Structural Basis for Agonistic Activity and Selectivity toward Melatonin Receptors hMT1 and hMT2
Abstract
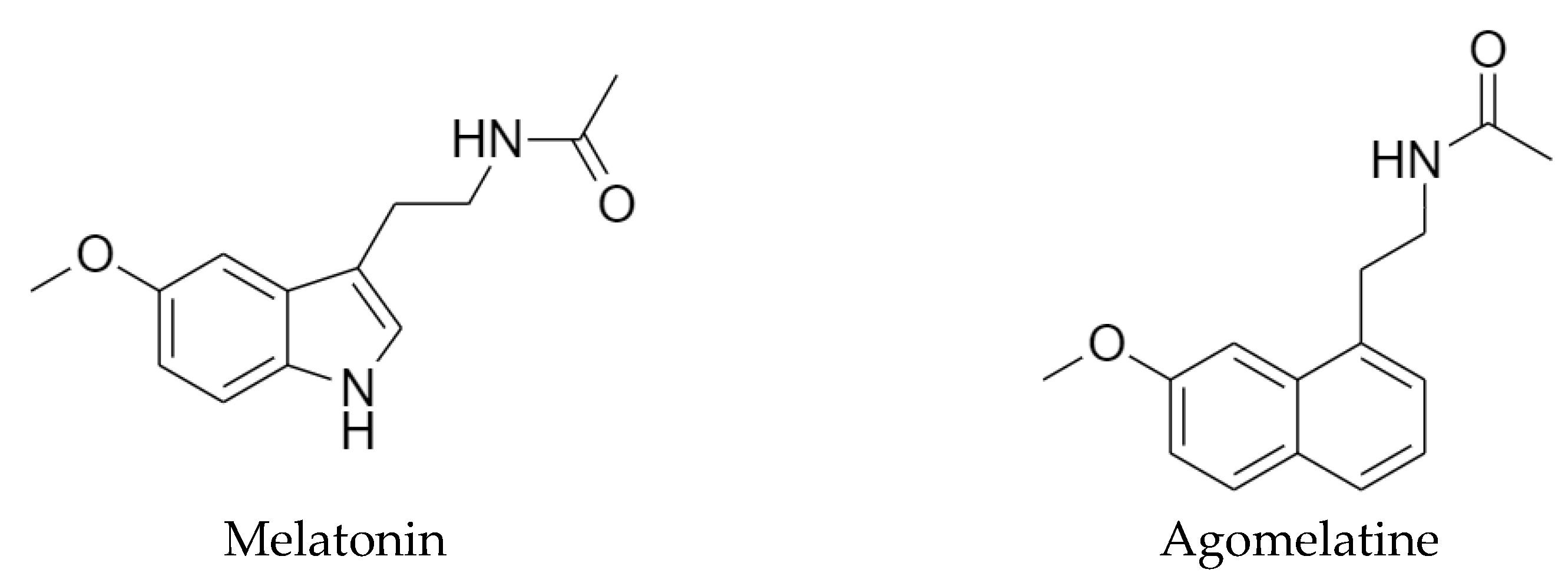
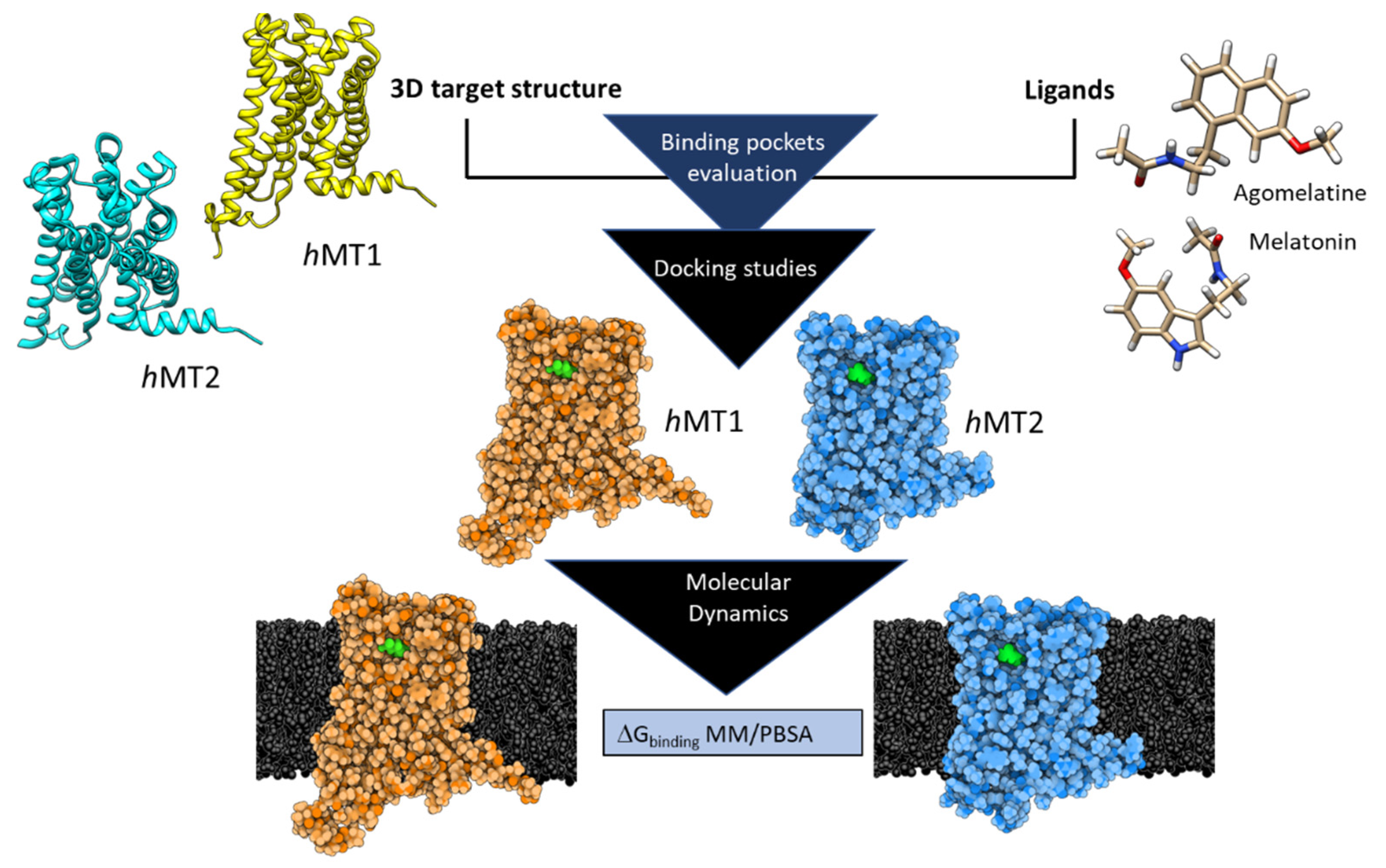
1. Introduction
2. Results and Discussion
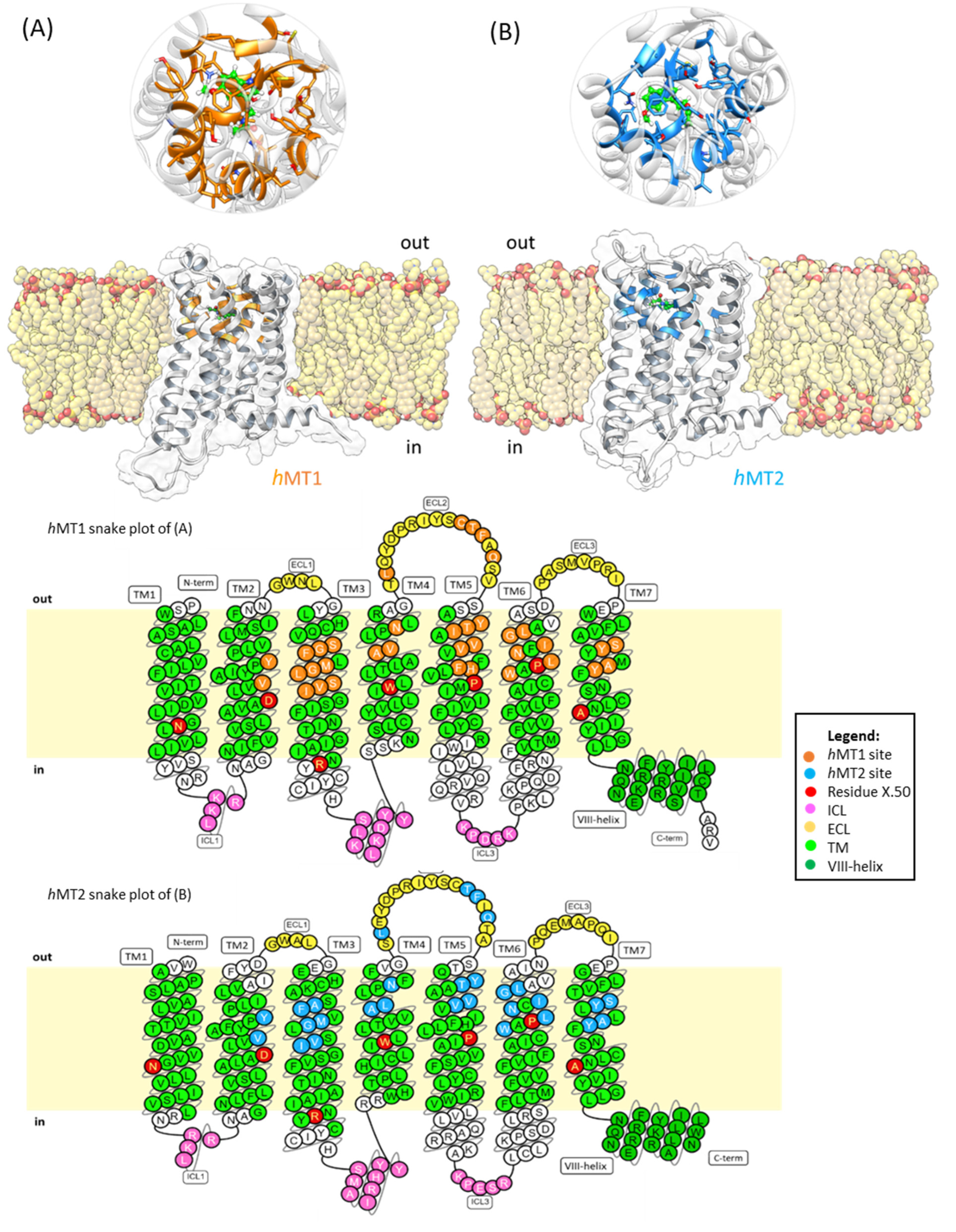
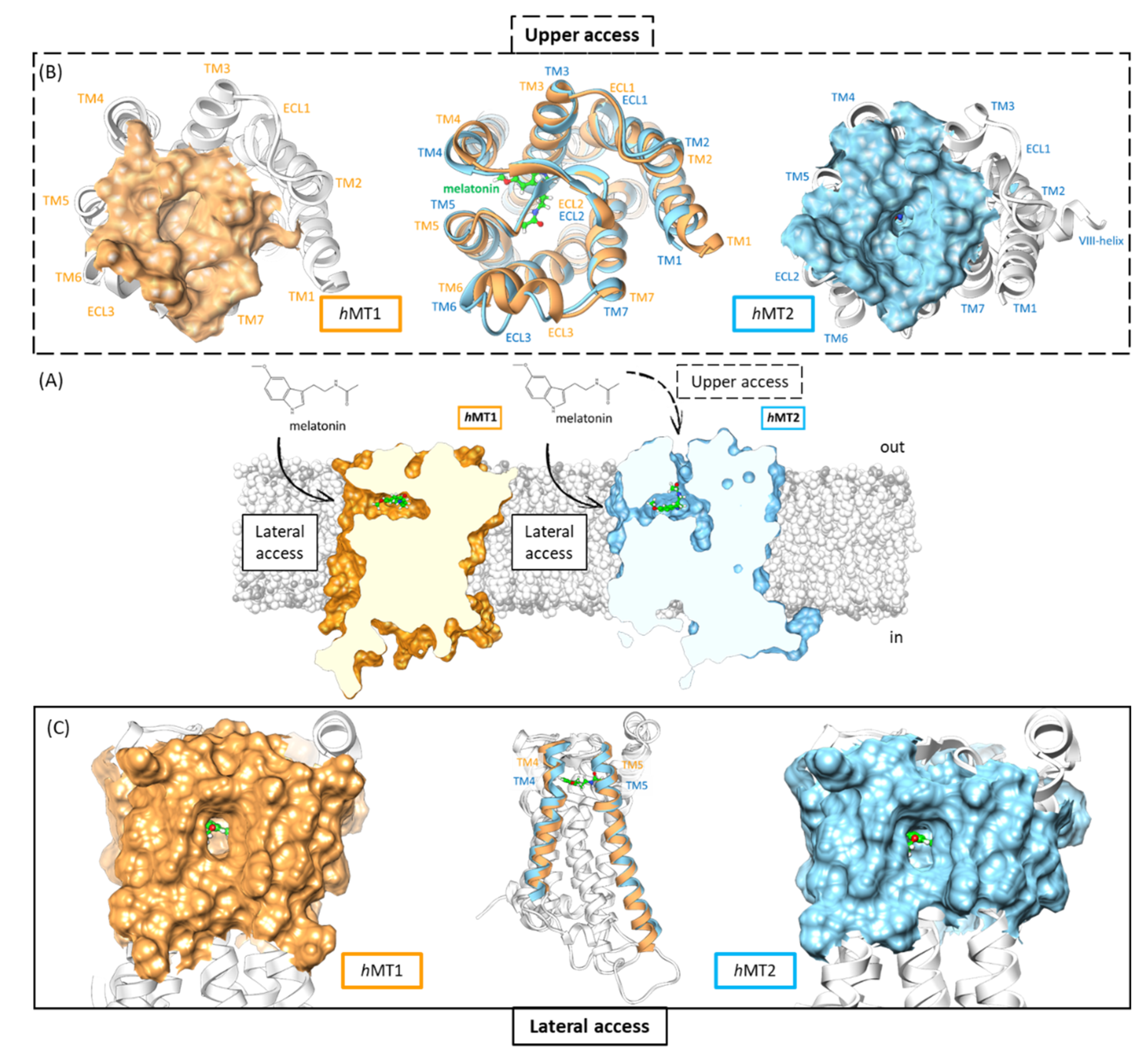
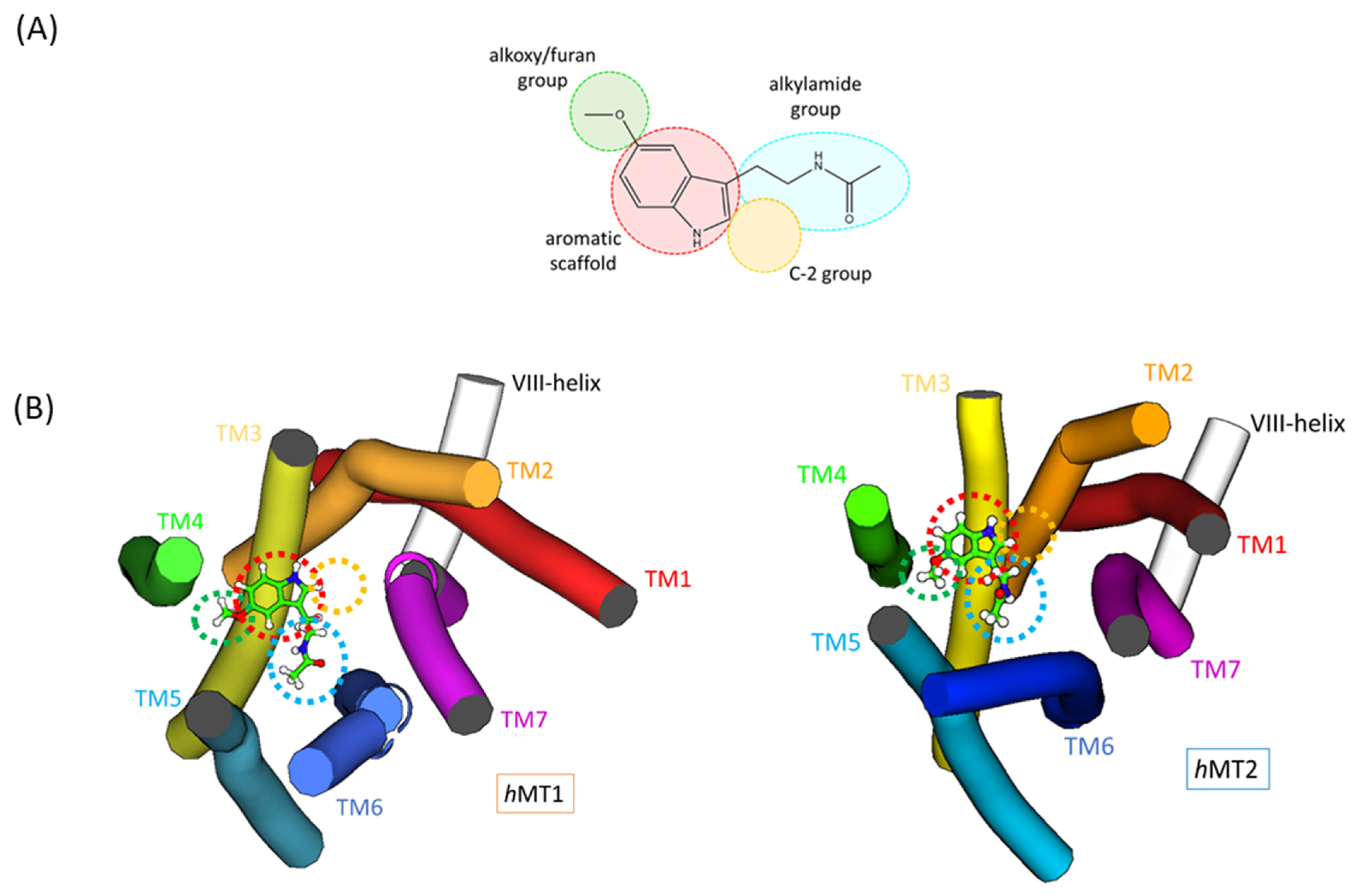
2.1. Structural Features of hMT1 and hMT2
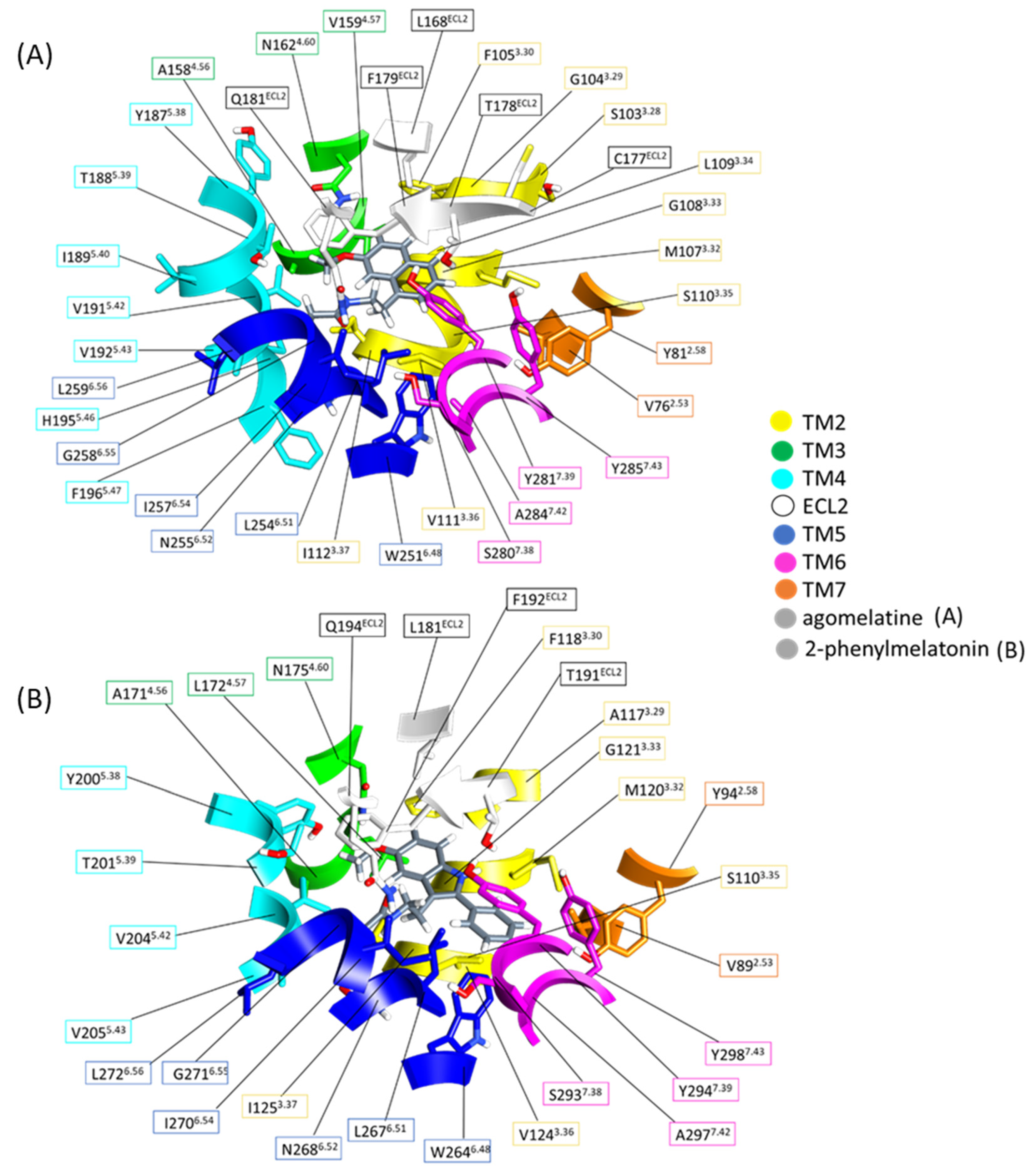
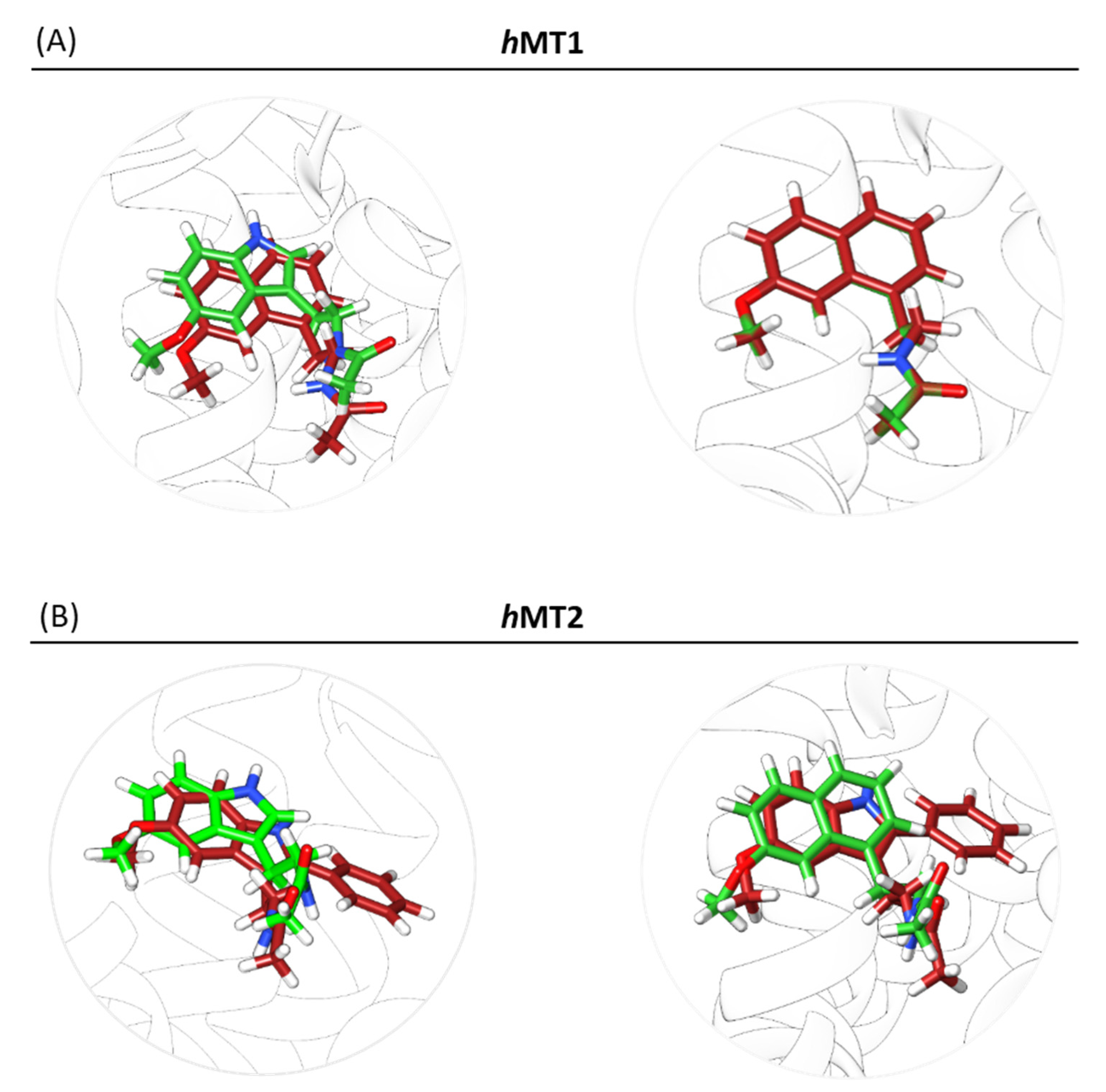
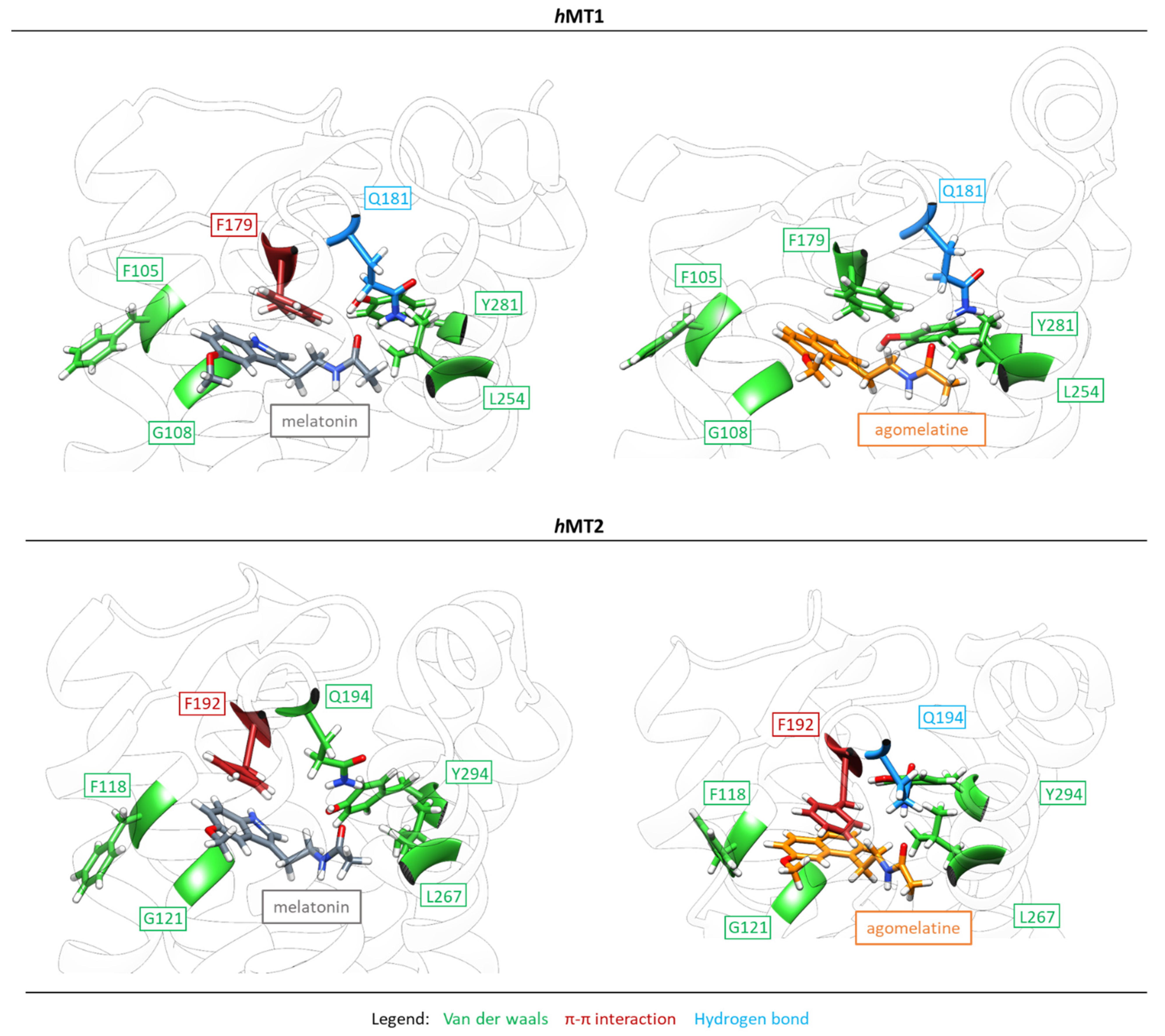
2.2. Exploration of Agonists’ Binding Poses within MTs Orthosteric Sites
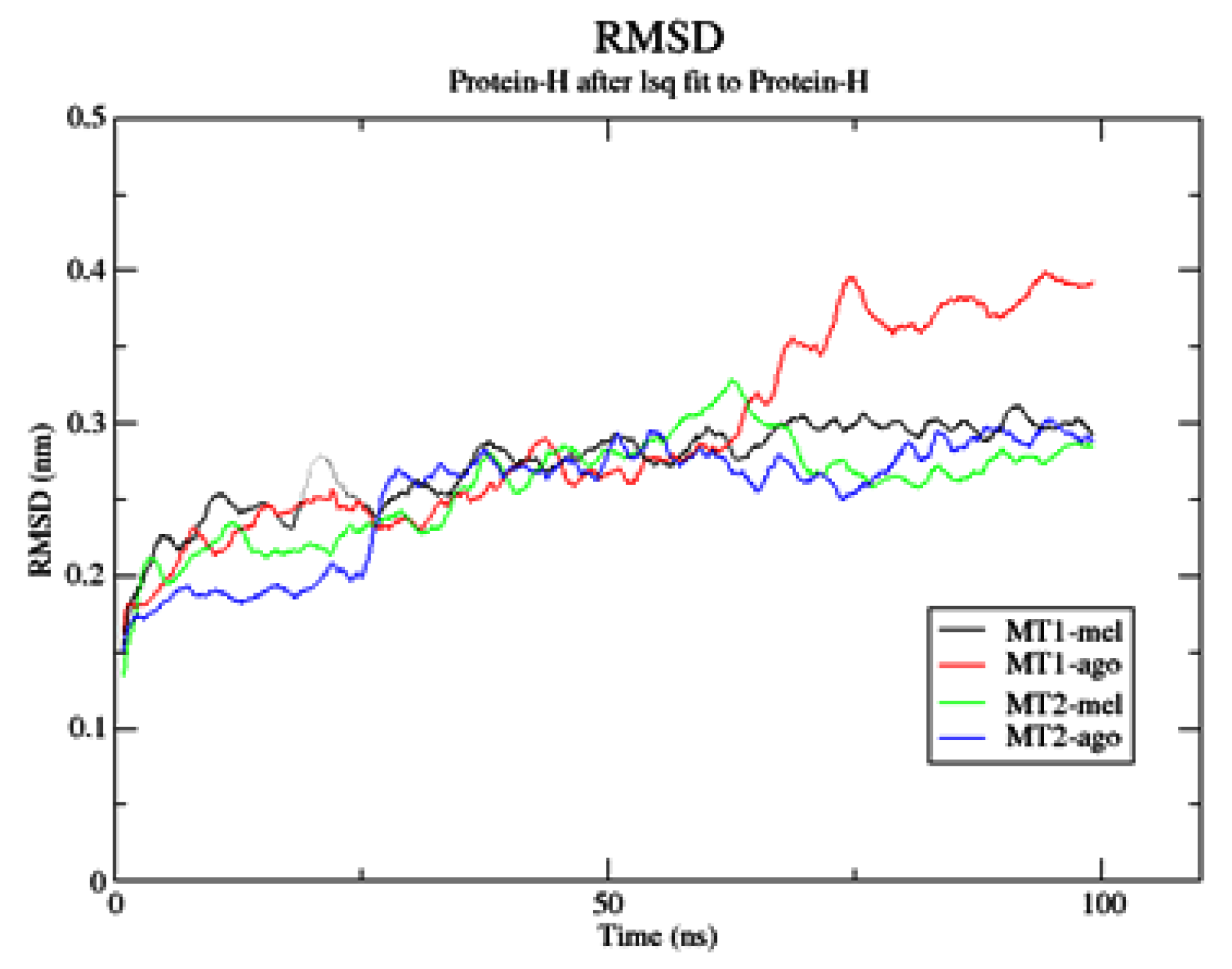
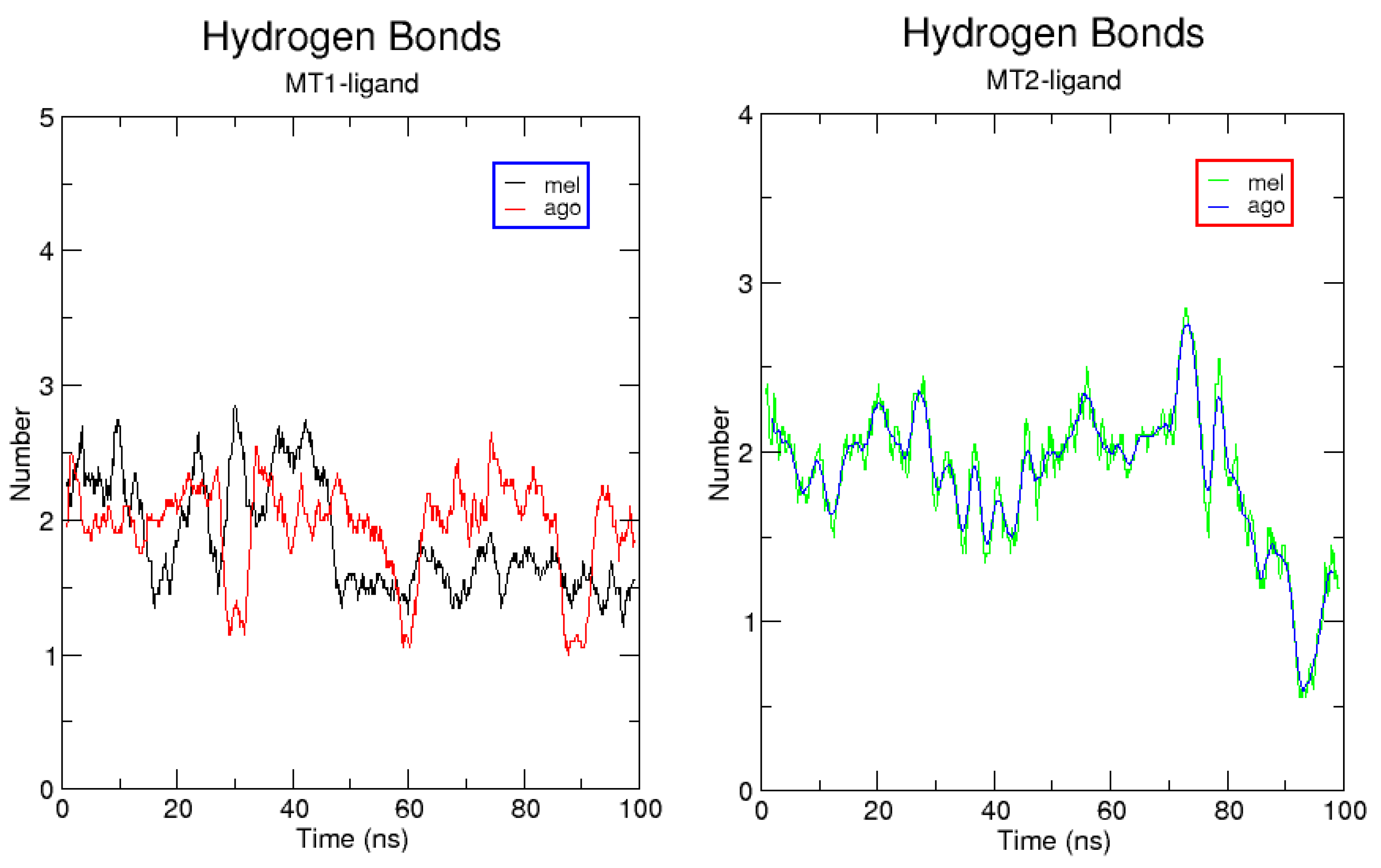
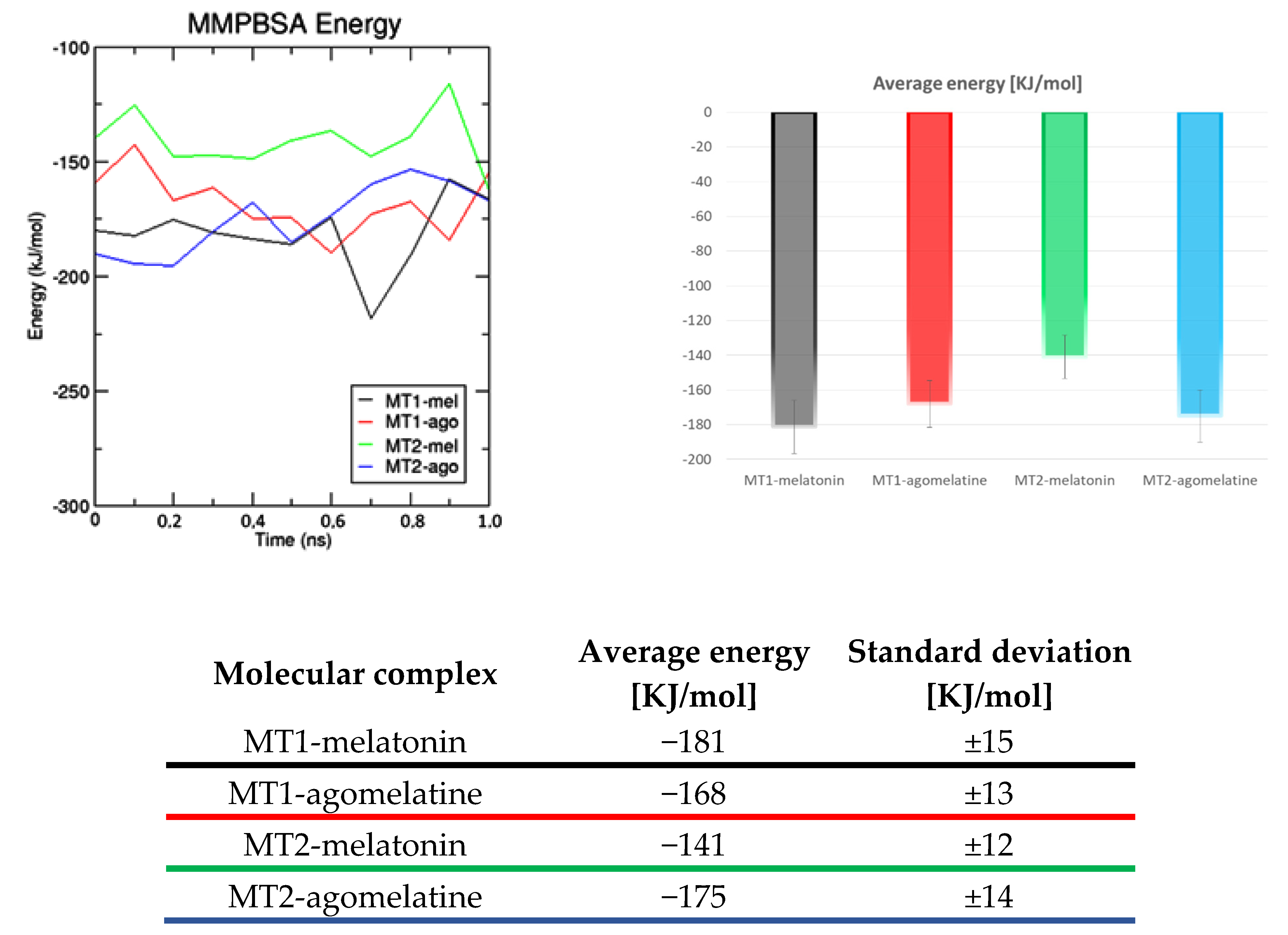
2.3. Complexes Conformational Stability: Molecular Dynamics Simulations
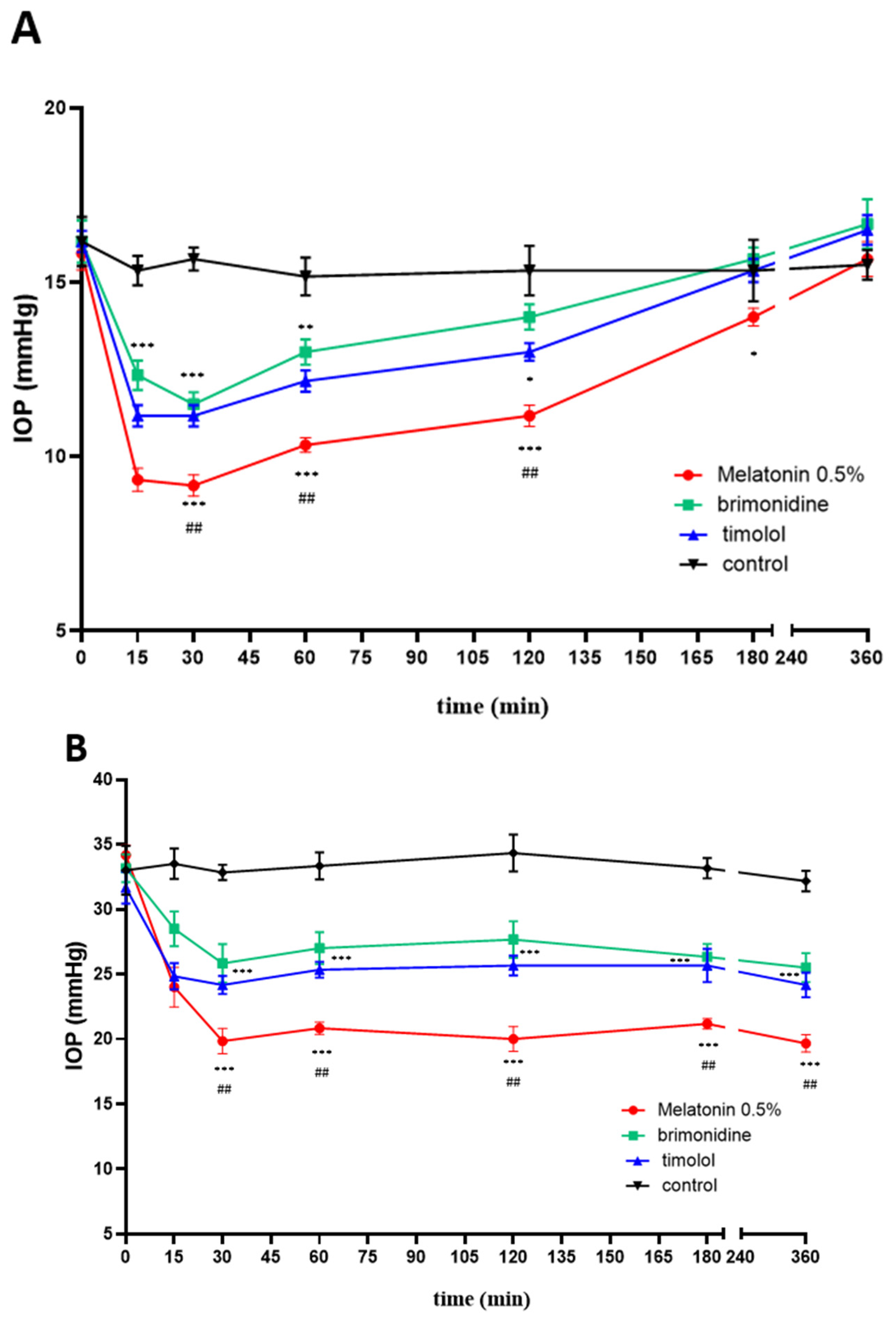
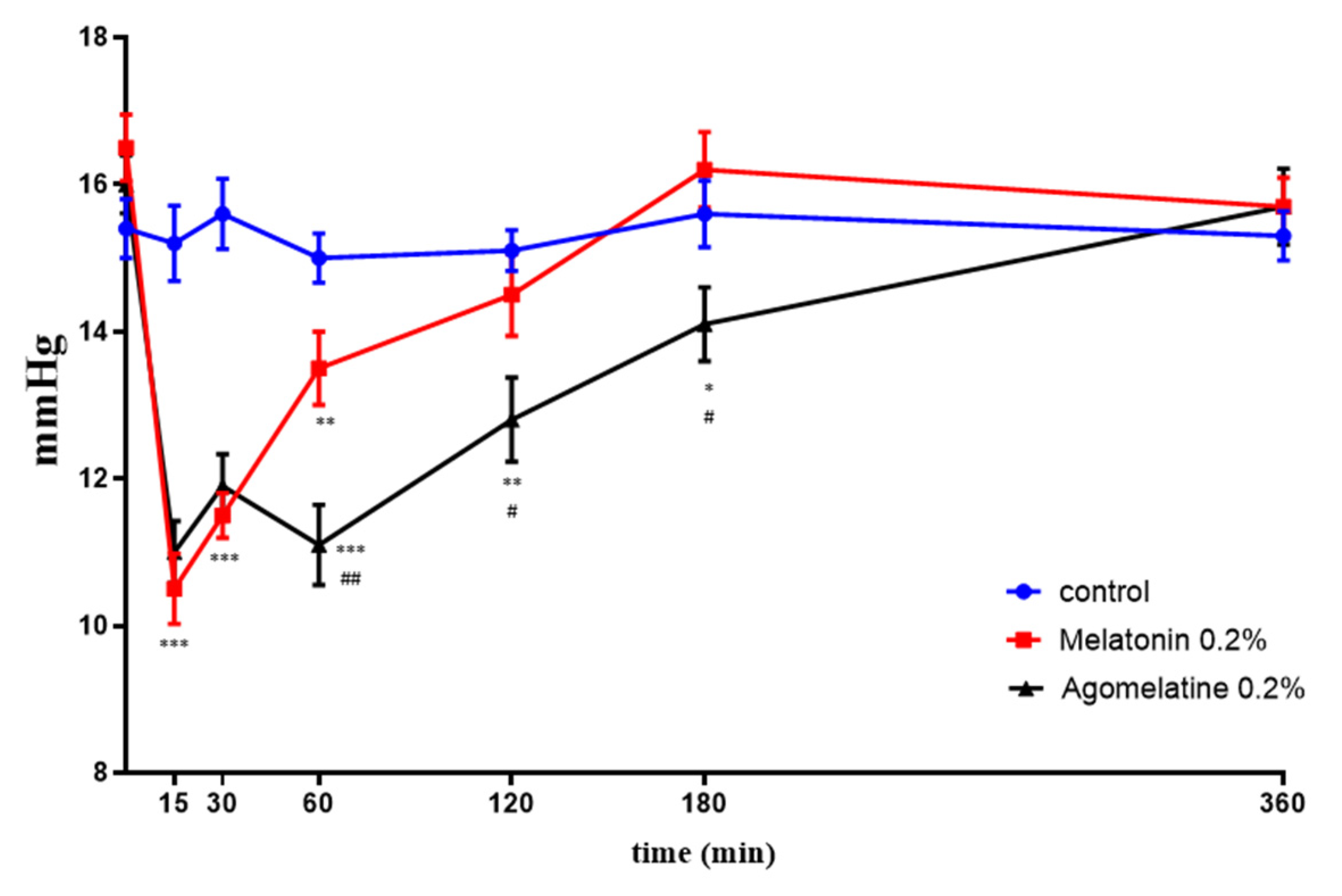
2.4. IOP Effects In Vivo: Melatonin and Agomelatine Relative Efficacy
2.5. Searching for Potential Melatonin Agonists: Test Case Studies
3. Material and Methods
3.1. D Protein Modeling
3.2. hMTs Ligands Docking
3.3. Molecular Dynamics Simulation of the hMTs-Ligands Complexes in Membrane
3.4. In Vivo IOP Reduction Effects of Melatonin and Agomelatine
3.5. In Silico Screening for Melatonin Agonists
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Weinreb, R.N.; Aung, T.; Medeiros, F.A. The pathophysiology and treatment of glaucoma: A review. JAMA J. Am. Med. Assoc. 2014, 311, 1901–1911. [Google Scholar] [CrossRef]
- Martínez-Águila, A.; Martín-Gil, A.; Carpena-Torres, C.; Pastrana, C.; Carracedo, G. Influence of circadian rhythm in the eye: Significance of melatonin in glaucoma. Biomolecules 2021, 11, 340. [Google Scholar] [CrossRef] [PubMed]
- Agorastos, A.; Skevas, C.; Matthaei, M.; Otte, C.; Klemm, M.; Richard, G.; Huber, C.G. Depression, anxiety, and disturbed sleep in glaucoma. J. Neuropsychiatry Clin. Neurosci. 2013, 25, 205–213. [Google Scholar] [CrossRef] [PubMed]
- Hardeland, R.; Madrid, J.A.; Tan, D.-X.; Reiter, R.J. Melatonin, the circadian multioscillator system and health: The need for detailed analyses of peripheral melatonin signaling. J. Pineal Res. 2012, 52, 139–166. [Google Scholar] [CrossRef] [PubMed]
- Agorastos, A.; Huber, C.G. The role of melatonin in glaucoma: Implications concerning pathophysiological relevance and therapeutic potential. J. Pineal Res. 2011, 50, 1–7. [Google Scholar] [CrossRef]
- Jean-Louis, G.; Zizi, F.; Lazzaro, D.R.; Wolintz, A.H. Circadian rhythm dysfunction in glaucoma: A hypothesis. J. Circadian Rhythm. 2008, 6, 1. [Google Scholar] [CrossRef]
