Differential Urinary Proteomic Analysis of High-Risk Cervical Intraepithelial Neoplasia
Abstract
1. Introduction
2. Results
2.1. Urine Proteomic Analysis Using the MALDI-TOF/MS and ESI-MS Ion Trap
2.2. MALDI-TOF/MS Detection Model Generation
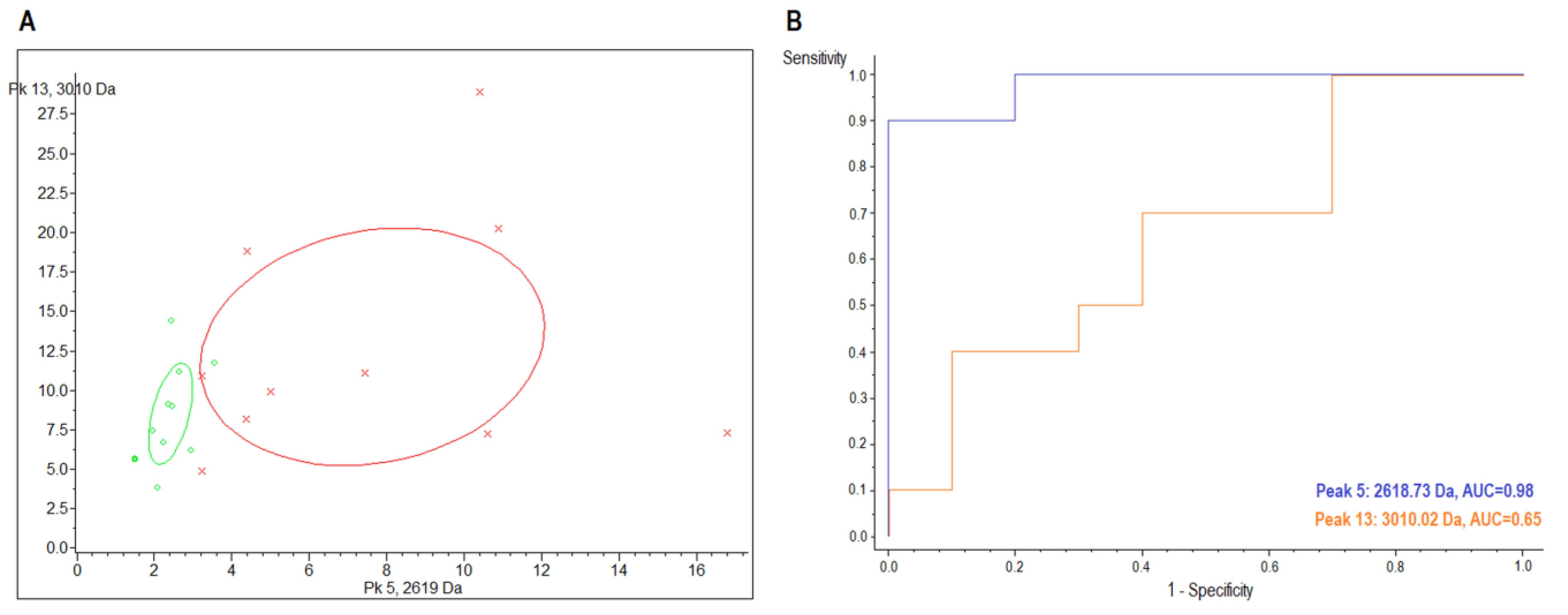
2.3. MALDI-TOF/MS Cluster Analysis
2.4. ESI-MS Analysis and Label-Free Quantitative Comparisons of Differentially Expressed Proteins in the Urine between CIN3 and Healthy Controls
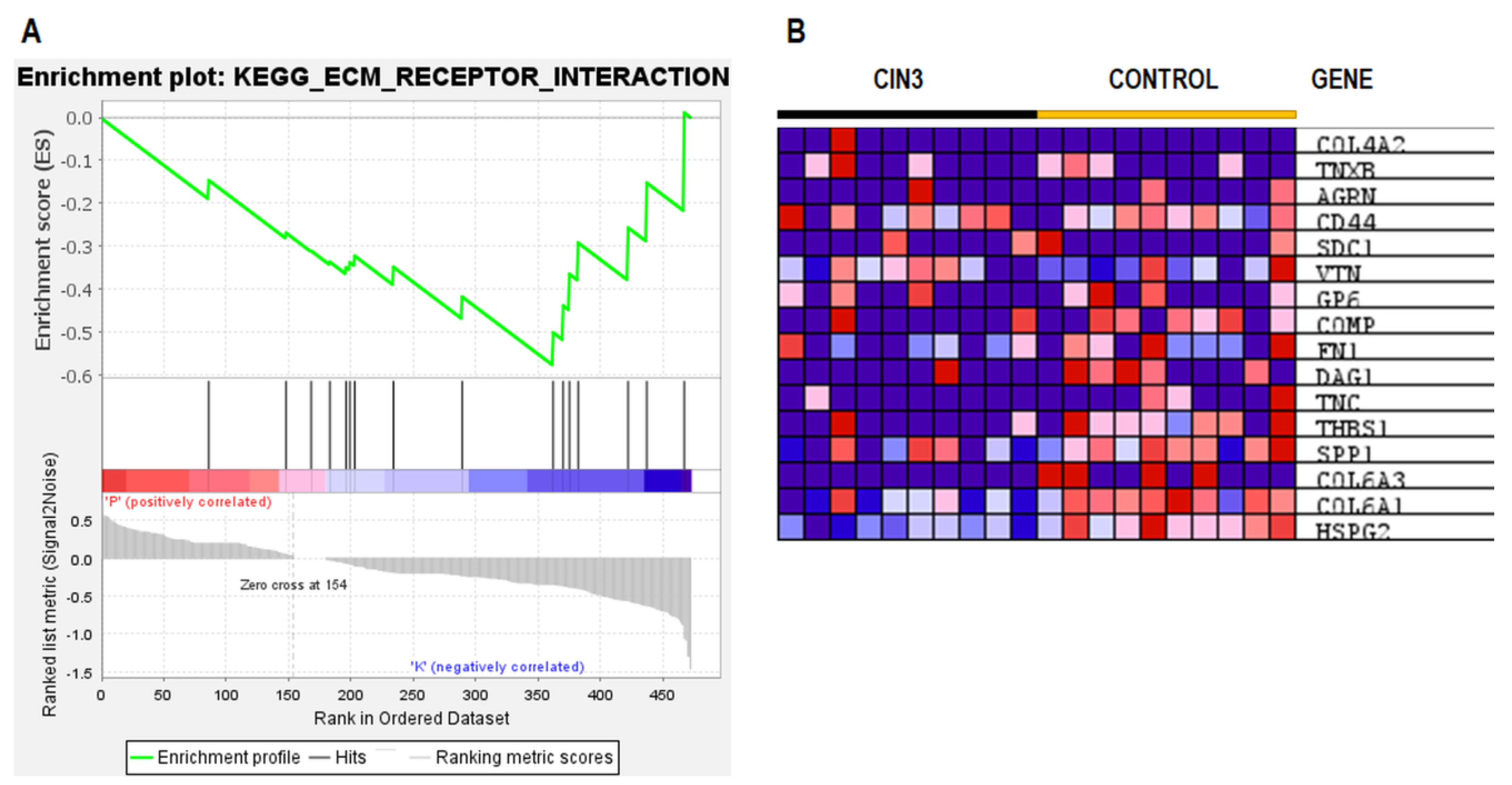
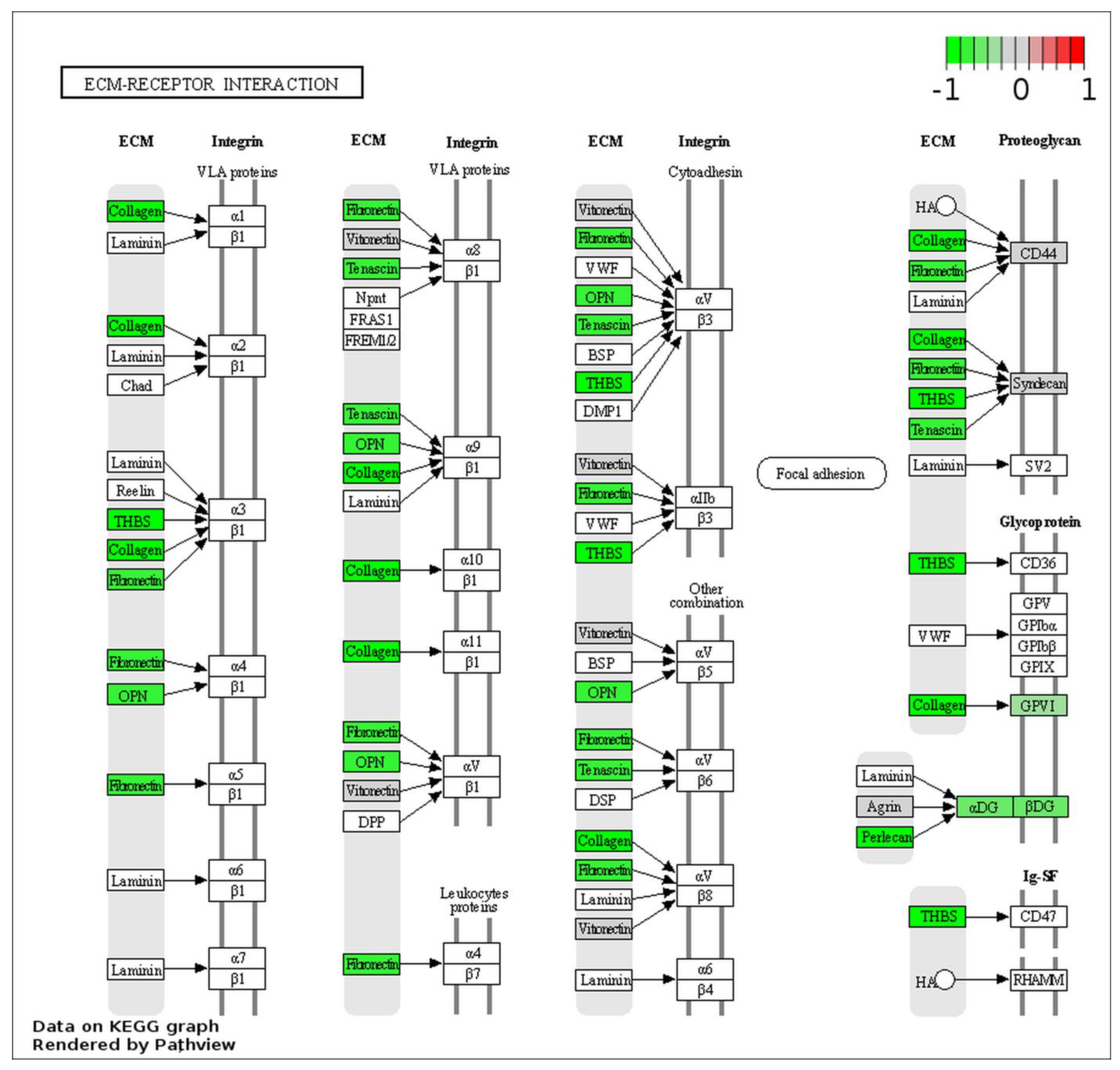
2.5. ESI–MS Analysis and Pathway Enrichment Analysis of the Differential Urine Proteins in CIN3 versus Control Samples
3. Discussion
4. Materials and Methods
4.1. Patient’s Selection
4.2. Urine Sampling
4.3. Sample and Matrix Preparation for MALDI-TOF/MS Analysis
4.4. Sample Processing for LC-MS/MS Analysis
4.5. LC–MS/MS Analysis and Database Search
4.6. Protein Identification and Label-Free Quantification
4.7. Pathway Analysis
4.8. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Bosch, F.X.; Lorincz, A.; Munoz, N.; Meijer, C.J.L.M.; Shah, K.V. The Causal Relation between Human Papillomavirus and Cervical Cancer. J. Clin. Pathol. 2002, 55, 244–265. [Google Scholar] [CrossRef] [PubMed]
- Ferlay, J.; Soerjomataram, I.; Dikshit, R.; Eser, S.; Mathers, C.; Rebelo, M.; Parkin, D.M.; Forman, D.; Bray, F. Cancer Incidence and Mortality Worldwide: Sources, Methods and Major Patterns in GLOBOCAN 2012. Int. J. Cancer 2014, 136, E359–E386. [Google Scholar] [CrossRef] [PubMed]
- Vergara, N.; Balanda, M.; Hidalgo, W.; Martín, H.S.; Aceituno, A.; Roldán, F.; Villalón, T.; Hott, M.; Espinoza, G.; Quiero, A.; et al. Detection and Genotyping of HPV in Urine Samples from Chilean Women Attending Primary Health Care Centers. Med. Microbiol. Immunol. 2017, 207, 95–103. [Google Scholar] [CrossRef] [PubMed]
- Crosbie, E.J.; Einstein, M.H.; Franceschi, S.; Kitchener, H.C. Human Papillomavirus and Cervical Cancer. Lancet 2013, 382, 889–899. [Google Scholar] [CrossRef] [PubMed]
