Abstract
Plant-specific SQUAMOSA promoter-binding protein-like (SPL) transcription factors play important regulatory roles during plant growth and development, fruit ripening, inflorescence branching, and biotic and abiotic stresses. However, there have been no identification or systematic studies of the SPL gene family in the sweet cherry. In this study, 12 SPL genes were identified in the sweet cherry reference genome, which were distributed over 6 chromosomes and classified into six groups according to phylogenetic relationships with other SPL gene families. Nine PavSPLs were highly expressed at green fruit stages and dramatically decreased at the onset of fruit ripening, which implied that they were important regulators during fruit development and ripening. The expression patterns of PavSPL genes under ABA, GA, and MeJA treatments showed that the PavSPLs were involved in the process of fruit ripening. A subcellular localization experiment proved that PavSPL4 and PavSPL7 proteins were localized in the nucleus. The genome-wide identification of the SPL gene family provided new insights while establishing an important foundation for sweet cherry studies.
1. Introduction
Transcription factors are an important class of regulatory proteins in gene expression that recognize and bind cis-acting elements to activate or repress the expression of downstream genes. Many plant-specific transcription factors have been identified in plants, such as NAC (NAM, ATAF, and CUC2) WRKY, ARF (auxin response factor), and SPL (SQUAMOSA promoter-binding protein-like), etc. [1].
SPL genes (AmSBP1 and AmSBP2) were first discovered in the floral meristem of Antirrhinum majus [2]. Then, cDNA cloning of the SBP gene family was performed in Arabidopsis thaliana [3]. Subsequently, the SPL gene family have been identified in many other plant species, such as 15 SPLs in the tomato (Solanum lycopersicum L.) [4], 14 SPLs in woodland the strawberry (Fragaria vesca) [5], 27 SPLs in the apple (Malus domestica Borkh.) [6], 16 SPLs in the Chinese jujube (Ziziphus jujuba Mill.) [7], 16 SPLs in Arabidopsis, and 19 SPLs in rice [8]. All SPLs contain a highly conserved DNA-binding domain (also known as SBP domain), which consists of two separate zinc finger structures, which contain Zn-1(Cys3His or Cys4) and Zn-2(Cys2HisCys) [9]. The C-terminus of the SBP domain is rich in basic amino acids, which provide a bidirectional nuclear localization signal (NLS) [10]. As an important transcription factor, the SPL proteins activate or inhibit the expression of target genes by recognizing and binding to a GTAC element in the promoter regions of target genes.
Recently, several functional studies have confirmed the function of SPLs family members among the many plant species. For example, AtSPL3 was reported to prevent early flowering in Arabidopsis [11], AtSPL9 and AtSPL15 controlled shoot maturation [12], and AtSPL9 regulated the vegetative phase change and cell elongation through the BR (brassinosteroid) signaling pathway [13]. AtSPL8 has recently been identified as affecting GA (gibberellins) signaling [14], while AtSPL9 negatively regulates anthocyanin accumulation through the destabilization of a MYB-bHLH-WD40 transcriptional activation complex [15]. SBP-box transcription factor tasselsheath-4 (TSH4) was found to regulate bract development and the establishment of meristem boundaries in maize [16]. SPL gene-CNR (colorless non-ripening) regulates fruit development and ripening in the tomato [17,18,19]. PySPL9 was found to regulate light-induced anthocyanin biosynthesis in the red pear (Pyrus pyrifolia L.) [20]. VcSPL12 regulates the accumulation of chlorophylls and anthocyanins during fruit ripening in the blueberry (Vaccinium corymbosum) [21]. CpSBP1 modulates fruit softening and carotenoid accumulation by repressing CpPME1/2 and CpPDS4 in the papaya (Carica papaya L.) [22]. Although SPLs have been shown to be involved in a variety of biological processes, there are limited studies on the function of SPLs in the sweet cherry.
Sweet cherry (Prunus avium L.) (2n = 2× = 16) is one of the most popular species, having high economic benefits and being widely grown around the world [23]. The sweet cherry shows no respiratory peaks during ripening and little ethylene release, making it a non-respiratory climacteric stone fruit [24,25]. Phytohormones play a vital role in fruit development and ripening. Abscisic acid (ABA) is one of the most important hormones in the ripening process of non-respiratory climacteric fruits [26]. In addition to ABA, other hormones such as auxin, GA, and MeJA (methyl jasmonate) have also been found to be involved in fruit development and ripening [27]. The sweet cherry is known for its excellent taste and various nutrients, such as vitamin C, sugars, organic acids, phenolics, and flavonoids [28]. However, the post-harvest life of the sweet cherry is relatively short, accompanied by darkening of the fruit color and shriveling [29]. Although major progress has been made in pre- and post-harvest treatments to improve the fruit quality and market access, more studies are still needed to obtain high-quality fruit that meets consumer expectations and improves growers’ returns on investment [28].
Although the SPL gene family has been identified in many plants, there have been no identification or systematic study reports on the SPL gene family in the sweet cherry (Prunus avium L.). In this study, we identified 12 characterized putative SPL gene family members in the sweet cherry genome. We built a phylogenetic tree of the SPL gene family from sweet cherry (PavSPL), Arabidopsis (AtSPL), tomato (SlySBP and CNR), apple (MdSBP), rice (OsSPL), and strawberry (FvSPL). In addition, we systematically analyzed the gene structure, chromosome location, conserved structural domains, cis-acting elements, and expression patterns of PavSPL genes during fruit development and ripening. Furthermore, we characterized the relationships among ABA, GA, and MeJA signaling with PavSPLs. These data provide a foundation for the functional analysis of SPL genes in sweet cherry and other species of Rosaceae.
2. Results
2.1. Identification and Classification of SPL Genes in Sweet Cherry
To identify the SPL genes in sweet cherry, all Arabidopsis thaliana SPL sequences were used to blast the sweet cherry genome and redundant sequences were removed. The candidate SPL genes were verified by the presence of the SBP domain using SMART and Pfam. A total of 12 SPL genes named PavSPL1 to PavSPL12 according to their chromosomal location by removing genes with incomplete SBP domains (Supplementary Figure S1). The detailed information for the PavSPL gene family is given in Table 1, including the protein lengths and accession numbers. The lengths of these 12 SPL proteins varied from 162 (PavSPL7) to 1069 (PavSPL8) amino acids. The predicted MWs ranged from 18 kDa (PavSPL3) to 118 kDa (PavSPL8) (Table 1).

Table 1.
The information on the SPL gene family in Prunus avium L.
2.2. Chromosome Distribution and Synteny Analysis of the PavSPL Family
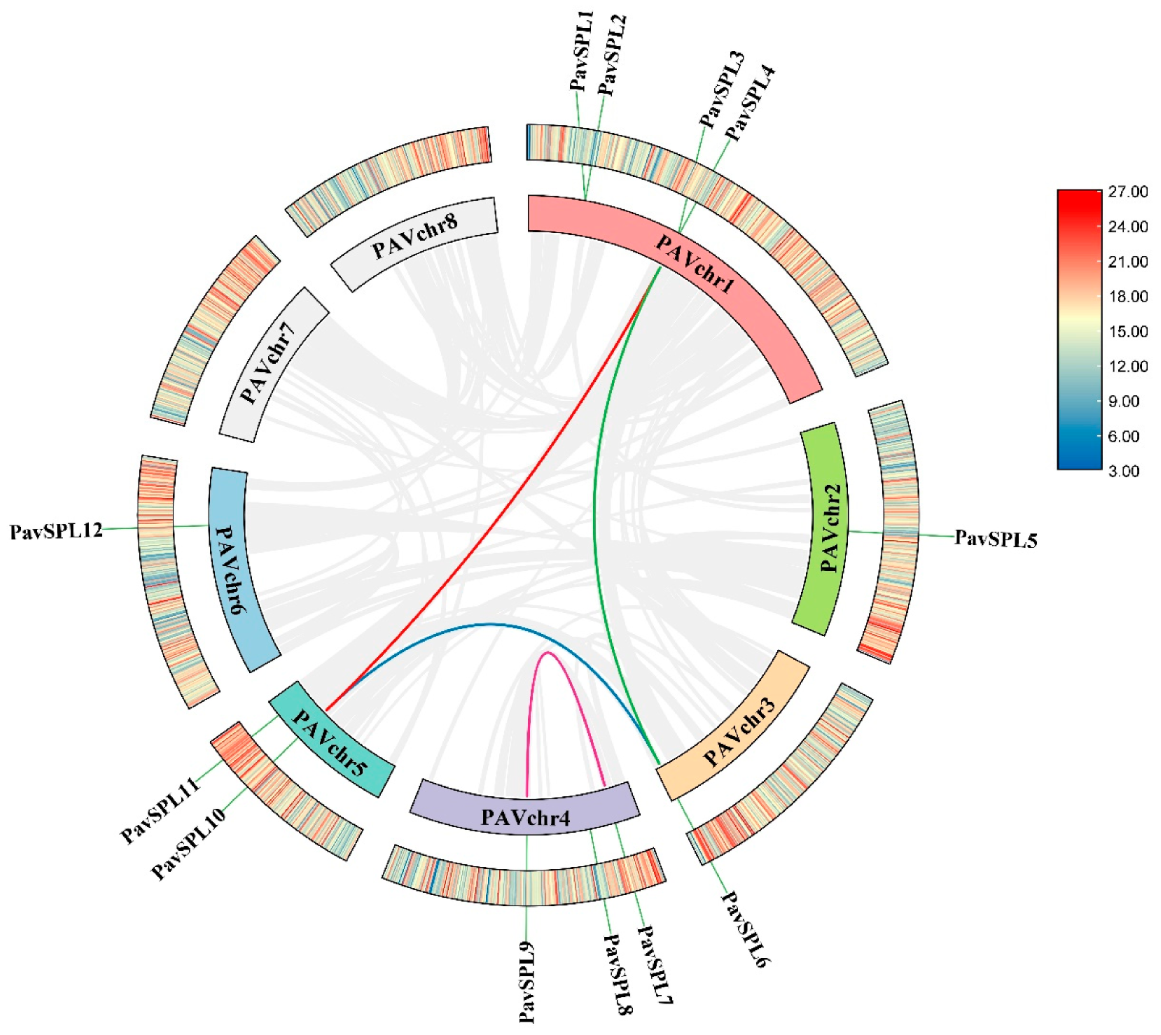
We constructed a chromosome location and collinearity plot of the sweet cherry SPL gene families to further explore the evolutionary relationships among the SPL genes. All 12 PavSPL genes were located on six of the eight sweet cherry chromosomes, chromosomes 7 and 8 did not contain any PavSPL gene, and the most PavSPL genes were mapped on chromosome 1 (Figure 1, Supplementary Figure S1). We also used TBtools software v1.106. https://github.com/CJ-Chen/TBtools/releases (accessed on 2 January 2023) to perform an intraspecies covariance analysis and found that the SPL gene family was replicated in the sweet cherry genome. The Ka/Ks ratios of all segmental duplicates were less than 1 (Table 2), indicating that the four gene pairs evolved under the effect of purifying selection.