- Alkozi, H.A.; Navarro, G.; Franco, R.; Pintor, J. Melatonin and the control of intraocular pressure. Prog. Retin. Eye Res. 2020, 75, 100798. [Google Scholar] [CrossRef]
- Pranil, T.; Moongngarm, A.; Loypimai, P. Influence of pH, temperature, and light on the stability of melatonin in aqueous solutions and fruit juices. Heliyon 2020, 6, e03648. [Google Scholar] [CrossRef]
- Audinot, V.; Mailliet, F.; Lahaye-Brasseur, C.; Bonnaud, A.; Le Gall, A.; Amossé, C.; Dromaint, S.; Rodriguez, M.; Nagel, N.; Galizzi, J.-P.; et al. New selective ligands of human cloned melatonin MT1 and MT2 receptors. Naunyn Schmiedebergs Arch. Pharmacol. 2003, 367, 553–561. [Google Scholar] [CrossRef]
- Stauch, B.; Johansson, L.C.; McCorvy, J.D.; Patel, N.; Han, G.W.; Huang, X.-P.; Gati, C.; Batyuk, A.; Slocum, S.T.; Ishchenko, A.; et al. Structural basis of ligand recognition at the human MT1 melatonin receptor. Nature 2019, 569, 284–288. [Google Scholar] [CrossRef]
- Manikandan, S. Agomelatine: A novel melatonergic antidepressant. J. Pharmacol. Pharmacother. 2010, 1, 122. [Google Scholar] [CrossRef] [PubMed]
- Attwood, T.K.; Findlay, J.B.C. Fingerprinting g-protein-coupled receptors. Protein Eng. Des. Sel. 1994, 7, 195–203. [Google Scholar] [CrossRef]
- Kolakowski, L.F. GCRDb: A G-protein-coupled receptor database. Recept. Channels 1994, 2, 1–7. [Google Scholar]
- Schiöth, H.B.; Fredriksson, R. The GRAFS classification system of G-protein coupled receptors in comparative perspective. Gen. Comp. Endocrinol. 2005, 142, 94–101. [Google Scholar] [CrossRef] [PubMed]
- Fredriksson, R.; Lagerström, M.C.; Lundin, L.G.; Schiöth, H.B. The G-protein-coupled receptors in the human genome form five main families. Phylogenetic analysis, paralogon groups, and fingerprints. Mol. Pharmacol. 2003, 63, 1256–1272. [Google Scholar] [CrossRef]
- Hu, G.-M.; Mai, T.-L.; Chen, C.-M. Visualizing the GPCR Network: Classification and Evolution. Sci. Rep. 2017, 7, 15495. [Google Scholar] [CrossRef] [PubMed]
- Rasmussen, S.G.F.; DeVree, B.T.; Zou, Y.; Kruse, A.C.; Chung, K.Y.; Kobilka, T.S.; Thian, F.S.; Chae, P.S.; Pardon, E.; Calinski, D.; et al. Crystal structure of the β 2 adrenergic receptor-Gs protein complex. Nature 2011, 477, 549–557. [Google Scholar] [CrossRef]
- Cherezov, V.; Rosenbaum, D.M.; Hanson, M.A.; Rasmussen, S.G.F.; Thian, F.S.; Kobilka, T.S.; Choi, H.-J.; Kuhn, P.; Weis, W.I.; Kobilka, B.K.; et al. High-resolution crystal structure of an engineered human β2-adrenergic G protein-coupled receptor. Science 2007, 318, 1258–1265. [Google Scholar] [CrossRef]