- Clifford, G.M.; Smith, J.S.; Aguado, T.; Franceschi, S. Comparison of HPV Type Distribution in High-Grade Cervical Lesions and Cervical Cancer: A Meta-Analysis. Br. J. Cancer 2003, 89, 101–105. [Google Scholar] [CrossRef]
- de Sanjose, S.; Quint, W.G.V.; Alemany, L.; Geraets, D.T.; Klaustermeier, J.E.; Lloveras, B.; Tous, S.; Felix, A.; Bravo, L.E.; Shin, H.R.; et al. Human Papillomavirus Genotype Attribution in Invasive Cervical Cancer: A Retrospective Cross-Sectional Worldwide Study. Lancet Oncol. 2010, 11, 1048–1056. [Google Scholar] [CrossRef]
- Greig, J.M.; Ellis, C.J. Biological Agents. In Occupational Hygiene; Blackwell Publishing Ltd.: Hoboken, NJ, USA, 2012; pp. 344–359. [Google Scholar]
- Herbster, S.; Paladino, A.; de Freitas, S.; Boccardo, E. Alterations in the Expression and Activity of Extracellular Matrix Components in HPV-Associated Infections and Diseases. Clinics 2018, 73, e551s. [Google Scholar] [CrossRef]
- Elgundi, Z.; Papanicolaou, M.; Major, G.; Cox, T.R.; Melrose, J.; Whitelock, J.M.; Farrugia, B.L. Cancer Metastasis: The Role of the Extracellular Matrix and the Heparan Sulfate Proteoglycan Perlecan. Front. Oncol. 2020, 9, 1482. [Google Scholar] [CrossRef]
- Sampaio, J.; Ferreira, J.; Santos, A.C.; Bicho, M.; Bicho, M.C. The Importance of the Extracellular Matrix in HPV-Associated Diseases. In Cervical Cancer—A Global Public Health Treatise; Rajkumar, R., Ed.; IntechOpen: London, UK, 2021; pp. 111–130. [Google Scholar]
- Senkomago, V.; Des Marais, A.C.; Rahangdale, L.; Vibat, C.R.T.; Erlander, M.G.; Smith, J.S. Comparison of Urine Specimen Collection Times and Testing Fractions for the Detection of High-Risk Human Papillomavirus and High-Grade Cervical Precancer. J. Clin. Virol. 2016, 74, 26–31. [Google Scholar] [CrossRef]
- Sahasrabuddhe, V.V.; Gravitt, P.E.; Dunn, S.T.; Robbins, D.; Brown, D.; Allen, R.A.; Eby, Y.J.; Smith, K.M.; Zuna, R.E.; Zhang, R.R.; et al. Evaluation of Clinical Performance of a Novel Urine-Based {HPV} Detection Assay among Women Attending a Colposcopy Clinic. J. Clin. Virol. 2014, 60, 414–417. [Google Scholar] [CrossRef]