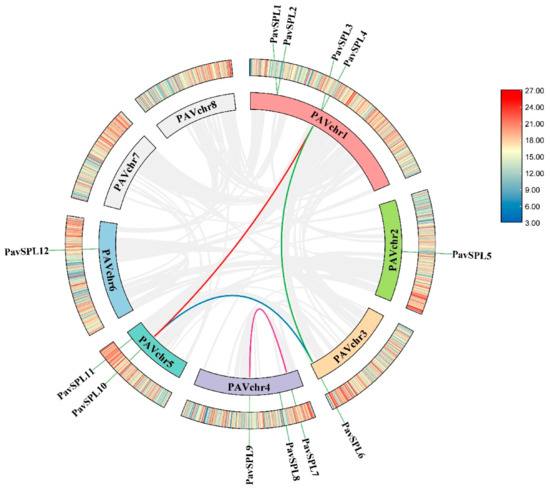
Figure 1.
Distribution of the intra-genomic duplications and collinear analysis of PavSPL genes in the sweet cherry. Different color lines represent the segmental duplicated PavSPL genes. The gray line represents all the covariates, and the chromatic line represents the pairwise replication of the SPL gene family.

Table 2.
Calculation of Ka and Ks ratios of duplicated PavSPL gene pairs.
2.3. Sequence Alignments and Phylogenetic Analysis of SPL Genes
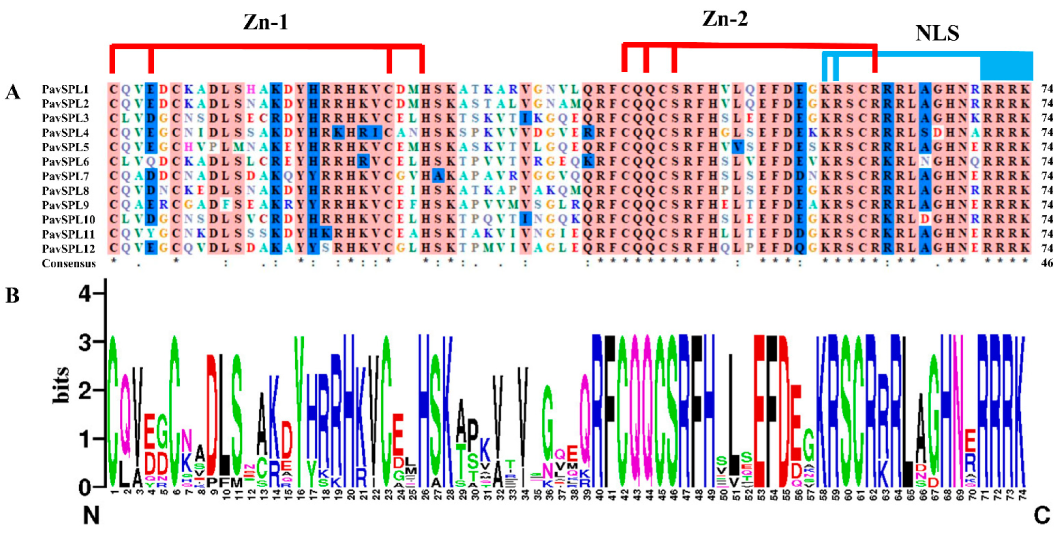
The multiple sequence alignment of full-length proteins was first performed using DNAMAN 9.0 software https://www.lynnon.com/dnaman.html (accessed on: 18 December 2022) to define the protein structure. Multiple alignments of conserved PavSPLs domains in length were completed (Figure 2A). The SBP domains were highly conserved at CQQC, SCR, and RRR sequences with about 74 amino acid residues (Figure 2B). These SPL domains have two conserved zinc finger structures (Zn-1, Zn-2) and a highly conserved nuclear localization signal (NLS).
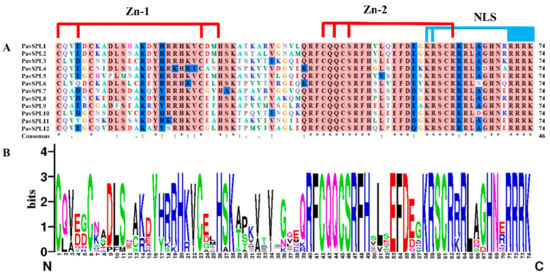
Figure 2.
SBP domain alignment of the PavSPL proteins: (A) multiple alignments of the SBP domains of the PavSPL proteins; two conserved zinc finger structures (Zn-1, Zn-2) and NLS are shown; (B) sequence logos of the SBP domain by the online WebLogo (http://weblogo.berkeley.edu/logo.cgi) (accessed on: 19 November 2022). The overall height of each stack represents the degree of conservation at this position, while the height of the letters within each stack indicates the relative frequency of the corresponding amino acids.
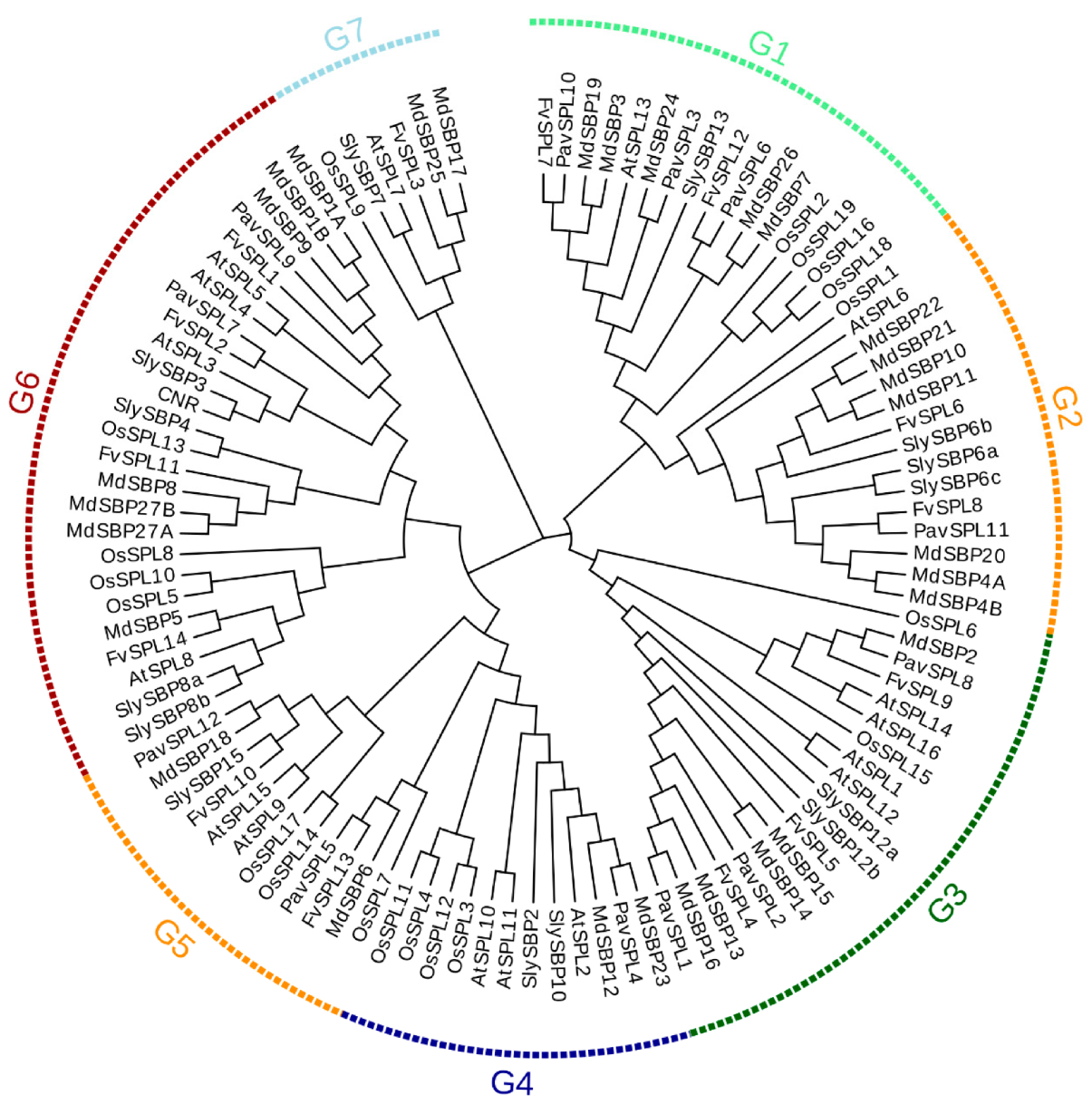
In order to further understand the evolutionary relationship of PavSPLs, we constructed a phylogenetic tree using the protein sequence of the 12 PavSPLs, 16 AtSPLs, 15 SlySBPs 27 MdSBPs, 19 OsSPLs, and 14 FvSPLs using MEGA 7.0 (Figure 3). All proteins were clustered into seven major groups (named G1 to G7). The 12 PavSPL proteins were clustered into six groups (G1 to G6), and the genes of sweet cherry were not clustered into the seven group. This result was consistent with the classification of SPL genes in Arabidopsis [3], strawberries [5], and tea (Camellia sinensis) [30]. Each group (G1 to G6) contained at least one PavSPL. PavSPL3, PavSPL6, and PavSPL10 were clustered into G1 together with AtSPL13. PavSPL11 was clustered into G2 together with AtSPL6. PavSPL1, PavSPL2, and PavSPL8 were grouped into G3 together with AtSPL1, AtSPL12, AtSPL14, and AtSPL16. PavSPL4 was grouped into G4 together with AtSPL2, AtSPL10, and AtSPL11. PavSPL5 and PavSPL12 were clustered into G5 together with AtSPL9 and AtSPL15. PavSPL7 and PavSPL9 were clustered into G6 together with AtSPL3, AtSPL4, AtSPL5, and AtSPL8.
