Microbiota-Associated HAF-EVs Regulate Monocytes by Triggering or Inhibiting Inflammasome Activation
Abstract
1. Introduction
2. Results and Discussion
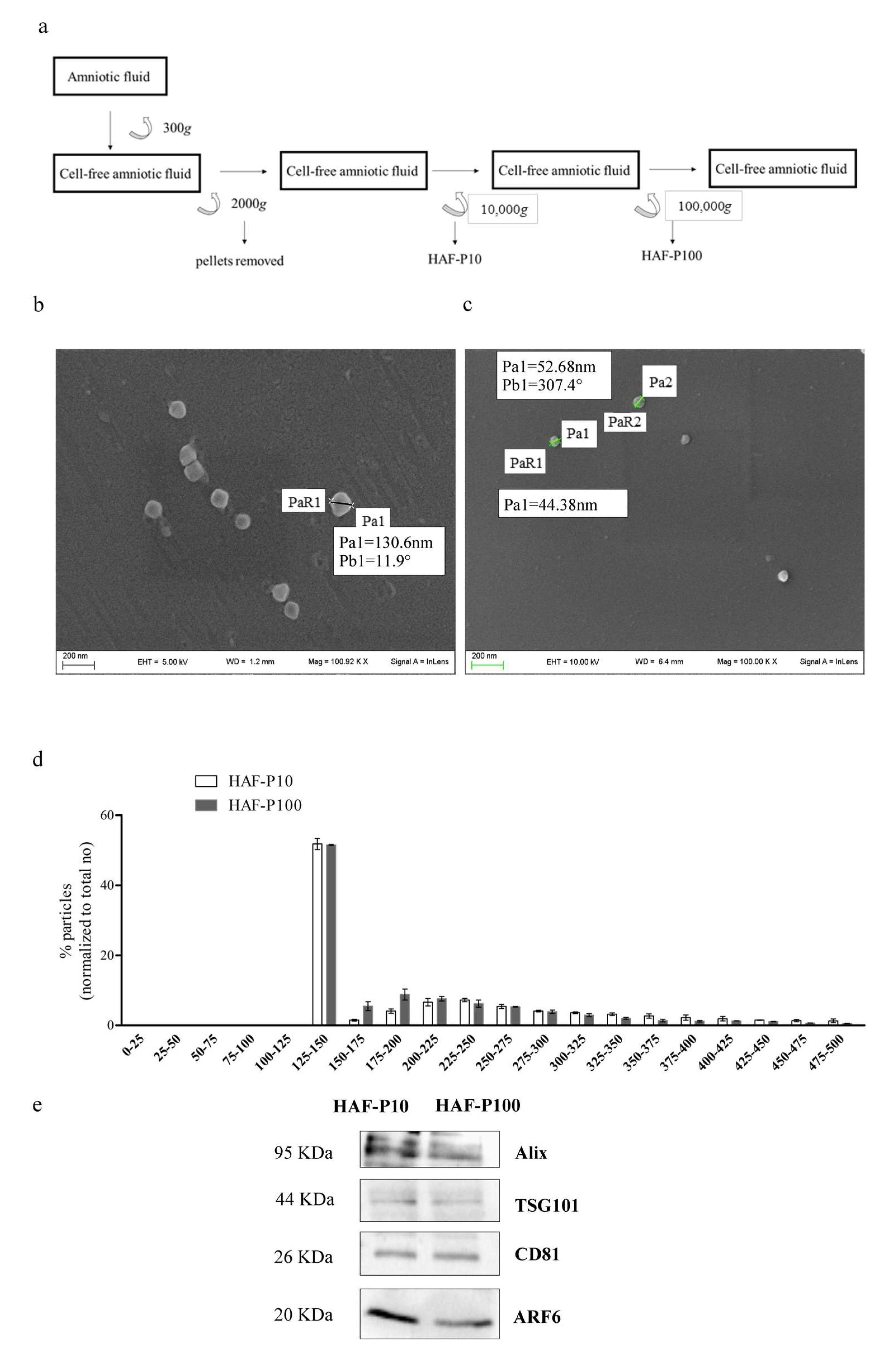