- Pattyn, J.; Van Keer, S.; Téblick, L.; Van Damme, P.; Vorsters, A. HPV DNA Detection in Urine Samples of Women: `an Efficacious and Accurate Alternative to Cervical Samples? Expert Rev. Anti. Infect. Ther. 2019, 17, 755–757. [Google Scholar] [CrossRef]
- Lefeuvre, C.; Pivert, A.; Le Guillou-Guillemette, H.; Lunel-Fabiani, F.; Veillon, P.; Duc-Banaszuk, A.-S.L.; Ducancelle, A. Urinary {HPV} {DNA} Testing as a Tool for Cervical Cancer Screening in Women Who Are Reluctant to Have a Pap Smear in France. J. Infect. 2020, 81, 248–254. [Google Scholar] [CrossRef]
- Asciutto, K.C.; Henningsson, A.J.; Borgfeldt, H.; Darlin, L.; Borgfeldt, C. Vaginal and urine self-sampling compared to cervical sampling for HPV-testing with the Cobas 4800 HPV test. Anticancer Res. 2017, 37, 4183–4187. [Google Scholar] [CrossRef]
- Sargent, A.; Fletcher, S.; Bray, K.; Kitchener, H.C.; Crosbie, E.J. Cross-sectional study of HPV testing in self-sampled urine and comparison with matched vaginal and cervical samples in women attending colposcopy for the management of abnormal cervical screening. BMJ Open 2019, 9, e025388. [Google Scholar] [CrossRef]
- Stanczuk, G.A.; Currie, H.; Baxter, G.; Foster, A.; Gibson, L.; Graham, C.; Cuschieri, K. Cobas 4800 HPV detection in the cervical, vaginal and urine samples of women with high-grade CIN before and after treatment. J. Clin. Pathol. 2015, 68, 567–570. [Google Scholar] [CrossRef]
- Cho, H.-W.; Ouh, Y.-T.; Hong, J.H.; Min, K.-J.; So, K.A.; Kim, T.J.; Paik, E.S.; Lee, J.; Moon, J.H.; Lee, J.K. Comparison of urine, self-collected vaginal swab, and cervical swab samples for detecting human papillomavirus (HPV) with Roche Cobas HPV, Anyplex II HPV, and RealTime HR-S HPV assay. J. Virol. Methods 2019, 269, 77–82. [Google Scholar] [CrossRef]