- Johansson, L.C.; Stauch, B.; McCorvy, J.D.; Han, G.W.; Patel, N.; Huang, X.-P.; Batyuk, A.; Gati, C.; Slocum, S.T.; Li, C.; et al. XFEL structures of the human MT2 melatonin receptor reveal the basis of subtype selectivity. Nature 2019, 569, 289–292. [Google Scholar] [CrossRef]
- Warne, T.; Edwards, P.C.; Doré, A.S.; Leslie, A.G.W.; Tate, C.G. Molecular basis for high-affinity agonist binding in GPCRs. Science 2019, 364, 775–778. [Google Scholar] [CrossRef]
- Patel, N.; Huang, X.P.; Grandner, J.M.; Johansson, L.C.; Stauch, B.; McCorvy, J.D.; Liu, Y.; Roth, B.; Katritch, V.; Department of Biological Sciences; et al. Structure-based discovery of potent and selective melatonin receptor agonists. Elife 2020, 9, e53779. [Google Scholar] [CrossRef] [PubMed]
- Ballesteros, J.A.; Weinstein, H. Integrated methods for the construction of three-dimensional models and computational probing of structure-function relations in G protein-coupled receptors. Methods Neurosci. 1995, 25, 366–428. [Google Scholar] [CrossRef]
- Berman, H.M.; Westbrook, J.; Feng, Z.; Gilliland, G.; Bhat, T.N.; Weissig, H.; Shindyalov, I.N.; Bourne, P.E. The Protein Data Bank. Nucleic Acids Res. 2000, 28, 235–242. [Google Scholar] [CrossRef] [PubMed]
- Isberg, V.; de Graaf, C.; Bortolato, A.; Cherezov, V.; Katritch, V.; Marshall, F.H.; Mordalski, S.; Pin, J.-P.; Stevens, R.C.; Vriend, G.; et al. Generic GPCR residue numbers—Aligning topology maps while minding the gaps. Trends Pharmacol. Sci. 2015, 36, 22–31. [Google Scholar] [CrossRef] [PubMed]
- Ishchenko, A.; Stauch, B.; Han, G.W.; Batyuk, A.; Shiriaeva, A.; Li, C.; Zatsepin, N.; Weierstall, U.; Liu, W.; Nango, E.; et al. Toward G protein-coupled receptor structure-based drug design using X-ray lasers. IUCrJ 2019, 6, 1106–1119. [Google Scholar] [CrossRef]
- Tian, W.; Chen, C.; Lei, X.; Zhao, J.; Liang, J. CASTp 3.0: Computed atlas of surface topography of proteins. Nucleic Acids Res. 2018, 46, W363–W367. [Google Scholar] [CrossRef]
- Edelsbrunner, H.; Mücke, E.P. Three-Dimensional Alpha Shapes. ACM Trans. Graph. 1994, 13, 43–72. [Google Scholar] [CrossRef]
- Ebalunode, J.O.; Ouyang, Z.; Liang, J.; Zheng, W. Novel approach to structure-based pharmacophore search using computational geometry and shape matching techniques. J. Chem. Inf. Model. 2008, 48, 889–901. [Google Scholar] [CrossRef]
- Tian, W.; Liang, J. On quantification of geometry and topology of protein pockets and channels for assessing mutation effects. In Proceedings of the 2018 IEEE EMBS International Conference on Biomedical and Health Informatics, BHI 2018, Las Vegas, NV, USA, 4–7 March 2018; Volume 2018, pp. 263–266. [Google Scholar] [CrossRef]