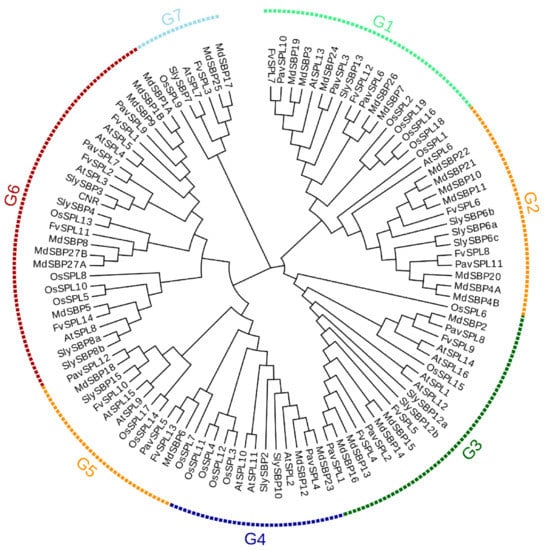
Figure 3.
Phylogenetic trees of SPL proteins are from sweet cherry (PavSPL), Arabidopsisa (AtSPL), tomato (SlySBP and CNR), apple (MdSBP), rice (OsSPL) and strawberry (FvSPL) made by neighbor-joining method with bootstrap values from 1000 replicates. G1, G2, G3, G4, G5, G6 and G7 mean Group1, Group2, Group3, Group4, Group5, Group6 and Group7. The sequence and origin of these plants are shown in Supplementary Table S1.
2.4. Conserved Motifs and Structure Analysis of the PavSPL Genes
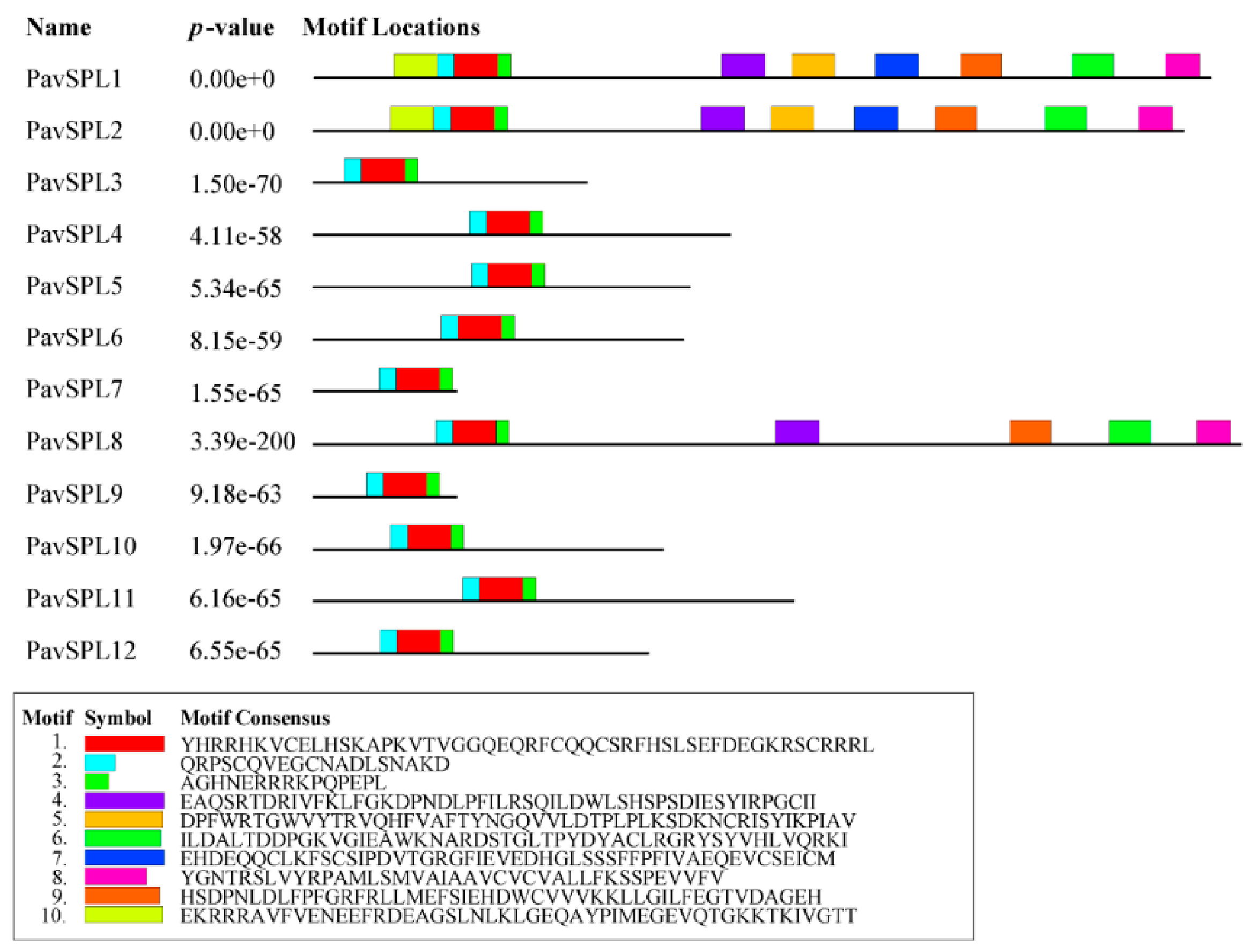
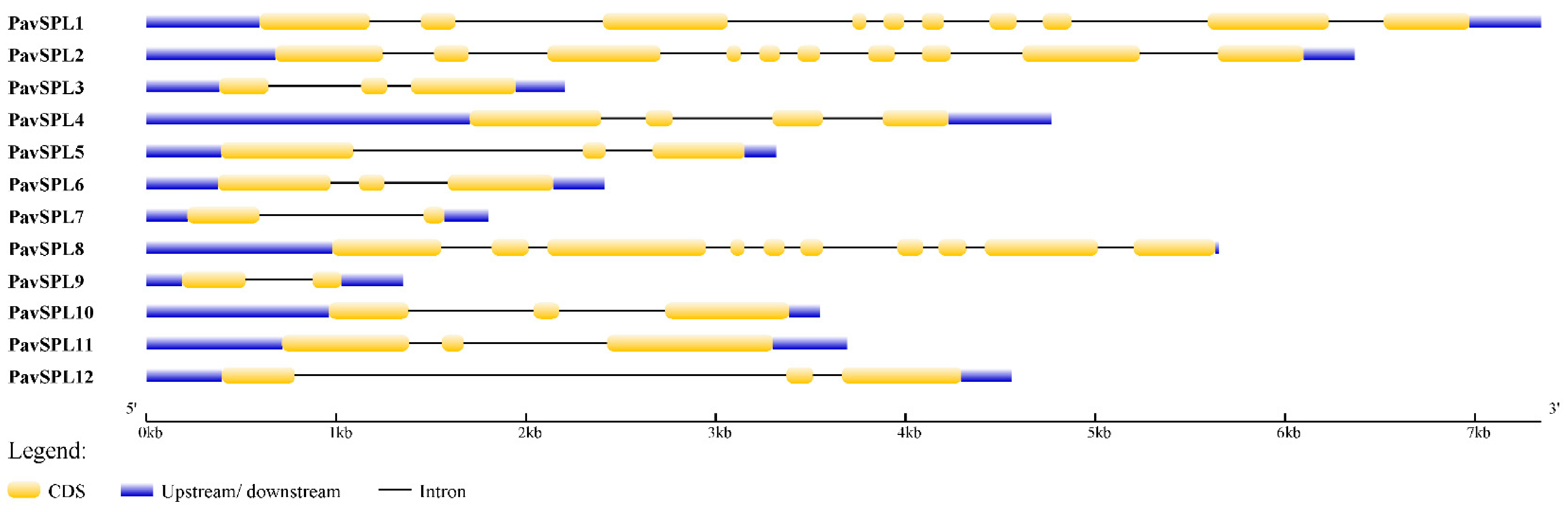
The lengths of PavSPL proteins vary widely, ranging from 162 to 1069 amino acids. The MEME motif analysis identified 10 motifs among the PavSPL members in the ‘Zaodaguo’ sweet cherry. Motifs 1, 2, and 3 were annotated as the basic motifs of PavSPL proteins and were present in all PavSPL proteins (Figure 4). This result revealed that motifs 1, 2, and 3 were conserved. Motifs 4, 6, 8, and 9 existed only in PavSPL1, -2, and -8, which were grouped into G3, and the coding sequence (CDS) lengths of these subfamilies were all over 3000 bp. The gene structure prediction showed that all PavSPL genes contained exons and introns (Figure 5). The numbers of exons and introns were not conserved among the genes and ranged from one to nine. For example, PavSPL7 had one intron, whereas the genes in group 3 had nine exons. PavSPL genes containing 2 introns were the most common type, accounting for 41.7%.