- Van Keer, S.; Tjalma, W.A.A.; Pattyn, J.; Biesmans, S.; Pieters, Z.; Van Ostade, X.; Ieven, M.; Van Damme, P.; Vorsters, A. Human papillomavirus genotype and viral load agreement between paired first-void urine and clinician-collected cervical samples. Eur. J. Clin. Microbiol. Infect. Dis. 2018, 37, 859–869. [Google Scholar] [CrossRef]
- Bernal, S.; Palomares, J.C.; Artura, A.; Parra, M.; Cabezas, J.L.; Robles, A.; Mazuelos, E.M. Comparison of urine and cervical samples for detecting human papillomavirus (HPV) with the Cobas 4800 HPV test. J. Clin. Virol. 2014, 61, 548–552. [Google Scholar] [CrossRef]
- Cuzick, J.; Cadman, L.; Ahmad, A.S.; Ho, L.; Terry, G.; Kleeman, M.; Lyons, D.; Austin, J.; Stoler, M.H.; Vibat, C.R.T.; et al. Performance and Diagnostic Accuracy of a Urine-Based Human Papillomavirus Assay in a Referral Population. Cancer Epidemiol. Biomark. Prev. 2017, 26, 1053–1059. [Google Scholar] [CrossRef]
- Cox, J.; Hein, M.Y.; Luber, C.A.; Paron, I.; Nagaraj, N.; Mann, M. Accurate Proteome-wide Label-free Quantification by Delayed Normalization and Maximal Peptide Ratio Extraction, Termed MaxLFQ. Mol. Cell. Proteom. 2014, 13, 2513–2526. [Google Scholar] [CrossRef]
- Wong, J.W.; Cagney, G. An overview of label-free quantita tion methods in proteomics by mass spectrometry. Methods Mol. Biol. 2010, 604, 273–283. [Google Scholar] [CrossRef] [PubMed]
- Manou, D.; Caon, I.; Bouris, P.; Triantaphyllidou, I.-E.; Giaroni, C.; Passi, A.; Karamanos, N.K.; Vigetti, D.; Theocharis, A.D. The Complex Interplay Between Extracellular Matrix and Cells in Tissues. Methods Mol. Biol. 2019, 1952, 1–20. [Google Scholar] [CrossRef] [PubMed]
- Vitale, D.L.; Katakam, S.K.; Greve, B.; Jang, B.; Oh, E.; Alaniz, L.; Götte, M. Proteoglycans and glycosaminoglycans as regulators of cancer stem cell function and therapeutic resistance. FEBS J. 2019, 286, 2870–2882. [Google Scholar] [CrossRef] [PubMed]
- Karousou, E.; Misra, S.; Ghatak, S.; Dobra, K.; Götte, M.; Vigetti, D.; Passi, A.; Karamanos, N.K.; Skandalis, S.S. Roles and targeting of the HAS/hyaluronan/CD44 molecular system in cancer. Matrix Biol. 2017, 59, 3–22. [Google Scholar] [CrossRef] [PubMed]