- Liang, J.; Woodward, C.; Edelsbrunner, H. Anatomy of protein pockets and cavities: Measurement of binding site geometry and implications for ligand design. Protein Sci. 1998, 7, 1884–1897. [Google Scholar] [CrossRef]
- Edelsbrunner, H.; Facello, M.; Liang, J. On the definition and the construction of pockets in macromolecules. Pac. Symp. Biocomput. 1996, 88, 272–287. [Google Scholar] [CrossRef]
- Pettersen, E.F.; Goddard, T.D.; Huang, C.C.; Couch, G.S.; Greenblatt, D.M.; Meng, E.C.; Ferrin, T.E. UCSF Chimera—A visualization system for exploratory research and analysis. J. Comput. Chem. 2004, 25, 1605–1612. [Google Scholar] [CrossRef] [PubMed]
- Salentin, S.; Schreiber, S.; Haupt, V.J.; Adasme, M.F.; Schroeder, M. PLIP: Fully automated protein-ligand interaction profiler. Nucleic Acids Res. 2015, 43, W443–W447. [Google Scholar] [CrossRef] [PubMed]
- Trzaskowski, B.; Latek, D.; Yuan, S.; Ghoshdastider, U.; Debinski, A.; Filipek, S. Action of Molecular Switches in GPCRs—Theoretical and Experimental Studies. Curr. Med. Chem. 2012, 19, 1090–1109. [Google Scholar] [CrossRef] [PubMed]
- Eddy, M.T.; Lee, M.-Y.; Gao, Z.-G.; White, K.L.; Didenko, T.; Horst, R.; Audet, M.; Stanczak, P.; McClary, K.M.; Han, G.W.; et al. Allosteric Coupling of Drug Binding and Intracellular Signaling in the A2A Adenosine Receptor. Cell 2018, 172, 68–80. [Google Scholar] [CrossRef]
- Filipek, S. Molecular switches in GPCRs. Curr. Opin. Struct. Biol. 2019, 55, 114–120. [Google Scholar] [CrossRef] [PubMed]
- Holst, B.; Nygaard, R.; Valentin-Hansen, L.; Bach, A.; Engelstoft, M.S.; Petersen, P.S.; Frimurer, T.M.; Schwartz, T.W. A conserved aromatic lock for the tryptophan rotameric switch in TM-VI of seven-transmembrane receptors. J. Biol. Chem. 2010, 285, 3973–3985. [Google Scholar] [CrossRef] [PubMed]
- Wescott, M.P.; Kufareva, I.; Paes, C.; Goodman, J.R.; Thaker, Y.; Puffer, B.A.; Berdougo, E.; Rucker, J.B.; Handel, T.M.; Doranz, B.J. Signal transmission through the CXC chemokine receptor 4 (CXCR4) transmembrane helices. Proc. Natl. Acad. Sci. USA 2016, 113, 9928–9933. [Google Scholar] [CrossRef]
- Tehan, B.G.; Bortolato, A.; Blaney, F.E.; Weir, M.P.; Mason, J.S. Unifying Family A GPCR Theories of Activation. Pharmacol. Ther. 2014, 143, 51–60. [Google Scholar] [CrossRef]
- Hofmann, K.P.; Scheerer, P.; Hildebrand, P.W.; Choe, H.-W.; Park, J.H.; Heck, M.; Ernst, O.P. A G protein-coupled receptor at work: The rhodopsin model. Trends Biochem. Sci. 2009, 34, 540–552. [Google Scholar] [CrossRef]