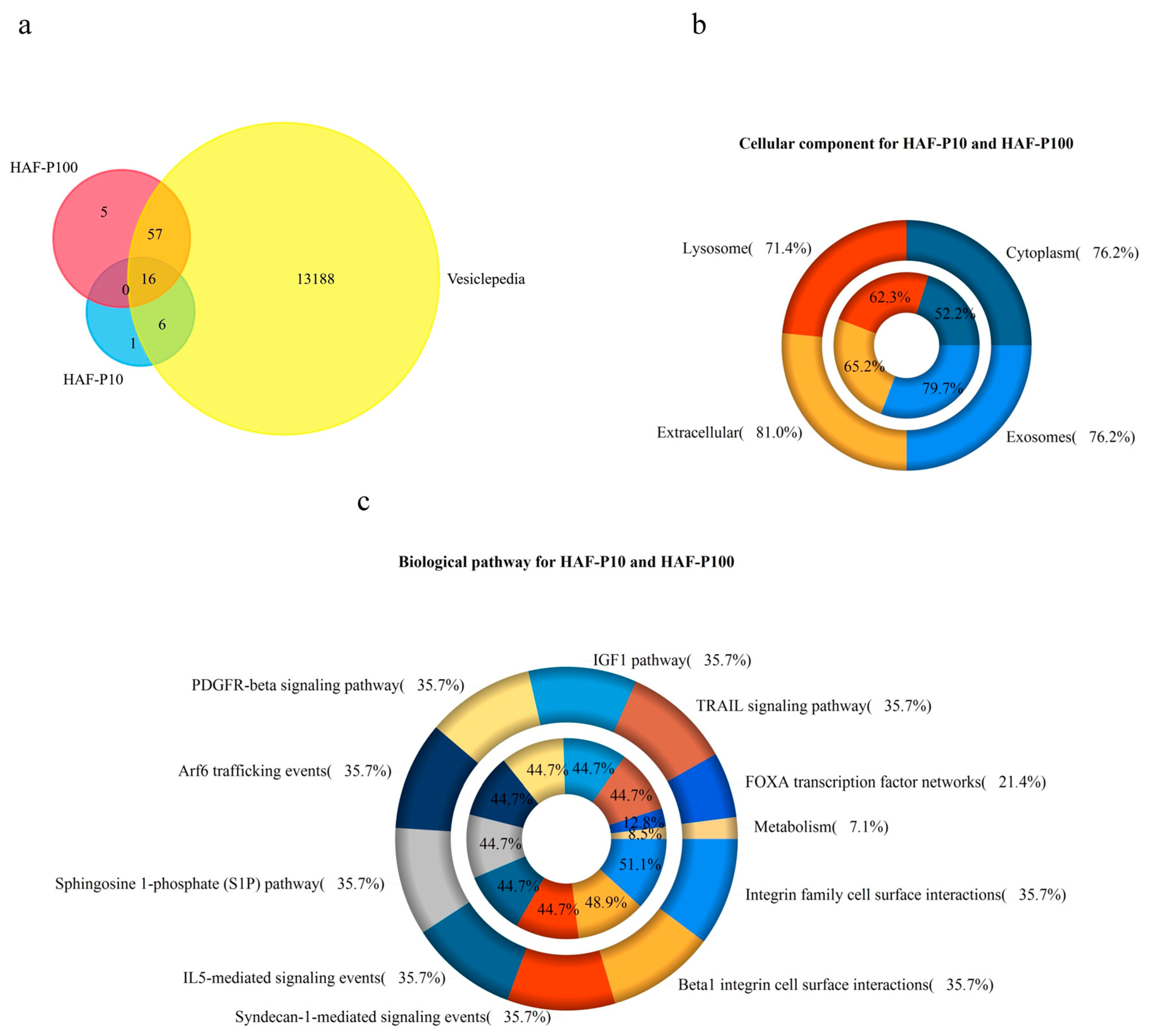
2.1. Characterization of HAF-EVs
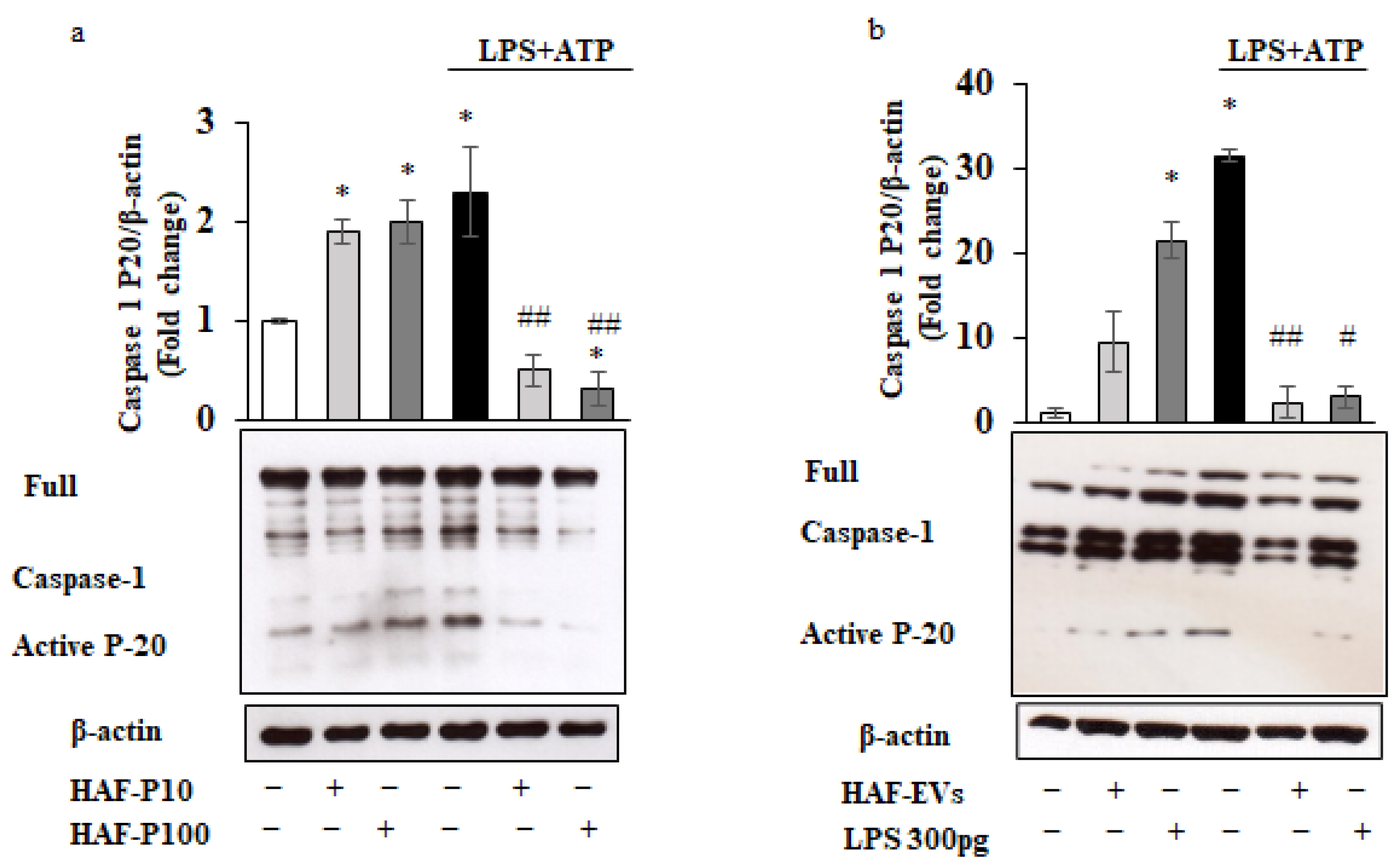