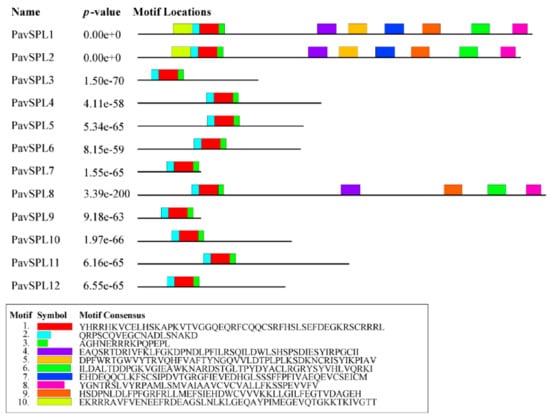
Figure 4.
Motif composition of PavSPL proteins. Motifs of PavSPL proteins were analyzed using s MEME analysis. The ten motifs are represented by different colors.
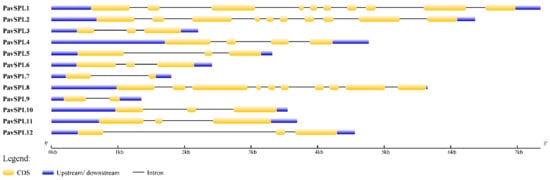
Figure 5.
Gene structure of PavSPL genes in the online Gene Structure Display Server (GSDS).
2.5. Cis-Acting Elements in the Promoters of PavSPL Genes
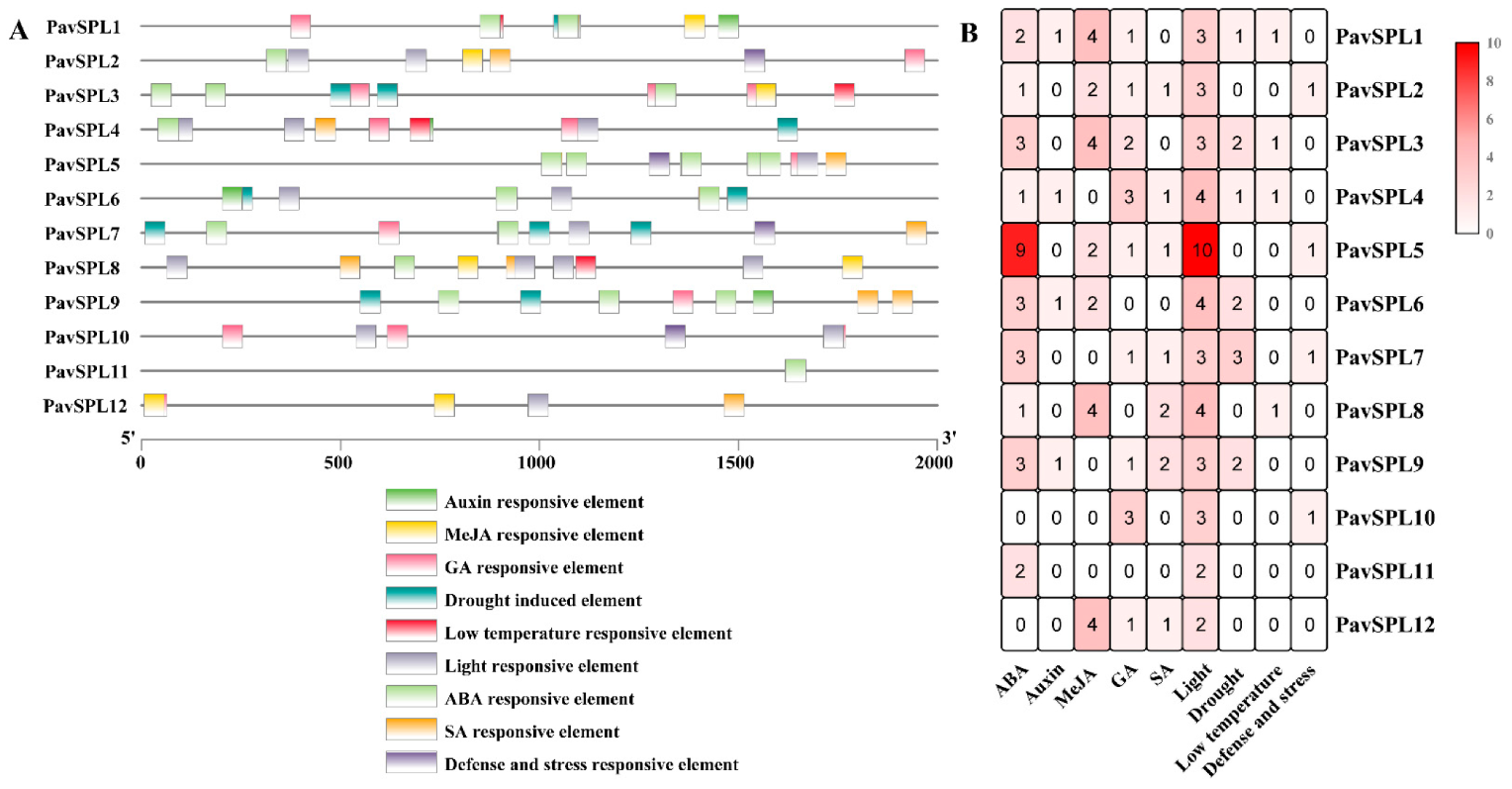
Many cis-acting elements on the promoter were distributed upstream of the gene coding sequence, which can regulate the expression characteristics of the genes. Predicting hormone response elements, stress response elements, and light response elements on the are 2000 bp upstream of the PavSPL promoter sequence (Figure 6). All 12 PavSPLs promoter sequences have hormone response elements, which may play a critical role in the hormone balance in sweet cherries. The element corresponded to five phytohormones, including ABA, MeJA, salicylic acid (SA), GA, and auxin. Most were ABA response elements, indicating that the PavSPL genes played a role in the ABA responses.
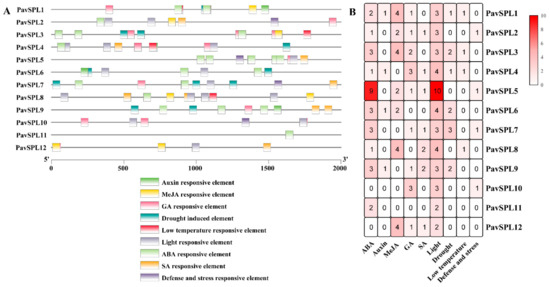
Figure 6.
Cis-element of the PavSPL genes in sweet cherry: (A) the cis-element analysis of the PavSPL genes, where colorful boxes represent different cis-elements; (B) numbers of cis-elements in the PavSPL genes.
2.6. Expression Analysis of the PavSPL Genes in Different Tissues and in Fruit Development
To further confirm the function of PavSPLs during vegetative and reproductive growth in sweet cherry, we detected the expression patterns of 12 PavSPL genes via qRT-PCR testing (Figure 7). The results indicated that 12 PavSPL genes displayed diverse expression patterns among the tissues and organs of the sweet cherries. The expression profiles of PavSPLs can be divided into two types. Parts of these genes exhibited highly tissue-specific expression patterns, while other parts were structurally expressed in all tissues and organs. For example, PavSPL1, PavSPL2, PavSPL4, PavSPL7, and PavSPL8 exhibited relatively high expression levels in all tissues. PavSPL9 was specifically expressed in mature leaves and flowers. PavSPL3, PavSPL10, and PavSPL11 exhibited higher relative expression levels in reproductive tissues than stems or mature leaves. PavSPL5, PavSPL8, and PavSPL12 exhibited higher relative expression levels in vegetative tissues than reproductive tissues.
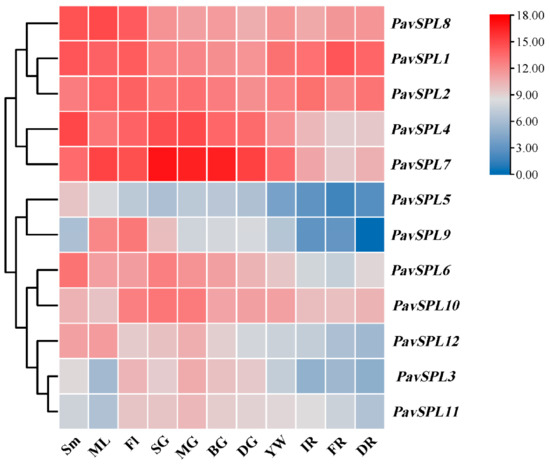
Figure 7.
The expression quantities of PavSPLs in different tissues as assessed using TBtools. Sm: stems; ML: mature leaves; SG: small green; MG: mid green; BG: big green fruit; DG: de-greening; YW: yellow fruit; IR: initial red; FR: full red fruit; DR: dark red. The relative mRNA levels are represented as the means ± SDs (n = 3). Statistically significant differences were assessed using Student’s t-test.