- Schaefer, L.; Dikic, I. Autophagy: Instructions from the extracellular matrix. Matrix Biol. 2021, 100–101, 1–8. [Google Scholar] [CrossRef] [PubMed]
- van der Flier, A.; Sonnenberg, A. Function and interactions of integrins. Cell Tissue Res. 2001, 305, 285–298. [Google Scholar] [CrossRef]
- Bao, Y.; Wang, L.; Shi, L.; Yun, F.; Liu, X.; Chen, Y.; Chen, C.; Ren, Y.; Jia, Y. Transcriptome profiling revealed multiple genes and ECM-receptor interaction pathways that may be associated with breast cancer. Cell. Mol. Biol. Lett. 2019, 24, 38. [Google Scholar] [CrossRef]
- Andersen, M.K.; Rise, K.; Giskeødegård, G.F.; Richardsen, E.; Bertilsson, H.; Størkersen; Bathen, T.F.; Rye, M.; Tessem, M.-B. Integrative metabolic and transcriptomic profiling of prostate cancer tissue containing reactive stroma. Sci. Rep. 2018, 8, 14269. [Google Scholar] [CrossRef]
- Yan, P.; He, Y.; Xie, K.; Kong, S.; Zhao, W. In silico analyses for potential key genes associated with gastric cancer. Peerj 2018, 6, e6092. [Google Scholar] [CrossRef]
- Wu, K.; Yi, Y.; Liu, F.; Wu, W.; Chen, Y.; Zhang, W. Identification of key pathways and genes in the progression of cervical cancer using bioinformatics analysis. Oncol. Lett. 2018, 16, 1003–1009. [Google Scholar] [CrossRef]
- Ramírez-Torres, A.; GIL, J.; Contreras, S.; Ramírez, G.; Valencia-González, H.A.; Salazar-Bustamante, E.; Gómez-Caudillo, L.; García-Carranca, A.; Encarnación-Guevara, S. Quantitative Proteomic Analysis of Cervical Cancer Tissues Identifies Proteins Associated With Cancer Progression. Cancer Genom. Proteom. 2022, 19, 241–258. [Google Scholar] [CrossRef]
- Giroglou, T.; Florin, L.; Schäfer, F.; Streeck, R.E.; Sapp, M. Human Papillomavirus Infection Requires Cell Surface Heparan Sulfate. J. Virol. 2001, 75, 1565–1570. [Google Scholar] [CrossRef]
- Buck, C.; Thompson, C.D.; Roberts, J.N.; Müller, M.; Lowy, D.R.; Schiller, J.T. Carrageenan Is a Potent Inhibitor of Papillomavirus Infection. PLOS Pathog. 2006, 2, e69. [Google Scholar] [CrossRef]