- Nygaard, R.; Frimurer, T.M.; Holst, B.; Rosenkilde, M.M.; Schwartz, T.W. Ligand binding and micro-switches in 7TM receptor structures. Trends Pharmacol. Sci. 2009, 30, 249–259. [Google Scholar] [CrossRef]
- Zhou, Q.; Yang, D.; Wu, M.; Guo, Y.; Guo, W.; Zhong, L.; Cai, X.; Dai, A.; Jang, W.; Shakhnovich, E.I.; et al. Common activation mechanism of class a GPCRs. eLife 2019, 8, e50279. [Google Scholar] [CrossRef] [PubMed]
- Pala, D.; Lodola, A.; Bedini, A.; Spadoni, G.; Rivara, S. Homology models of melatonin receptors: Challenges and recent advances. Int. J. Mol. Sci. 2013, 14, 8093–8121. [Google Scholar] [CrossRef] [PubMed]
- Frisch, M.J.; Trucks, G.W.; Schlegel, H.B.; Scuseria, G.E.; Robb, M.A.; Cheeseman, J.R.; Scalmani, G.; Barone, V.; Petersson, G.A.; Nakatsuji, H.; et al. Gaussian 16, Revision C.01; Gaussian Inc.: Wallingford, CT, USA, 2016. [Google Scholar]
- Legros, C.; Brasseur, C.; Delagrange, P.; Ducrot, P.; Nosjean, O.; Boutin, J.A. Alternative radioligands for investigating the molecular pharmacology of melatonin receptors. J. Pharmacol. Exp. Ther. 2016, 356, 681–692. [Google Scholar] [CrossRef] [PubMed]
- Legros, C.; Matthey, U.; Grelak, T.; Pedragona-Moreau, S.; Hassler, W.; Yous, S.; Thomas, E.; Suzenet, F.; Folleas, B.; Lefoulon, F.; et al. New radioligands for describing the molecular pharmacology of MT1 and MT2 melatonin receptors. Int. J. Mol. Sci. 2013, 14, 8948–8962. [Google Scholar] [CrossRef] [PubMed]
- Lavedan, C.; Forsberg, M.; Gentile, A.J. Tasimelteon: A selective and unique receptor binding profile. Neuropharmacology 2015, 91, 142–147. [Google Scholar] [CrossRef]
- Sargsyan, K.; Grauffel, C.; Lim, C. How Molecular Size Impacts RMSD Applications in Molecular Dynamics Simulations. J. Chem. Theory Comput. 2017, 13, 1518–1524. [Google Scholar] [CrossRef]
- Beresford, I.J.; Browning, C.; Starkey, S.J.; Brown, J.; Foord, S.M.; Coughlan, J.; North, P.C.; Dubocovich, M.L.; Hagan, R.M. GR196429: A nonindolic agonist at high-affinity melatonin receptors. J. Pharmacol. Exp. Ther. 1998, 285, 1239–1245. [Google Scholar]
- Dubocovich, M.L. Characterization of a retinal melatonin receptor. J. Pharmacol. Exp. Ther. 1985, 234, 395–401. [Google Scholar] [PubMed]
- Dubocovich, M.L.; Masana, M.I.; Iacob, S.; Sauri, D.M. Melatonin receptor antagonists that differentiate hetween the human Mel(1a) and Mel(1b) recombinant subtypes are used to assess the pharmacological profile of the rabbit retina ML(1) presynaptic heteroreceptor. Naunyn Schmiedebergs Arch. Pharmacol. 1997, 355, 365–375. [Google Scholar] [CrossRef]