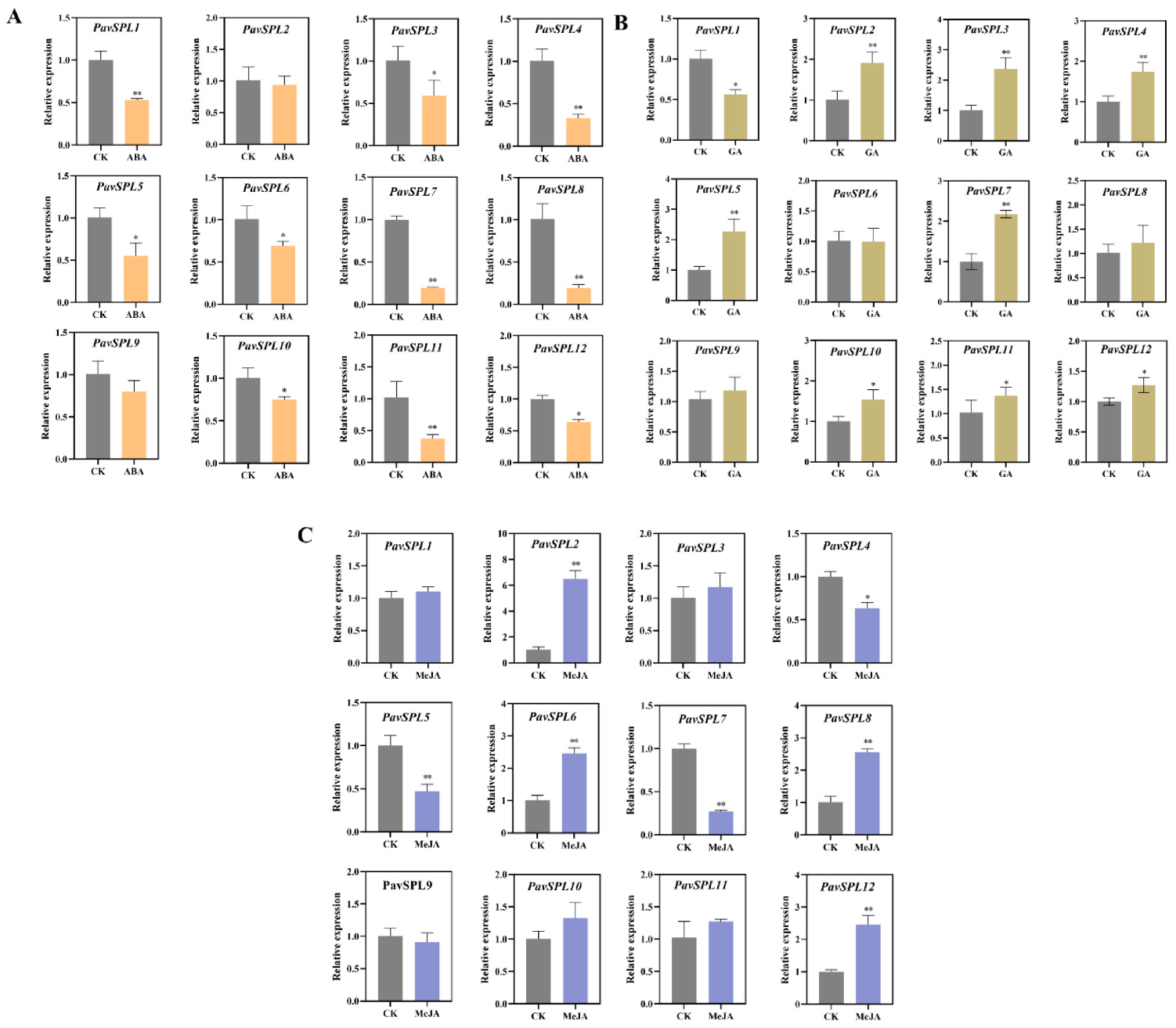
With the development and ripening of the sweet cherry fruit, the expression levels of PavSPL3, PavSPL4, PavSPL5, PavSPL6, PavSPL7, PavSPL9, PavSPL10, PavSPL11, and PavSPL12 decreased gradually; the levels were the highest in the green stage. The expression levels of PavSPL1 were the highest in the full red stage, and the expression levels of PavSPL2 and PavSPL8 remained stable throughout the development stages.
2.7. Expression Analysis of the PavSPL Genes during Hormone Treatment
ABA, MeJA, and GA are the main hormones involved in fruit development and ripening. Therefore, we treated sweet cherry fruits with these hormones and examined the expression levels of PavSPL genes. The expression level of PavSPL1, PavSPL3, PavSPL4, PavSPL5, PavSPL6, PavSPL7, PavSPL8, PavSPL10, PavSPL11, and PavSPL12 were significantly downregulated under the ABA treatment (Figure 8A). Under the GA treatment, PavSPL2, PavSPL3, PavSPL4, PavSPL5, PavSPL7, PavSPL10, PavSPL11, and PavSPL12 were significantly induced, while only PavSPL1 was significantly upregulated (Figure 8B). Under the MeJA treatment, PavSPL4, PavSPL5, and PavSPL7 were significantly reduced, while PavSPL2, PavSPL6, PavSPL8, and PavSPL12 were significantly upregulated (Figure 8C).
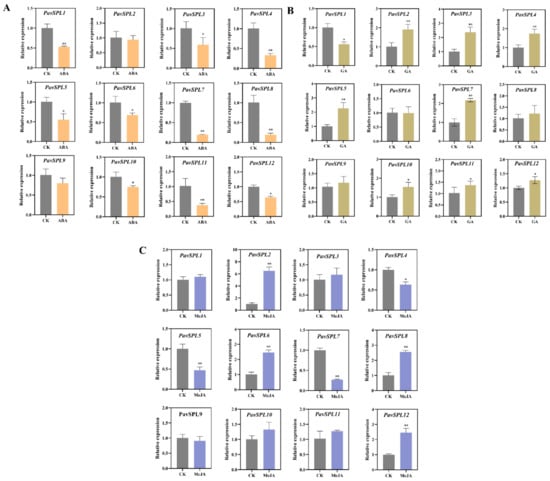
Figure 8.
(A) ABA treatment, (B) GA treatment, (C) MeJA treatment. Expression patterns of PavSPL genes under ABA, GA, and MeJA treatments. The relative mRNA levels are represented as the means ± SDs (n = 3). Statistically significant differences were assessed using Student’s t-test (* p < 0.05, ** p < 0.01).
2.8. Subcellular Localization of PavSPLs
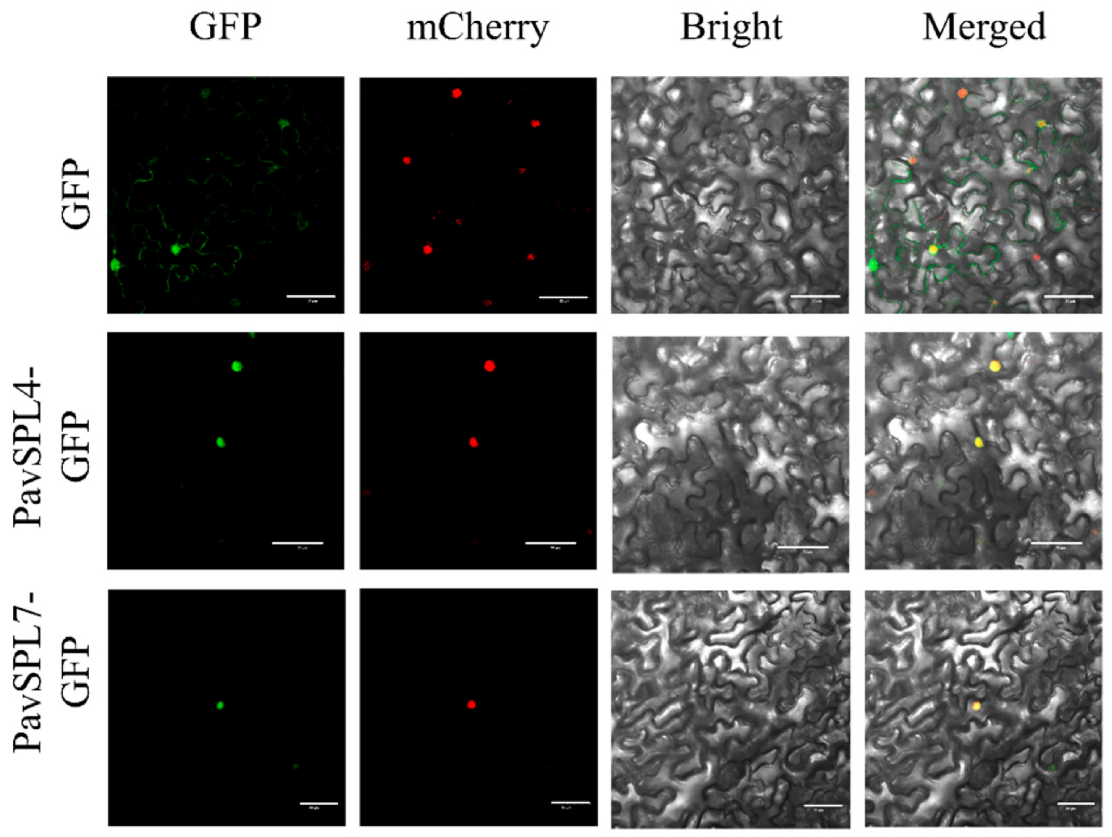
The nuclear localization of transcription factors is a very important signature. In other species such as VpSBP11 in grape [31] and CpSBP1 in papaya [22], they are localized in the nucleus. In order to investigate the location of PavSPL genes in sweet cherry, we examined the subcellular localization of PavSPL4 and PavSPL7 through transient expression in tobacco (N. benthamiana) leaves. We found that the green fluorescent protein (GFP) signals of PavSPL4 and PavSPL7 overlapped with the nuclear marker mCherry signal, which indicated that the PavSPL4 and PavSPL7 proteins were localized in the nucleus (Figure 9). These results demonstrated that PavSPL4 and PavSPL7 are nuclear proteins that could serve as transcription factors.
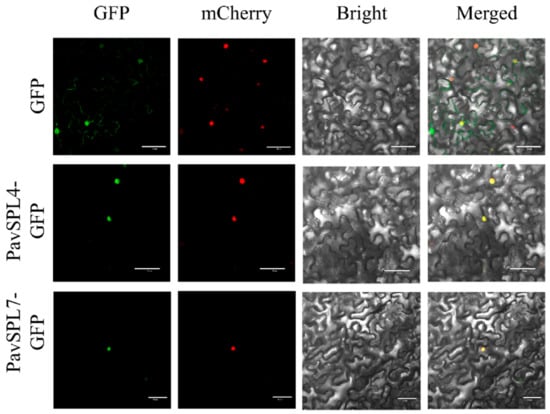
Figure 9.