- Winder, S.J. The complexities of dystroglycan. Trends Biochem. Sci. 2001, 26, 118–124. [Google Scholar] [CrossRef]
- Sgambato, A.; Tarquini, E.; Resci, F.; De Paola, B.; Faraglia, B.; Camerini, A.; Rettino, A.; Migaldi, M.; Cittadini, A.; Zannoni, G.F. Aberrant expression of α-dystroglycan in cervical and vulvar cancer. Gynecol. Oncol. 2006, 103, 397–404. [Google Scholar] [CrossRef]
- Zhang, Z.; Zhang, Y.; Ma, Y.; Xiang, S.; Shen, J.; Zhao, Y.; Wu, X.; Li, M.; Yang, X.; Du, F.; et al. Identification of Pathways and Genes Associated with Chemoradiotherapy in Locally Advanced Cervical Cancer. Research Square (preprint), Version 1. Res. Artic. 2021, 1–19. [Google Scholar] [CrossRef]
- Inki, P.; Stenbäck, F.; Grenman, S.; Jalkanen, M. Immunohistochemical localization of syndecan-1 in normal and pathological human uterine cervix. J. Pathol. 1994, 172, 349–355. [Google Scholar] [CrossRef]
- Shinyo, Y.; Kodama, J.; Hasengaowa; Kusumoto, T.; Hiramatsu, Y. Loss of cell-surface heparan sulfate expression in both cervical intraepithelial neoplasm and invasive cervical cancer. Gynecol. Oncol. 2005, 96, 776–783. [Google Scholar] [CrossRef]
- Davidson, B.; Goldberg, I.; Gotlieb, W.H.; Ben-Baruch, G.; Kopolovic, J. CD44 expression in uterine cervical intraepithelial neoplasia and squamous cell carcinoma: An immunohistochemical study. Eur. J. Gynaecol. Oncol. 1998, 19, 46–49. [Google Scholar]
- Rey, O.; Lee, S.; Park, N.-H. Human Papillomavirus Type 16 E7 Oncoprotein Represses Transcription of Human Fibronectin. J. Virol. 2000, 74, 4912–4918. [Google Scholar] [CrossRef]
- Toussaint-Smith, E.; Donner, D.B.; Roman, A. Expression of human papillomavirus type 16 E6 and E7 oncoproteins in primary foreskin keratinocytes is sufficient to alter the expression of angiogenic factors. Oncogene 2004, 23, 2988–2995. [Google Scholar] [CrossRef] [PubMed]
- Fermin, D.; Basrur, V.; Yocum, A.K.; Nesvizhskii, A.I. Abacus: A computational tool for extracting and pre-processing spectral count data for label-free quantitative proteomic analysis. Proteomics 2011, 11, 1340–1345. [Google Scholar] [CrossRef] [PubMed]