- Wang, C.; Nguyen, P.H.; Pham, K.; Huynh, D.; Le, T.-B.N.; Wang, H.; Ren, P.; Luo, R. Calculating protein-ligand binding affinities with MMPBSA: Method and error analysis. J. Comput. Chem. 2016, 37, 2436–2446. [Google Scholar] [CrossRef]
- Sharif, N.A. Ocular Hypotension: Involvement of Serotonergic 5-HT2 Receptors. In 5-HT2C Receptors in the Pathophysiology of CNS Disease; Di Giovanni, G., Esposito, E., Di Matteo, V., Eds.; The Receptors; Humana Press: Totowa, NJ, USA, 2011; Volume 22. [Google Scholar] [CrossRef]
- UniProt. UniProt: A worldwide hub of protein knowledge. Nucleic Acids Res. 2021, 47, D506–D515. [Google Scholar]
- Lomize, M.A.; Pogozheva, I.D.; Joo, H.; Mosberg, H.I.; Lomize, A.L. OPM database and PPM web server: Resources for positioning of proteins in membranes. Nucleic Acids Res. 2012, 40, D370–D376. [Google Scholar] [CrossRef]
- Lomize, A.L.; Pogozheva, I.D.; Lomize, M.A.; Mosberg, H.I. Positioning of proteins in membranes: A computational approach. Protein Sci. 2006, 15, 1318–1333. [Google Scholar] [CrossRef] [PubMed]
- Biovia. Dassault Systemes BIOVIA, Discovery Studio Modelling Environment, Release 4.5; Accelrys Softw Inc.: San Diego, CA, USA, 2015. [Google Scholar]
- Zisapel, N. New perspectives on the role of melatonin in human sleep, circadian rhythms and their regulation. Br. J. Pharmacol. 2018, 175, 3190–3199. [Google Scholar] [CrossRef]
- Gioia, D.; Bertazzo, M.; Recanatini, M.; Masetti, M.; Cavalli, A. Dynamic docking: A paradigm shift in computational drug discovery. Molecules 2017, 22, 2029. [Google Scholar] [CrossRef]
- Salmaso, V.; Moro, S. Bridging molecular docking to molecular dynamics in exploring ligand-protein recognition process: An overview. Front. Pharmacol. 2018, 9, 923. [Google Scholar] [CrossRef]
- Morris, G.M.; Huey, R.; Lindstrom, W.; Sanner, M.F.; Belew, R.K.; Goodsell, D.S.; Olson, A.J. Software news and updates AutoDock4 and AutoDockTools4: Automated docking with selective receptor flexibility. J. Comput. Chem. 2009, 30, 2785–2791. [Google Scholar] [CrossRef]
- Serrano Cardona, L.; Muñoz Mata, E. Paraninfo Digital. Early Hum. Dev. 2013, 83, 1–11. [Google Scholar] [CrossRef]
- Massaccesi, L.; Laudadio, E.; Mobbili, G.; Minnelli, C.; Galeazzi, R. Cholesterol-mediated oligomerization pathways of serotonin G-coupled receptor 5-HT2C. Int. J. Biol. Macromol. 2020, 160, 1090–1100. [Google Scholar] [CrossRef]
- Abraham, M.J.; Murtola, T.; Schulz, R.; Páll, S.; Smith, J.C.; Hess, B.; Lindahl, E. Gromacs: High performance molecular simulations through multi-level parallelism from laptops to supercomputers. SoftwareX 2015, 1–2, 19–25. [Google Scholar] [CrossRef]