Subcellular localization of PavSPL4 and PavSPL7 proteins. The PavSPL–GFP fusion proteins were transiently expressed in N. benthamiana leaves and the GFP fluorescence signal was observed after 48 h. Scale bar, 50 µm.
3. Discussion
3.1. Bioinformatics Analysis of SPL Gene Family in Sweet Cherry
SPL transcription factors have been attracting attention in the scientific community due to their powerful functions. In recent years, although the function of SPL genes has been increasingly studied in many plants, it has not been studied in the sweet cherry. In this study, we performed a comprehensive analysis of the SPL gene family in the sweet cherry. Firstly, we identified 18 PavSPL sequences from the Prunus avium genome, which was released in 2017. Among them, Pav_sc0001411.1_g180.1.mk, Pav_sc0001280.1_g590.1.mk, Pav_sc0000492.1_g530.1.mk, Pav_co4005735.1_g010.1.mk, Pav_sc0000815.1_g180.1.mk, and Pav_sc0001974.1_g370.1.mk were excluded for their incomplete SBP domains. The numbers of SPL family members vary greatly among species. For example, there are 16, 25, 24, 29, and 59 members in Arabidopsis thaliana [8], tea [30], tartary buckwheat (Fagopyrum tataricum) [32], Chinese cabbage (Brassica rapa subsp. pekinensis) [33], and cotton (Gossypium hirsutum) [34], respectively. In this study, 12 SPL gene family members were identified from the sweet cherry genome, which was less than for other species, suggesting that SPL genes have undergone extensive changes during their genome evolution, which has led to the diversification of the gene functions.
Based on a phylogenetic analysis (Figure 3), the PavSPL genes were clustered into six groups, with at least one gene from Arabidopsis and the tomato in each group, implying the functional conservation of SPL genes between different plants. Members of each subfamily contained a typical SBP domain, which suggested that the SBP domains were conserved. The analysis of the gene structure of the PavSPL gene family showed that the maximum numbers of exons and introns were present in G3 (Figure 4 and Figure 5). The minimum numbers of exons and introns were present in G6. These results indicated that the evolution of the SPL gene family may be closely related to the diversification of the gene structures [7]. Therefore, we speculate that different SPL groups probably perform similar functions within Arabidopsis, due to the presence of similar structures and conserved motifs.
3.2. Expression Profiles of PavSPLs in Sweet Cherry Development
SPL genes have been reported to be involved in reproductive growth and vegetative growth, but no studies have been performed in sweet cherry. We predicted the function of the PavSPL genes based on expression patterns among the tissues and organs of sweet cherry and the functions of homologous genes (Figure 7). G1 contained PavSPL3, PavSPL6, PavSPL10, and AtSPL13. These genes had relatively high transcript levels in the stems and flowers, suggesting that they may play vital roles in shoot and flower development. In G3, AtSPL16, PavSPL1, PavSPL2, and PavSPL8 are similar to AtSPL1, AtSPL12, and AtSPL14 and are long genes. Similar to Arabidopsis, these three PavSPLs were highly constitutively expressed in all tissues and organs [3]. AtSPL9 and AtSPL15 clustered in G5 and have been proved to control shoot maturation redundantly [12]. PavSPL5 and PavSPL12 were clustered in G5 and may play the same roles as their presumed orthologs.
Sweet cherry is highly nutritious and the fruit is rich in anthocyanins, with antioxidant activity [35]. The growth and development of sweet cherry is regulated by a variety of transcription factors [35,36,37]. Therefore, studying the expression patterns of PavSPL genes in sweet cherries at different stages of development and ripening can provide a basis for screening potential transcription factors that regulate fruit growth and development. The qRT-PCR assay revealed that 12 PavSPL genes were expressed in different patterns in the processes of fruit development and ripening (Figure 7), suggesting that PavSPL genes may be critical during fruit growth, development, and ripening.
The SPL genes play key roles in the fruit development and ripening processes, which have been demonstrated in other species. Different members of the SPL family show functional differences. For example, VcSPL12 (classified into the same subfamily as AtSPL2, AtSPL10, and AtSPL11) regulated the accumulation of chlorophylls and anthocyanins during fruit ripening in blueberries [21]. Accordingly, PavSPL4 clustered in G4 with AtSPL2, AtSPL10, and AtSPL11 and may have important functions in leaf development or fruit ripening. In papaya, CpSBP1 (classified into the same subfamily with SlSPL-CNR, AtSPL3, and AtSPL4) modulated fruit softening and carotenoid accumulation [22]. SlSPL-CNR is a key gene that controls tomato fruit ripening. The epigenetic variation in the promoter region of SlSPL-CNR affects the ripening of fruit [4,19,38]. SlSPL-CNR clustered in G6 with PavSPL7 and PavSPL9. Therefore, PavSPL7 and PavSPL9 may have an important function in fruit ripening. They had the same motif compositions, but the specific functions of these PavSPL genes need to be verified in further experiments in the future. In our study, we demonstrated that most SPL genes such as PavSPL4 and PavSPL7 showed a downregulation trend, consistent with the results presented in grapes [39]. This means that these genes are negatively regulated during fruit development and ripening in sweet cherry.
3.3. Expression Profiles of PavSPLs in Plant Responses to Multiple Hormones
The hormone signals and environmental changes are critical to fruit development and ripening [40,41,42,43]. In our study, the analysis of cis-acting elements in the 2000 bp sequence upstream of the PavSPLs promoter revealed that most members of this family respond to a variety of hormones and stresses, indicating that PavSPLs may be regulated by light, stresses, and phytohormones (Figure 6). The roles of SPL genes in hormone regulation are not well defined, but there are some relevant reports in other species. For example, SPL8 acted as a local regulator in a subset of GA-mediated developmental processes in Arabidopsis [14]. SPL9 interacted with ABI5 to enhance the ABA responses in Arabidopsis [44]. ELONGATED HYPOCOTYL5 (HY5) interacted with SPL7 to mediate coordinated responses to light and copper [45]. BRASSINAZOLE-RESISTANT 1 (BZR1), the master transcription factor of the BR signaling pathway, interacted with AtSPL9 to cooperatively regulate the vegetative phase change and cell elongation [13]. In previous studies, an exogenous ABA treatment promoted the fruit ripening process and the ABA content increased significantly during fruit ripening in ‘Hong Deng’ and ‘Rainier’ sweet cherries [35,46]. The exogenous ABA treatment could regulate strawberry fruit ripening [47]. In our study, we treated ‘Zaodaguo’ sweet cherries using ABA, GA, and MeJA exogenous hormones. The gene expression pattern showed that the SPL genes were responsive to many hormone treatments (Figure 8). PavSPL4, PavSPL5, PavSPL7, and PavSPL12 were regulated by many hormone treatments, suggesting that the PavSPL genes may be involved in the crosstalk of multiple different hormone signals regulating fruit development and ripening.
4. Materials and Methods
4.1. Plant Materials and Hormone Treatments
The sweet cherry (Prunus avium L.) trees of the cultivar ‘Zaodaguo’ were 10 years old, and the cherries were harvested from the Beijing Academy of Agriculture and Forestry Sciences, located in Beijing. The fruit samples from the different developmental periods were selected according to the anthesis time, being harvested in the small green (SG), mid-green (MG), big green (BG), de-greening (DG), yellow (YW), initial red (IR), full red (FR), and dark red (DR) phases. For the tissue-specific expression analysis, the stems, leaves and flowers were collected from same ‘Zaodaguo’ trees. The mature leaves and stem segments were collected when the leaves were dark green and fully unfolded. The flowers were sampled when they were fully blooming. The samples were transported to the laboratory, then immediately frozen in liquid nitrogen and stored at −80 °C.
The ABA, MeJA, and GA treatments were applied to fruit samples 19–21 days after anthesis. Fruit samples of the same size as the ‘Zaodaguo’ samples were taken back to the laboratory for hydroponic treatment and incubated in the dark for 1 day to eliminate field heat. Then, these fruits treated with exogenous hormones. The concentration of ABA was 100 μmol/L, the MeJA concentration was 200 μmol/L, and the GA concentration was 200 μmol/L. Water was used for the control treatment. The hormone stock solution was prepared using anhydrous ethanol, and the same volume of anhydrous ethanol was added to the control treatment to eliminate errors. After 5 days of the above treatment, the samples were then immediately frozen in liquid nitrogen and stored at −80°C. All treatments were performed with three biological replicates.
4.2. Identification of SPL Genes in Sweet Cherry
The protein sequence of 16 AtSPLs were downloaded from the Arabidopsis genome database (https://www.arabidopsis.org/)(accessed on: 12 February 2022). PavSPL protein sequences were searched in the sweet cherry genome database (http://cherry.kazusa.or.jp/) (accessed on: 2 December 2022). The molecular weights, isoelectric points (pIs), and grand average of hydropathicity (GRAVY) values of the PavSPL proteins were calculated using the ExPASy website (https://web.expasy.org/protparam/) (accessed on: 21 June 2022).
4.3. Sequence Alignments, Phylogenetic Analyses, and Gene Structure Analysis
We used ExPASy (https://web.expasy.org/protparam/) (accessed on: 21 June 2022) to compute the physical and chemical parameters of each protein sequence. Multiple sequence alignment was carried out using DNAMAN V9.0. https://www.lynnon.com/dnaman.html (accessed on: 18 December 2022). The phylogenetic trees were constructed using MEGA 7.0 software with the neighbor-joining (NJ) method and the bootstrap test was replicated 1000 times. Domains were identified with the SMART (http://smart.embl-heidelberg.de) (accessed on: 19 June 2022) and Pfam (http://pfam.xfam.org) (accessed on: 13 November 2022) programs. The sequence logos of the conserved domains were generated using WebLogo (http://weblogo.berkeley.edu/logo.cgi) (accessed on: 19 November 2022). Furthermore, MEME (http://meme-suite.org/) (accessed on: 13 October 2022) was used to search for motifs in all SPL genes. A structure analysis of the PavSPL genes was performed using the Gene Structure Display Server (GSDS) (http://gsds.gao-lab.org/) (accessed on: 1 October 2022).