- Pham, T.V.; Piersma, S.R.; Warmoes, M.; Jimenez, C.R. On the beta-binomial model for analysis of spectral count data in label-free tandem mass spectrometry-based proteomics. Bioinformatics 2009, 26, 363–369. [Google Scholar] [CrossRef] [PubMed]


| Samples | Model | Generated Peaks | Cross-Validation (%) | Recognition Capability (%) |
|---|---|---|---|---|
| urine | GA | 10 | 51.56 | 95.00 |
| SNN | 15 | 51.02 | 90.00 | |
| QC | 3 | 62.66 | 85.00 |
| Sample | Index Peak | Mass (m/z) | Dave 1 | PTTA 2 | PWKW 3 | PAD 4 | Control (avg 5 ± SD 6) | CIN3 (avg ± SD) | AUC |
|---|---|---|---|---|---|---|---|---|---|
| urine | 5 | 2618.73 | 5.79 | 0.289 | 0.0026 | 0.000219 | 8.48 ± 4.94 | 2.69 ± 0.63 | 0.98 |
| 13 | 3010.02 | 4.71 | 0.59 | 0.672 | 0.00872 | 14.17 ± 8.36 | 9.46 ± 3.55 | 0.65 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Bober, P.; Tkáčiková, S.; Talian, I.; Urdzík, P.; Toporcerová, S.; Sabo, J. Differential Urinary Proteomic Analysis of High-Risk Cervical Intraepithelial Neoplasia. Int. J. Mol. Sci. 2023, 24, 2531. https://doi.org/10.3390/ijms24032531
Bober P, Tkáčiková S, Talian I, Urdzík P, Toporcerová S, Sabo J. Differential Urinary Proteomic Analysis of High-Risk Cervical Intraepithelial Neoplasia. International Journal of Molecular Sciences. 2023; 24(3):2531. https://doi.org/10.3390/ijms24032531
Chicago/Turabian StyleBober, Peter, Soňa Tkáčiková, Ivan Talian, Peter Urdzík, Silvia Toporcerová, and Ján Sabo. 2023. "Differential Urinary Proteomic Analysis of High-Risk Cervical Intraepithelial Neoplasia" International Journal of Molecular Sciences 24, no. 3: 2531. https://doi.org/10.3390/ijms24032531
APA StyleBober, P., Tkáčiková, S., Talian, I., Urdzík, P., Toporcerová, S., & Sabo, J. (2023). Differential Urinary Proteomic Analysis of High-Risk Cervical Intraepithelial Neoplasia. International Journal of Molecular Sciences, 24(3), 2531. https://doi.org/10.3390/ijms24032531






