrossRef] [PubMed]
- Tesi, N.; van der Lee, S.J.; Hulsman, M.; Jansen, I.E.; Stringa, N.; van Schoor, N.M.; Scheltens, P.; van der Flier, W.M.; Huisman, M.; Reinders, M.J.T.; et al. Immune response and endocytosis pathways are associated with the resilience against Alzheimer’s disease. Transl. Psychiatry 2020, 10, 332. [Google Scholar] [CrossRef] [PubMed]
- Jeong, A.; Suazo, K.F.; Wood, W.G.; Distefano, M.D.; Li, L. Isoprenoids and protein prenylation: Implications in the pathogenesis and therapeutic intervention of Alzheimer’s disease. Crit. Rev. Biochem. Mol. Biol. 2018, 53, 279–310. [Google Scholar] [CrossRef] [PubMed]
- Gordon, L.B.; Kleinman, M.E.; Miller, D.T.; Neuberg, D.S.; Giobbie-Hurder, A.; Gerhard-Herman, M.; Smoot, L.B.; Gordon, C.M.; Cleveland, R.; Snyder, B.D.; et al. Clinical trial of a farnesyltransferase inhibitor in children with Hutchinson-Gilford progeria syndrome. Proc. Natl. Acad. Sci. USA 2012, 109, 16666–16671. [Google Scholar] [CrossRef] [PubMed]
- Muiras, M.L.; Muller, M.; Schachter, F.; Burkle, A. Increased poly(ADP-ribose) polymerase activity in lymphoblastoid cell lines from centenarians. J. Mol. Med. 1998, 76, 346–354. [Google Scholar] [CrossRef] [PubMed]
- Chevanne, M.; Calia, C.; Zampieri, M.; Cecchinelli, B.; Caldini, R.; Monti, D.; Bucci, L.; Franceschi, C.; Caiafa, P. Oxidative DNA damage repair and parp 1 and parp 2 expression in Epstein-Barr virus-immortalized B lymphocyte cells from young subjects, old subjects, and centenarians. Rejuvenation Res. 2007, 10, 191–204. [Google Scholar] [CrossRef]
- Kang, M.; Park, S.; Park, S.H.; Lee, H.G.; Park, J.H. A Double-Edged Sword: The Two Faces of PARylation. Int. J. Mol. Sci. 2022, 23, 9826. [Google Scholar] [CrossRef]
- Mangerich, A.; Burkle, A. Pleiotropic cellular functions of PARP1 in longevity and aging: Genome maintenance meets inflammation. Oxid. Med. Cell. Longev. 2012, 2012, 321653. [Google Scholar] [CrossRef]
- Sanchez-Roman, I.; Ferrando, B.; Holst, C.M.; Mengel-From, J.; Rasmussen, S.H.; Thinggaard, M.; Bohr, V.A.; Christensen, K.; Stevnsner, T. Molecular markers of DNA repair and brain metabolism correlate with cognition in centenarians. Geroscience 2022, 44, 103–125. [Google Scholar] [CrossRef]
- Castellani, R.J. The Significance of Tau Aggregates in the Human Brain. Brain Sci. 2020, 10, 972. [Google Scholar] [CrossRef]
- Kang, D.; Baek, Y.; Lee, J.S. Mechanisms of RNA and Protein Quality Control and Their Roles in Cellular Senescence and Age-Related Diseases. Cells 2022, 11, 4062. [Google Scholar] [CrossRef]
- Sebastiani, P.; Federico, A.; Morris, M.; Gurinovich, A.; Tanaka, T.; Chandler, K.B.; Andersen, S.L.; Denis, G.; Costello, C.E.; Ferrucci, L.; et al. Protein signatures of centenarians and their offspring suggest centenarians age slower than other humans. Aging Cell 2021, 20, e13290. [Google Scholar] [CrossRef]
- Siino, V.; Ali, A.; Accardi, G.; Aiello, A.; Ligotti, M.E.; Mosquim Junior, S.; Candore, G.; Caruso, C.; Levander, F.; Vasto, S. Plasma proteome profiling of healthy individuals across the life span in a Sicilian cohort with long-lived individuals. Aging Cell 2022, 21, e13684. [Google Scholar] [CrossRef] [PubMed]
- Miura, Y.; Sato, Y.; Arai, Y.; Abe, Y.; Takayama, M.; Toda, T.; Hirose, N.; Endo, T. Proteomic analysis of plasma proteins in Japanese semisuper centenarians. Exp. Gerontol. 2011, 46, 81–85. [Google Scholar] [CrossRef] [PubMed]