4.4. Cis-Acting Element Prediction and Chromosomal Localization Analysis
Cis-acting elements within the PavSPL gene promoters were analyzed using the PlantCARE online program (http://bioinformatics.psb.ugent.be/webtools/plantcare/html/) (accessed on: 30 October 2022). TBtools v1.106. https://github.com/CJ-Chen/TBtools/releases (accessed on 2 January 2023) [48] was used to map the genes to chromosomes and visualize these results.
4.5. Expression Analysis of PavSPLs
The total RNA was extracted using the RNA extraction kit (Huayueyang Biotechnology, Beijing, China). One microgram of the total RNA was used for the cDNA synthesis using a reverse transcription kit (TaKaRa Biotechnology, Dalian, China). The primers for SPL genes were designed using DNAMAN software and tested for specificity using BLAST on the genomics website. All primers of PavSPL genes used for the quantitative RT-PCR (qRT-PCR) assay are listed in Supplementary Table S2. The qRT-PCR assay was performed in the presence of SYBR Premix Ex Taq (Kangwei Century Biotechnology, Beijing, China). The reactions were incubated in a Rotor Gene Q Machine (Qiagen, Germany). PavActin was screened as internal reference gene for sweet cherry [46]. The relative expression value was quantified using the 2−∆∆CT method [49].
4.6. Subcellular Localization Analysis
For the colocalization of PavSPL4 and PavSPL7, the full-length PavSPL4 and PavSPL7 sequences without the termination codon were amplified from cDNA from ‘Zaodaguo’ fruit. Each amplification product was a fusion expression with a green fluorescence protein (GFP) label inserted into the pCAMBIA1302 vector under the control of theCaMV35S promoter. The vectors were transformed into the Agrobacterium tumefaciens strain GV3101. The primers are listed in Supplementary Table S3.
N. benthamiana plants were grown in a plant growth chamber at 24 °C under 16 h/8 h light/dark conditions. These plants can be used as experimental materials at about two months of age. The Agrobacterium microbial concentrations were transiently transformed in N. benthamiana leaves. The agroinfiltrated leaves were photographed 48 h after transformation. Then, observations with an Olympus laser-scanning fluorescence instrument were performed as described in [50].
5. Conclusions
In our study, we identified 12 PavSPL genes in the sweet cherry genome database. We systematically studied the gene structures and expression patterns of PavSPL genes at different developmental periods and with ABA, GA, and MeJA hormone treatments in sweet cherry fruit. The expression levels of several PavSPL genes increased or decreased in response to various hormone treatments. Our results demonstrated that the PavSPL genes play important roles in fruit development and ripening. In conclusion, our genome-wide study analysis of the SPL gene family lays the foundation for the study of the ripening mechanism in non-respiratory climacteric fruit.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/ijms24032880/s1.
Author Contributions
Y.S. and T.L. conceived and designed the experiments. Y.S., Y.W. and Y.X. prepared the plant materials and gene expression analysis. Y.S., M.T. and X.Z. conducted the bioinformatic analysis. C.W., S.T., and S.G. performed the transcriptional activation activity and subcellular localization analysis. J.H., Q.Y. and B.D. conducted the data analysis. Y.S. wrote the manuscript. T.L. revised the manuscript. All authors have read and agreed to the published version of the manuscript.
Funding
This research was supported by the National Natural Science Foundation of China (grant No. 31972390, 32272686), the Construction of Beijing Science and Technology Innovation and Service Capacity in Top Subjects (Grant Number CEFF–PXM2019_014207_000032), and the 2115 Talent Development Program of China Agricultural University.
Data Availability Statement
Not applicable.
Acknowledgments
We are grateful to Kaichun Zhang (Beijing Institute of Forestry and Pomology, Beijing Academy of Agricultural and Forestry Sciences, Beijing, China) for the material support.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Riechmann, J.L.; Ratcliffe, O.J. A genomic perspective on plant transcription factors. Curr Opin Plant. Biol. 2000, 3, 423–434. [Google Scholar] [CrossRef] [PubMed]
- Klein, J.; Saedler, H.; Huijser, P. A new family of DNA binding proteins includes putative transcriptional regulators of the Antirrhinum majus floral meristem identity gene SQUAMOSA. Mol. Gen. Genet. 1996, 250, 7–16. [Google Scholar] [CrossRef] [PubMed]
- Cardon, G.; Hohmann, S.; Klein, J.; Nettesheim, K.; Saedler, H.; Huijser, P. Molecular characterisation of the Arabidopsis SBP-box genes. Gene 1999, 237, 91–104. [Google Scholar] [CrossRef] [PubMed]
- Salinas, M.; Xing, S.; Hohmann, S.; Berndtgen, R.; Huijser, P. Genomic organization, phylogenetic comparison and differential expression of the SBP-box family of transcription factors in tomato. Planta 2012, 235, 1171–1184. [Google Scholar] [CrossRef]
- Xiong, J.; Zheng, D.; Zhu, H.; Chen, J.; Na, R.; Cheng, Z. Genome-wide identification and expression analysis of the SPL gene family in woodland strawberry Fragaria vesca. Genome 2018, 61, 675–683. [Google Scholar] [CrossRef]
- Li, J.; Hou, H.; Li, X.; Xiang, J.; Yin, X.; Gao, H.; Zheng, Y.; Bassett, C.L.; Wang, X. Genome-wide identification and analysis of the SBP-box family genes in apple (Malus × domestica Borkh.). Plant. Physiol. Biochem. 2013, 70, 100–114. [Google Scholar] [CrossRef]
- Song, S.; Zhou, H.; Sheng, S.; Cao, M.; Li, Y.; Pang, X. Genome-wide organization and expression profiling of the SBP-Box gene family in Chinese jujube (Ziziphus jujuba Mill.). Int J. Mol. Sci. 2017, 18, 1734. [Google Scholar] [CrossRef]
- Yang, Z.; Wang, X.; Gu, S.; Hu, Z.; Xu, H.; Xu, C. Comparative study of SBP-box gene family in Arabidopsis and rice. Gene 2008, 407, 1–11. [Google Scholar] [CrossRef]
- Yamasaki, K.; Kigawa, T.; Inoue, M.; Tateno, M.; Yamasaki, T.; Yabuki, T.; Aoki, M.; Seki, E.; Matsuda, T.; Nunokawa, E.; et al. A novel zinc-binding motif revealed by solution structures of DNA-binding domains of Arabidopsis SBP-family transcription factors. J. Mol. Biol. 2004, 337, 49–63. [Google Scholar] [CrossRef]
- Birkenbihl, R.P.; Jach, G.; Saedler, H.; Huijser, P. Functional dissection of the plant-specific SBP-domain: Overlap of the DNA-binding and nuclear localization domains. J. Mol. Biol. 2005, 352, 585–596. [Google Scholar] [CrossRef]
- Gandikota, M.; Birkenbihl, R.P.; Hohmann, S.; Cardon, G.H.; Saedler, H.; Huijser, P. The miRNA156/157 recognition element in the 3’ UTR of the Arabidopsis SBP box gene SPL3 prevents early flowering by translational inhibition in seedlings. Plant. J. 2007, 49, 683–693. [Google Scholar] [CrossRef]
- Schwarz, S.; Grande, A.V.; Bujdoso, N.; Saedler, H.; Huijser, P. The microRNA regulated SBP-box genes SPL9 and SPL15 control shoot maturation in Arabidopsis. Plant. Mol. Biol. 2008, 67, 183–195. [Google Scholar] [CrossRef]
- Wang, L.; Yu, P.; Lyu, J.; Hu, Y.; Han, C.; Bai, M.Y.; Fan, M. BZR1 physically interacts with SPL9 to regulate the vegetative phase change and cell elongation in Arabidopsis. Int. J. Mol. Sci. 2021, 22, 415. [Google Scholar] [CrossRef]
- Zhang, Y.; Schwarz, S.; Saedler, H.; Huijser, P. SPL8, a local regulator in a subset of gibberellin-mediated developmental processes in Arabidopsis. Plant. Mol. Biol. 2007, 63, 429–439. [Google Scholar] [CrossRef]
- Gou, J.; Felippes, F.F.; Liu, C.; Detlef, W.; Wang, J. Negative regulation of anthocyanin biosynthesis in Arabidopsis by a miR156-targeted SPL transcription factor. Plant. Cell 2011, 23, 1512–1522. [Google Scholar] [CrossRef]
- Chuck, G.; Whipple, C.; Jackson, D.; Hake, S. The maize SBP-box transcription factor encoded by tasselsheath4 regulates bract development and the establishment of meristem boundaries. Development 2010, 137, 1243–1250. [Google Scholar] [CrossRef]
- Manning, K.; Tor, M.; Poole, M.; Hong, Y.; Thompson, A.J.; King, G.J.; Giovannoni, J.J.; Seymour, G.B. A naturally occurring epigenetic mutation in a gene encoding an SBP-box transcription factor inhibits tomato fruit ripening. Nat. Genet. 2006, 38, 948–952. [Google Scholar] [CrossRef]