- Jo, S.; Kim, T.; Iyer, V.G.; Im, W. CHARMM-GUI: A web-based graphical user interface for CHARMM. J. Comput. Chem. 2008, 29, 1859–1865. [Google Scholar] [CrossRef]
- Wu, E.L.; Cheng, X.; Jo, S.; Rui, H.; Song, K.C.; Dávila-Contreras, E.M.; Qi, Y.; Lee, J.; Monje-Galvan, V.; Venable, R.M.; et al. CHARMM-GUI membrane builder toward realistic biological membrane simulations. J. Comput. Chem. 2014, 35, 1997–2004. [Google Scholar] [CrossRef]
- Jo, S.; Kim, T.; Im, W. Automated builder and database of protein/membrane complexes for molecular dynamics simulations. PLoS ONE 2007, 2, e880. [Google Scholar] [CrossRef]
- Maier, J.A.; Martinez, C.; Kasavajhala, K.; Wickstrom, L.; Hauser, K.E.; Simmerling, C. ff14SB: Improving the Accuracy of Protein Side Chain and Backbone Parameters from ff99SB. J. Chem. Theory Comput. 2015, 11, 3696–3713. [Google Scholar] [CrossRef]
- Darden, T.; York, D.; Pedersen, L. Particle mesh Ewald: An N·log(N) method for Ewald sums in large systems. J. Chem. Phys. 1993, 98, 10089–10092. [Google Scholar] [CrossRef]
- Hess, B.; Bekker, H.; Berendsen, H.J.C.; Fraaije, J.G.E.M. LINCS: A Linear Constraint Solver for molecular simulations. J. Comput. Chem. 1997, 18, 1463–1472. [Google Scholar] [CrossRef]
- Wardi, Y. A stochastic steepest-descent algorithm. J. Optim. Theory Appl. 1988, 59, 307–323. [Google Scholar] [CrossRef]
- Berendsen, H.J.C.; Postma, J.P.M.; Van Gunsteren, W.F.; DiNola, A.; Haak, J.R. Molecular dynamics with coupling to an external bath. J. Chem. Phys. 1984, 81, 3684–3690. [Google Scholar] [CrossRef]
- Nosé, S. A unified formulation of the constant temperature molecular dynamics methods. J. Chem. Phys. 1984, 81, 511–519. [Google Scholar] [CrossRef]
- Hoover, W.G. Canonical dynamics: Equilibrium phase-space distributions. Phys. Rev. A 1985, 31, 1695–1697. [Google Scholar] [CrossRef] [PubMed]
- Parrinello, M.; Rahman, A. Polymorphic transitions in single crystals: A new molecular dynamics 73.method. J. Appl. Phys. 1981, 52, 7182–7190. [Google Scholar] [CrossRef]
- Wang, C.; Greene, D.; Xiao, L.; Qi, R.; Luo, R. Recent developments and applications of the MMPBSA method. Front. Mol. Biosci. 2018, 4, 87. [Google Scholar] [CrossRef] [PubMed]
- Laudadio, E.; Mobbili, G.; Sorci, L.; Galeazzi, R.; Minnelli, C. Mechanistic insight toward EGFR activation induced by ATP: Role of mutations and water in ATP binding patterns. J. Biomol. Struct. Dyn. 2022, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Monte, O.D.; Chu, C.C.J.; Fagan, N.A.; Chang, S.W.C. Specialized medial prefrontal-amygdala coordination in other-regarding decision preference. Nat. Neurosci. 2020, 23, 565–574. [Google Scholar] [CrossRef]