- Deutsch, E.W.; Omenn, G.S.; Sun, Z.; Maes, M.; Pernemalm, M.; Palaniappan, K.K.; Letunica, N.; Vandenbrouck, Y.; Brun, V.; Tao, S.C.; et al. Advances and Utility of the Human Plasma Proteome. J. Proteome Res. 2021, 20, 5241–5263. [Google Scholar] [CrossRef]
- Rothman, S. How is the balance between protein synthesis and degradation achieved? Theor. Biol. Med. Model. 2010, 7, 25. [Google Scholar] [CrossRef] [PubMed]
- Ruano, D. Proteostasis Dysfunction in Aged Mammalian Cells. The Stressful Role of Inflammation. Front. Mol. Biosci. 2021, 8, 658742. [Google Scholar] [CrossRef] [PubMed]
- Navon, A.; Ciechanover, A. The 26 S proteasome: From basic mechanisms to drug targeting. J. Biol. Chem. 2009, 284, 33713–33718. [Google Scholar] [CrossRef]
- Finley, D.; Chen, X.; Walters, K.J. Gates, Channels, and Switches: Elements of the Proteasome Machine. Trends Biochem. Sci. 2016, 41, 77–93. [Google Scholar] [CrossRef]
- Kaushik, S.; Cuervo, A.M. Proteostasis and aging. Nat. Med. 2015, 21, 1406–1415. [Google Scholar] [CrossRef]
- Ferrington, D.A.; Kapphahn, R.J. Catalytic site-specific inhibition of the 20S proteasome by 4-hydroxynonenal. FEBS Lett. 2004, 578, 217–223. [Google Scholar] [CrossRef] [PubMed]
- Ponnappan, U.; Zhong, M.; Trebilcock, G.U. Decreased proteasome-mediated degradation in T cells from the elderly: A role in immune senescence. Cell Immunol. 1999, 192, 167–174. [Google Scholar] [CrossRef]
- Ferrington, D.A.; Husom, A.D.; Thompson, L.V. Altered proteasome structure, function, and oxidation in aged muscle. FASEB J. 2005, 19, 644–646. [Google Scholar] [CrossRef] [PubMed]
- Chondrogianni, N.; Petropoulos, I.; Franceschi, C.; Friguet, B.; Gonos, E.S. Fibroblast cultures from healthy centenarians have an active proteasome. Exp. Gerontol. 2000, 35, 721–728. [Google Scholar] [CrossRef]
- Martinez de Toda, I.; Rattan, S.I.S.; De la Fuente, M.; Arranz, L. Female Mice Reaching Exceptionally High Old Age Have Preserved 20S Proteasome Activities. Antioxidants 2021, 10, 1397. [Google Scholar] [CrossRef] [PubMed]
- Harris, L.D.; Jasem, S.; Licchesi, J.D.F. The Ubiquitin System in Alzheimer’s Disease. Adv. Exp. Med. Biol. 2020, 1233, 195–221. [Google Scholar] [CrossRef]
- Carrano, A.C.; Liu, Z.; Dillin, A.; Hunter, T. A conserved ubiquitination pathway determines longevity in response to diet restriction. Nature 2009, 460, 396–399. [Google Scholar] [CrossRef]
- Scrofano, M.M.; Shang, F.; Nowell, T.R., Jr.; Gong, X.; Smith, D.E.; Kelliher, M.; Dunning, J.; Mura, C.V.; Taylor, A. Aging, calorie restriction and ubiquitin-dependent proteolysis in the livers of Emory mice. Mech. Ageing Dev. 1998, 101, 277–296. [Google Scholar] [CrossRef]
- Pavel, M.; Rubinsztein, D.C. Mammalian autophagy and the plasma membrane. FEBS J. 2017, 284, 672–679. [Google Scholar] [CrossRef]
- Franceschi, C.; Garagnani, P.; Vitale, G.; Capri, M.; Salvioli, S. Inflammaging and ‘Garb-aging’. Trends Endocrinol. Metab. 2017, 28, 199–212. [Google Scholar] [CrossRef]
- Arbogast, F.; Gros, F. Lymphocyte Autophagy in Homeostasis, Activation, and Inflammatory Diseases. Front. Immunol. 2018, 9, 1801. [Google Scholar] [CrossRef] [PubMed]