- Eriksson, E.M.; Bovy, A.; Manning, K.; Harrison, L.; Andrews, J.; De Silva, J.; Tucker, G.A.; Seymour, G.B. Effect of the colorless non-ripening mutation on cell wall biochemistry and gene expression during tomato fruit development and ripening. Plant. Physiol. 2004, 136, 4184–4197. [Google Scholar] [CrossRef]
- Lai, T.; Wang, X.; Ye, B.; Jin, M.; Chen, W.; Wang, Y.; Zhou, Y.; Blanks, A.M.; Gu, M.; Zhang, P.; et al. Molecular and functional characterization of the SBP-box transcription factor SPL-CNR in tomato fruit ripening and cell death. J. Exp. Bot. 2020, 71, 2995–3011. [Google Scholar] [CrossRef]
- Liu, H.; Shu, Q.; Lin-Wang, K.; Allan, A.C.; Espley, R.V.; Su, J.; Pei, M.; Wu, J. The PyPIF5-PymiR156a-PySPL9-PyMYB114/MYB10 module regulates light-induced anthocyanin biosynthesis in red pear. Mol. Hortic. 2021, 1. [Google Scholar] [CrossRef]
- Li, X.; Hou, Y.; Xie, X.; Li, H.; Li, X.; Zhu, Y.; Zhai, L.; Zhang, C.; Bian, S. A blueberry MIR156a-SPL12 module coordinates the accumulation of chlorophylls and anthocyanins during fruit ripening. J. Exp. Bot. 2020, 71, 5976–5989. [Google Scholar] [CrossRef] [PubMed]
- Han, Y.; Gao, H.; Chen, H.; Fu, C. The involvement of papaya CpSBP1 in modulating fruit softening and carotenoid accumulation by repressing CpPME1/2 and CpPDS4. Sci. Hortic. 2019, 256. [Google Scholar] [CrossRef]
- Zhao, X.; Qu, D.; Wang, L.; Gao, Y.; An, N.; Wang, A.; Li, Y.; Yang, J.; Wu, F.; Su, H. Genome-wide identification of cysteine-rich receptor-like kinases in sweet cherry reveals that PaCRK1 enhances sweet cherry resistance to salt stress. Plant. Cell Rep. 2022, 41, 2037–2088. [Google Scholar] [CrossRef] [PubMed]
- Li, Q.; Chen, P.; Dai, S.; Sun, Y.; Yuan, B.; Kai, W.; Pei, Y.; He, S.; Liang, B.; Zhang, Y.; et al. PacCYP707A2 negatively regulates cherry fruit ripening while PacCYP707A1 mediates drought tolerance. J. Exp. Bot. 2015, 66, 3765–3774. [Google Scholar] [CrossRef] [PubMed]
- Gong, Y.; Fan, X.; Mattheis, J.P. Responses of ‘Bing’ and ‘Rainier’ sweet cherries to ethylene and 1-methylcyclopropene. J. Amer. Soc. Hort. Sci. 2002, 127, 831–835. [Google Scholar] [CrossRef]
- Tijero, V.; Teribia, N.; Munoz, P.; Munne-Bosch, S. Implication of abscisic acid on ripening and quality in sweet cherries: Differential effects during pre- and post-harvest. Front. Plant. Sci. 2016, 7, 602. [Google Scholar] [CrossRef]
- Kumar, R.; Khurana, A.; Sharma, A.K. Role of plant hormones and their interplay in development and ripening of fleshy fruits. J. Exp. Bot. 2014, 65, 4561–4575. [Google Scholar] [CrossRef]
- Chockchaisawasdee, S.; Golding, J.B.; Vuong, Q.V.; Papoutsis, K.; Stathopoulos, C.E. Sweet cherry: Composition, postharvest preservation, processing and trends for its future use. Trends Food Sci. Tech. 2016, 55, 72–83. [Google Scholar] [CrossRef]
- Correia, S.; Schouten, R.; Silva, A.P.; Goncalves, B. Factors affecting quality and health promoting compounds during growth and postharvest life of sweet cherry (Prunus avium L.). Front. Plant. Sci 2017, 8, 2166. [Google Scholar] [CrossRef]
- Zhang, D.; Han, Z.; Li, J.; Qin, H.; Zhou, L.; Wang, Y.; Zhu, X.; Ma, Y.; Fang, W. Genome-wide analysis of the SBP-box gene family transcription factors and their responses to abiotic stresses in tea (Camellia sinensis). Genomics 2020, 112, 2194–2202. [Google Scholar] [CrossRef]
- Hou, H.; Yan, X.; Sha, T.; Yan, Q.; Wang, X. The SBP-box gene VpSBP11 from Chinese wild Vitis is involved in floral transition and affects leaf development. Int J. Mol. Sci. 2017, 18, 1493. [Google Scholar] [CrossRef]
- Liu, M.; Sun, W.; Ma, Z.; Huang, L.; Wu, Q.; Tang, Z.; Bu, T.; Li, C.; Chen, H. Genome-wide identification of the SPL gene family in tartary buckwheat (Fagopyrum tataricum) and expression analysis during fruit development stages. BMC Plant. Biol. 2019, 19, 299. [Google Scholar] [CrossRef]
- Tan, H.; Song, X.; Duan, W.; Wang, Y.; Hou, X. Genome-wide analysis of the SBP-box gene family in Chinese cabbage (Brassica rapa subsp. pekinensis). Genome 2015, 58, 463–477. [Google Scholar] [CrossRef]
- Cai, C.; Guo, W.; Zhang, B. Genome-wide identification and characterization of SPL transcription factor family and their evolution and expression profiling analysis in cotton. Sci Rep. 2018, 8, 762. [Google Scholar] [CrossRef]
- Shen, X.; Zhao, K.; Liu, L.; Zhang, K.; Yuan, H.; Liao, X.; Wang, Q.; Guo, X.; Li, F.; Li, T. A role for PacMYBA in ABA-regulated anthocyanin biosynthesis in red-colored sweet cherry cv. Hong Deng (Prunus avium L.). Plant. Cell Physiol. 2014, 55, 862–880. [Google Scholar] [CrossRef]
- Qi, X.; Liu, C.; Song, L.; Li, M. PaMADS7, a MADS-box transcription factor, regulates sweet cherry fruit ripening and softening. Plant. Sci. 2020, 301, 110634. [Google Scholar] [CrossRef]
- Jin, W.; Wang, H.; Li, M.; Wang, J.; Yang, Y.; Zhang, X.; Yan, G.; Zhang, H.; Liu, J.; Zhang, K. The R2R3 MYB transcription factor PavMYB10.1 involves in anthocyanin biosynthesis and determines fruit skin colour in sweet cherry (Prunus avium L.). Plant. Biotechnol. J. 2016, 14, 2120–2133. [Google Scholar] [CrossRef]
- Chen, W.; Yu, Z.; Kong, J.; Wang, H.; Li, Y.; Zhao, M.; Wang, X.; Zheng, Q.; Shi, N.; Zhang, P.; et al. Comparative WGBS identifies genes that influence non-ripe phenotype in tomato epimutant Colourless non-ripening. Sci. China Life Sci. 2018, 61, 244–252. [Google Scholar] [CrossRef]
- Cui, M.; Wang, C.; Zhang, W.; Pervaiz, T.; Haider, M.S.; Tang, W.; Fang, J. Characterization of Vv-miR156: Vv-SPL pairs involved in the modulation of grape berry development and ripening. Mol. Genet. Genom. 2018, 293, 1333–1354. [Google Scholar] [CrossRef]
- Leng, P.; Yuan, B.; Guo, Y. The role of abscisic acid in fruit ripening and responses to abiotic stress. J. Exp. Bot. 2014, 65, 4577–4588. [Google Scholar] [CrossRef]
- Guo, J.; Wang, S.; Yu, X.; Dong, R.; Li, Y.; Mei, X.; Shen, Y. Polyamines regulate strawberry fruit ripening by abscisic acid, auxin, and ethylene. Plant. Physiol. 2018, 177, 339–351. [Google Scholar] [CrossRef] [PubMed]
- Santner, A.; Calderon-Villalobos, L.I.; Estelle, M. Plant hormones are versatile chemical regulators of plant growth. Nat. Chem. Biol. 2009, 5, 301–307. [Google Scholar] [CrossRef] [PubMed]
- Kuhn, N.; Ponce, C.; Arellano, M.; Time, A.; Sagredo, B.; Donoso, J.M.; Meisel, L.A. Gibberellic acid modifies the transcript abundance of ABA pathway orthologs and modulates sweet cherry (Prunus avium) fruit ripening in early- and mid-season varieties. Plants (Basel) 2020, 9, 1796. [Google Scholar] [CrossRef] [PubMed]
- Dong, H.; Yan, S.; Jing, Y.; Yang, R.; Zhang, Y.; Zhou, Y.; Zhu, Y.; Sun, J. MIR156-targeted SPL9 is phosphorylated by SnRK2s and interacts with ABI5 to enhance ABA responses in Arabidopsis. Front. Plant. Sci. 2021, 12, 708573. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H.; Zhao, X.; Li, J.; Cai, H.; Deng, X.W.; Li, L. MicroRNA408 is critical for the HY5-SPL7 gene network that mediates the coordinated response to light and copper. Plant. Cell 2014, 26, 4933–4953. [Google Scholar] [CrossRef]
- Wang, Y.; Zhai, Z.; Sun, Y.; Feng, C.; Peng, X.; Zhang, X.; Xiao, Y.; Zhou, X.; Wang, W.; Jiao, J.; et al. Genome-wide identification of the B-BOX genes that respond to multiple ripening related signals in sweet cherry fruit. Int. J. Mol. Sci. 2021, 22, 1622. [Google Scholar] [CrossRef]
- Jia, H.F.; Chai, Y.M.; Li, C.L.; Lu, D.; Luo, J.J.; Qin, L.; Shen, Y.Y. Abscisic acid plays an important role in the regulation of strawberry fruit ripening. Plant. Physiol. 2011, 157, 188–199. [Google Scholar] [CrossRef]
- Chen, C.; Chen, H.; Zhang, Y.; Thomas, H.R.; Frank, M.H.; He, Y.; Xia, R. TBtools: An integrative toolkit developed for interactive analyses of big biological data. Mol. Plant. 2020, 13, 1194–1202. [Google Scholar] [CrossRef]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef]
- Sparkes, I.A.; Runions, J.; Kearns, A.; Hawes, C. Rapid, transient expression of fluorescent fusion proteins in tobacco plants and generation of stably transformed plants. Nat. Protoc. 2006, 1, 2019–2025. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).