- Eberhardt, J.; Santos-Martins, D.; Tillack, A.F.; Forli, S. AutoDock Vina 1.2.0: New Docking Methods, Expanded Force Field, and Python Bindings. J. Chem. Inf. Model. 2021, 61, 3891–3898. [Google Scholar] [CrossRef]
- Trott, O.; Olson, A.J. AutoDock Vina: Improving the speed and accuracy of docking with a new scoring function, efficient optimization, and multithreading. J. Comput. Chem. 2010, 31, 455–461. [Google Scholar] [CrossRef] [PubMed]

| Receptor | Compound | Cluster Conformation | ΔGbind (Kcal/mol) |
|---|---|---|---|
| hMT1 | Melatonin | 1 of 1 | −6.42 |
| Agomelatine | 2 of 7 | −6.81 | |
| hMT2 | Melatonin | 1 of 1 | −7.12 |
| Agomelatine | 1 of 1 | −7.52 |
| Receptor | Ligand | Molecular Docking ΔGbind | Receptor | Ligand | Molecular Docking ΔGbind |
|---|---|---|---|---|---|
| hMT1 | Melatonin | −6.42 Kcal/mol | hMT2 | Melatonin | −7.12 Kcal/mol |
| Agomelatine | −6.81 Kcal/mol | Agomelatine | −7.52 Kcal/mol | ||
| Ramelteon | −7.93 Kcal/mol | Ramelteon | −8.66 Kcal/mol | ||
| Clozapine | −8.47 Kcal/mol | Clozapine | −8.33 Kcal/mol | ||
| Flumazenil | −7.43 Kcal/mol | Flumazenil | −7.71 Kcal/mol |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Cantarini, M.; Rusciano, D.; Amato, R.; Canovai, A.; Cammalleri, M.; Monte, M.D.; Minnelli, C.; Laudadio, E.; Mobbili, G.; Giorgini, G.; et al. Structural Basis for Agonistic Activity and Selectivity toward Melatonin Receptors hMT1 and hMT2. Int. J. Mol. Sci. 2023, 24, 2863. https://doi.org/10.3390/ijms24032863
Cantarini M, Rusciano D, Amato R, Canovai A, Cammalleri M, Monte MD, Minnelli C, Laudadio E, Mobbili G, Giorgini G, et al. Structural Basis for Agonistic Activity and Selectivity toward Melatonin Receptors hMT1 and hMT2. International Journal of Molecular Sciences. 2023; 24(3):2863. https://doi.org/10.3390/ijms24032863
Chicago/Turabian StyleCantarini, Mattia, Dario Rusciano, Rosario Amato, Alessio Canovai, Maurizio Cammalleri, Massimo Dal Monte, Cristina Minnelli, Emiliano Laudadio, Giovanna Mobbili, Giorgia Giorgini, and et al. 2023. "Structural Basis for Agonistic Activity and Selectivity toward Melatonin Receptors hMT1 and hMT2" International Journal of Molecular Sciences 24, no. 3: 2863. https://doi.org/10.3390/ijms24032863
APA StyleCantarini, M., Rusciano, D., Amato, R., Canovai, A., Cammalleri, M., Monte, M. D., Minnelli, C., Laudadio, E., Mobbili, G., Giorgini, G., & Galeazzi, R. (2023). Structural Basis for Agonistic Activity and Selectivity toward Melatonin Receptors hMT1 and hMT2. International Journal of Molecular Sciences, 24(3), 2863. https://doi.org/10.3390/ijms24032863