- Terman, A.; Brunk, U.T. Autophagy in cardiac myocyte homeostasis, aging, and pathology. Cardiovasc. Res. 2005, 68, 355–365. [Google Scholar] [CrossRef]
- Merker, K.; Sitte, N.; Grune, T. Hydrogen peroxide-mediated protein oxidation in young and old human MRC-5 fibroblasts. Arch. Biochem. Biophys. 2000, 375, 50–54. [Google Scholar] [CrossRef] [PubMed]
- Terman, A. The effect of age on formation and elimination of autophagic vacuoles in mouse hepatocytes. Gerontology 1995, 41 (Suppl. S2), 319–326. [Google Scholar] [CrossRef] [PubMed]
- Bektas, A.; Schurman, S.H.; Gonzalez-Freire, M.; Dunn, C.A.; Singh, A.K.; Macian, F.; Cuervo, A.M.; Sen, R.; Ferrucci, L. Age-associated changes in human CD4(+) T cells point to mitochondrial dysfunction consequent to impaired autophagy. Aging 2019, 11, 9234–9263. [Google Scholar] [CrossRef]
- Phadwal, K.; Alegre-Abarrategui, J.; Watson, A.S.; Pike, L.; Anbalagan, S.; Hammond, E.M.; Wade-Martins, R.; McMichael, A.; Klenerman, P.; Simon, A.K. A novel method for autophagy detection in primary cells: Impaired levels of macroautophagy in immunosenescent T cells. Autophagy 2012, 8, 677–689. [Google Scholar] [CrossRef]
- Thompson, J.S.; Wekstein, D.R.; Rhoades, J.L.; Kirkpatrick, C.; Brown, S.A.; Roszman, T.; Straus, R.; Tietz, N. The immune status of healthy centenarians. J. Am. Geriatr. Soc. 1984, 32, 274–281. [Google Scholar] [CrossRef]
- Sansoni, P.; Cossarizza, A.; Brianti, V.; Fagnoni, F.; Snelli, G.; Monti, D.; Marcato, A.; Passeri, G.; Ortolani, C.; Forti, E.; et al. Lymphocyte subsets and natural killer cell activity in healthy old people and centenarians. Blood 1993, 82, 2767–2773. [Google Scholar] [CrossRef]
- Hashimoto, K.; Kouno, T.; Ikawa, T.; Hayatsu, N.; Miyajima, Y.; Yabukami, H.; Terooatea, T.; Sasaki, T.; Suzuki, T.; Valentine, M.; et al. Single-cell transcriptomics reveals expansion of cytotoxic CD4 T cells in supercentenarians. Proc. Natl. Acad. Sci. USA 2019, 116, 24242–24251. [Google Scholar] [CrossRef]
- Cossarizza, A.; Ortolani, C.; Paganelli, R.; Barbieri, D.; Monti, D.; Sansoni, P.; Fagiolo, U.; Castellani, G.; Bersani, F.; Londei, M.; et al. CD45 isoforms expression on CD4+ and CD8+ T cells throughout life, from newborns to centenarians: Implications for T cell memory. Mech. Ageing Dev. 1996, 86, 173–195. [Google Scholar] [CrossRef]
- Raz, Y.; Guerrero-Ros, I.; Maier, A.; Slagboom, P.E.; Atzmon, G.; Barzilai, N.; Macian, F. Activation-Induced Autophagy Is Preserved in CD4+ T-Cells in Familial Longevity. J. Gerontol. Ser. A 2017, 72, 1201–1206. [Google Scholar] [CrossRef]
- Xiao, F.H.; Chen, X.Q.; Yu, Q.; Ye, Y.; Liu, Y.W.; Yan, D.; Yang, L.Q.; Chen, G.; Lin, R.; Yang, L.; et al. Transcriptome evidence reveals enhanced autophagy-lysosomal function in centenarians. Genome Res. 2018, 28, 1601–1610. [Google Scholar] [CrossRef] [PubMed]
- Matarrese, P.; Tinari, A.; Ascione, B.; Gambardella, L.; Remondini, D.; Salvioli, S.; Tenedini, E.; Tagliafico, E.; Franceschi, C.; Malorni, W. Survival features of EBV-stabilized cells from centenarians: Morpho-functional and transcriptomic analyses. Age 2012, 34, 1341–1359. [Google Scholar] [CrossRef] [PubMed]
- Nakamura, S.; Yoshimori, T. Autophagy and Longevity. Mol. Cells 2018, 41, 65–72. [Google Scholar] [CrossRef]
- Emanuele, E.; Minoretti, P.; Sanchis-Gomar, F.; Pareja-Galeano, H.; Yilmaz, Y.; Garatachea, N.; Lucia, A. Can enhanced autophagy be associated with human longevity? Serum levels of the autophagy biomarker beclin-1 are increased in healthy centenarians. Rejuvenation Res. 2014, 17, 518–524. [Google Scholar] [CrossRef] [PubMed]
- Peinado-Ruiz, I.C.; Burgos-Molina, A.M.; Sendra-Portero, F.; Ruiz-Gomez, M.J. Relationship between heat shock proteins and cellular resistance to drugs and ageing. Exp. Gerontol. 2022, 167, 111896. [Google Scholar] [CrossRef]
- Martinez de Toda, I.; De la Fuente, M. The role of Hsp70 in oxi-inflamm-aging and its use as a potential biomarker of lifespan. Biogerontology 2015, 16, 709–721. [Google Scholar] [CrossRef]
- Rea, I.M.; McNerlan, S.; Pockley, A.G. Serum heat shock protein and anti-heat shock protein antibody levels in aging. Exp. Gerontol. 2001, 36, 341–352. [Google Scholar] [CrossRef]
- Terry, D.F.; Wyszynski, D.F.; Nolan, V.G.; Atzmon, G.; Schoenhofen, E.A.; Pennington, J.Y.; Andersen, S.L.; Wilcox, M.A.; Farrer, L.A.; Barzilai, N.; et al. Serum heat shock protein 70 level as a biomarker of exceptional longevity. Mech. Ageing Dev. 2006, 127, 862–868. [Google Scholar] [CrossRef]
- Marini, M.; Lapalombella, R.; Canaider, S.; Farina, A.; Monti, D.; De Vescovi, V.; Morellini, M.; Bellizzi, D.; Dato, S.; De Benedictis, G.; et al. Heat shock response by EBV-immortalized B-lymphocytes from centenarians and control subjects: A model to study the relevance of stress response in longevity. Exp. Gerontol. 2004, 39, 83–90. [Google Scholar] [CrossRef]
- Ono, Y.; Sorimachi, H. Calpains: An elaborate proteolytic system. Biochim. Biophys. Acta 2012, 1824, 224–236. [Google Scholar] [CrossRef]
- Sorimachi, H.; Hata, S.; Ono, Y. Calpain chronicle--an enzyme family under multidisciplinary characterization. Proc. Jpn. Acad. Ser. B Phys. Biol. Sci. 2011, 87, 287–327. [Google Scholar] [CrossRef] [PubMed]
- Lopatniuk, P.; Witkowski, J.M. Conventional calpains and programmed cell death. Acta Biochim. Pol. 2011, 58, 287–296. [Google Scholar] [CrossRef] [PubMed]
- Mikosik, A.; Jasiulewicz, A.; Daca, A.; Henc, I.; Frackowiak, J.E.; Ruckemann-Dziurdzinska, K.; Foerster, J.; Le Page, A.; Bryl, E.; Fulop, T.; et al. Roles of calpain-calpastatin system (CCS) in human T cell activation. Oncotarget 2016, 7, 76479–76495. [Google Scholar] [CrossRef] [PubMed]
- Mikosik, A.; Foerster, J.; Jasiulewicz, A.; Frackowiak, J.; Colonna-Romano, G.; Bulati, M.; Buffa, S.; Martorana, A.; Caruso, C.; Bryl, E.; et al. Expression of calpain-calpastatin system (CCS) member proteins in human lymphocytes of young and elderly individuals; pilot baseline data for the CALPACENT project. Immun. Ageing 2013, 10, 27. [Google Scholar] [CrossRef] [PubMed]
- Miyata, T.; Kokame, K.; Matsumoto, M.; Fujimura, Y. ADAMTS13 activity and genetic mutations in Japan. Hamostaseologie 2013, 33, 131–137. [Google Scholar] [CrossRef]
 —no significant difference;
—no significant difference;  —better/higher than in compared age group;
—better/higher than in compared age group;  —worse/lower than in compared age group; ?—no or inconclusive data.
—worse/lower than in compared age group; ?—no or inconclusive data.
 —no significant difference;
—no significant difference;  —better/higher than in compared age group;
—better/higher than in compared age group;  —worse/lower than in compared age group; ?—no or inconclusive data.
—worse/lower than in compared age group; ?—no or inconclusive data.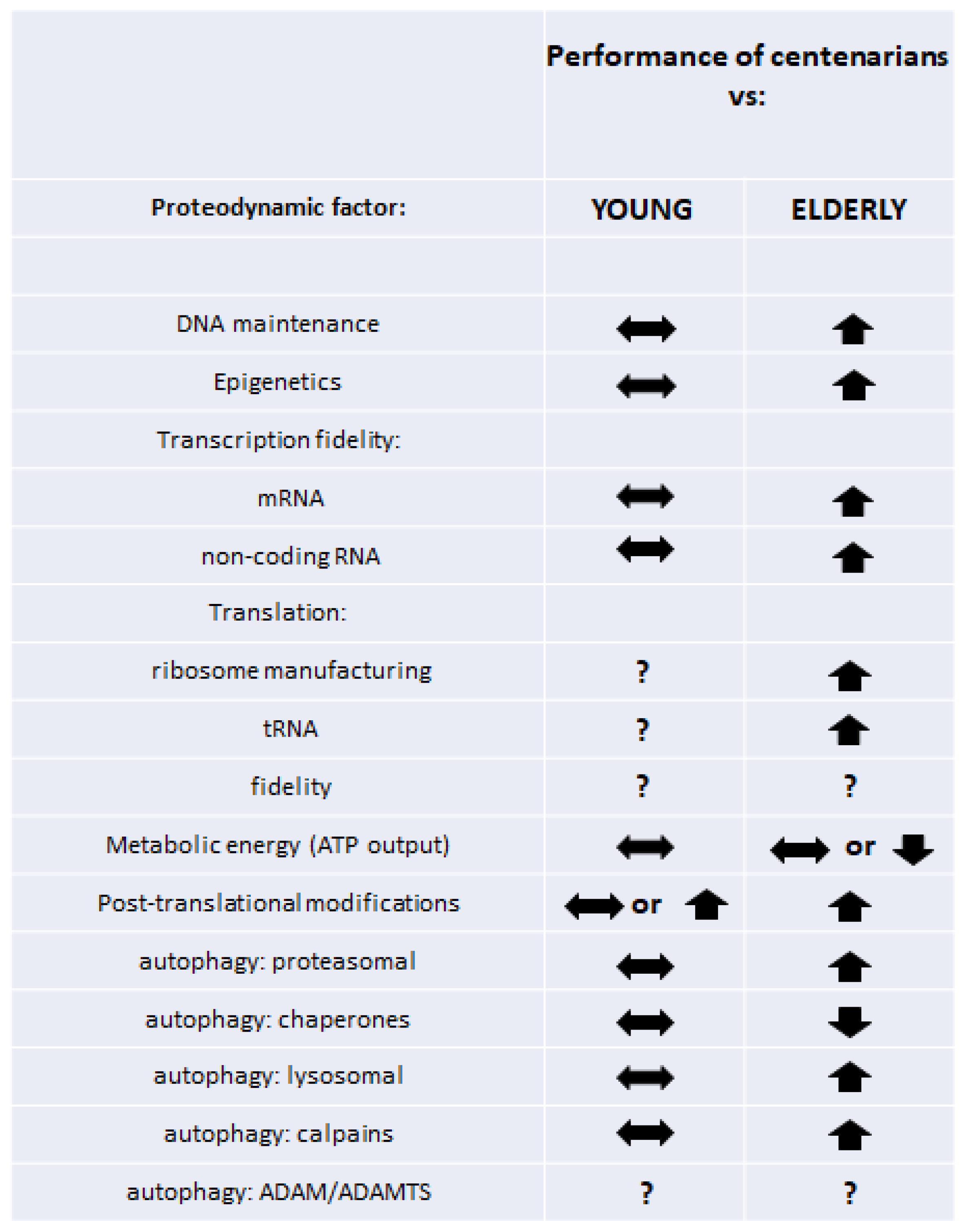
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Frankowska, N.; Bryl, E.; Fulop, T.; Witkowski, J.M. Longevity, Centenarians and Modified Cellular Proteodynamics. Int. J. Mol. Sci. 2023, 24, 2888. https://doi.org/10.3390/ijms24032888
Frankowska N, Bryl E, Fulop T, Witkowski JM. Longevity, Centenarians and Modified Cellular Proteodynamics. International Journal of Molecular Sciences. 2023; 24(3):2888. https://doi.org/10.3390/ijms24032888
Chicago/Turabian StyleFrankowska, Natalia, Ewa Bryl, Tamas Fulop, and Jacek M. Witkowski. 2023. "Longevity, Centenarians and Modified Cellular Proteodynamics" International Journal of Molecular Sciences 24, no. 3: 2888. https://doi.org/10.3390/ijms24032888
APA StyleFrankowska, N., Bryl, E., Fulop, T., & Witkowski, J. M. (2023). Longevity, Centenarians and Modified Cellular Proteodynamics. International Journal of Molecular Sciences, 24(3), 2888. https://doi.org/10.3390/ijms24032888