2.2. Bimodal Regulation of the Inflammasome by HAF-EVs in THP-1 Cells
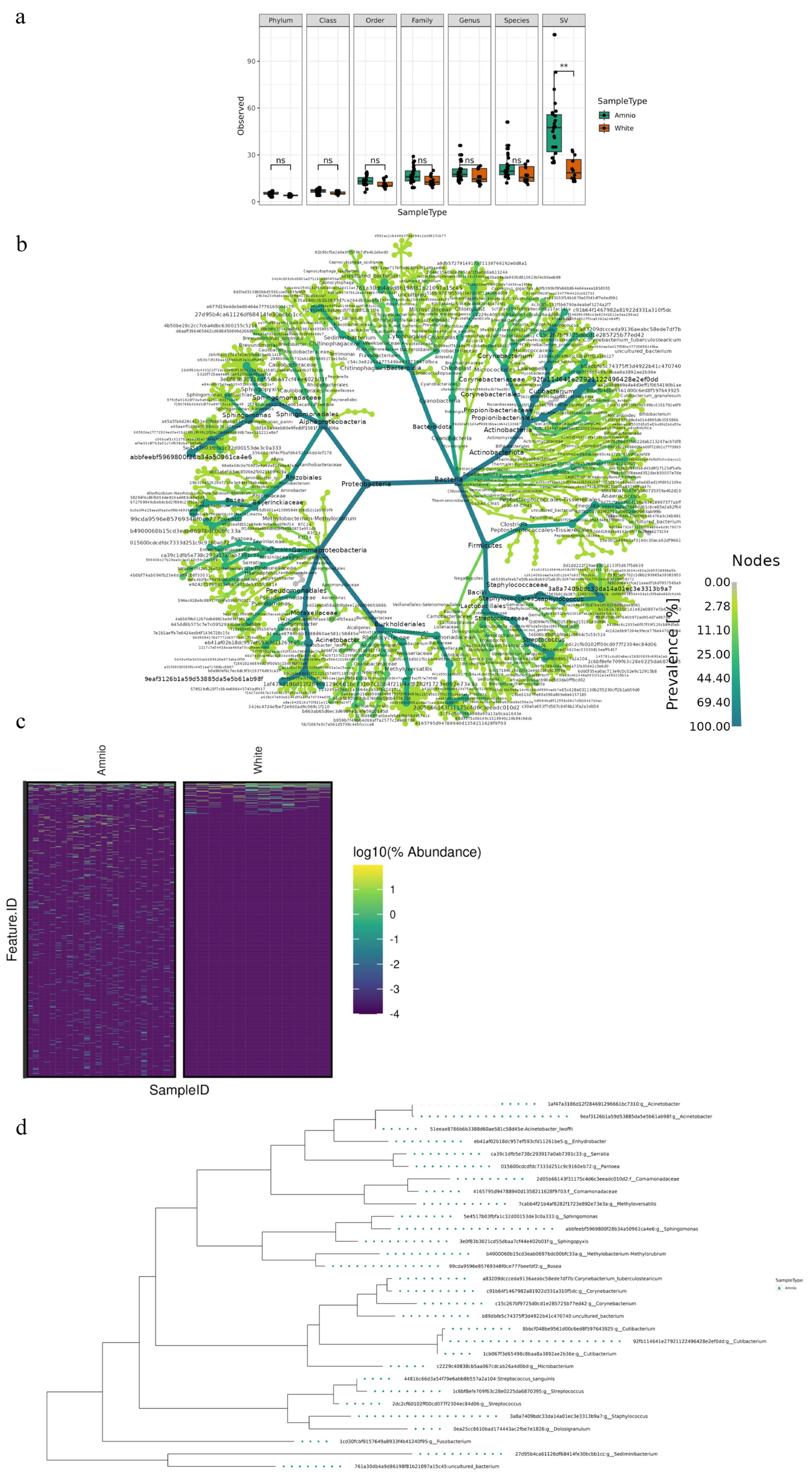
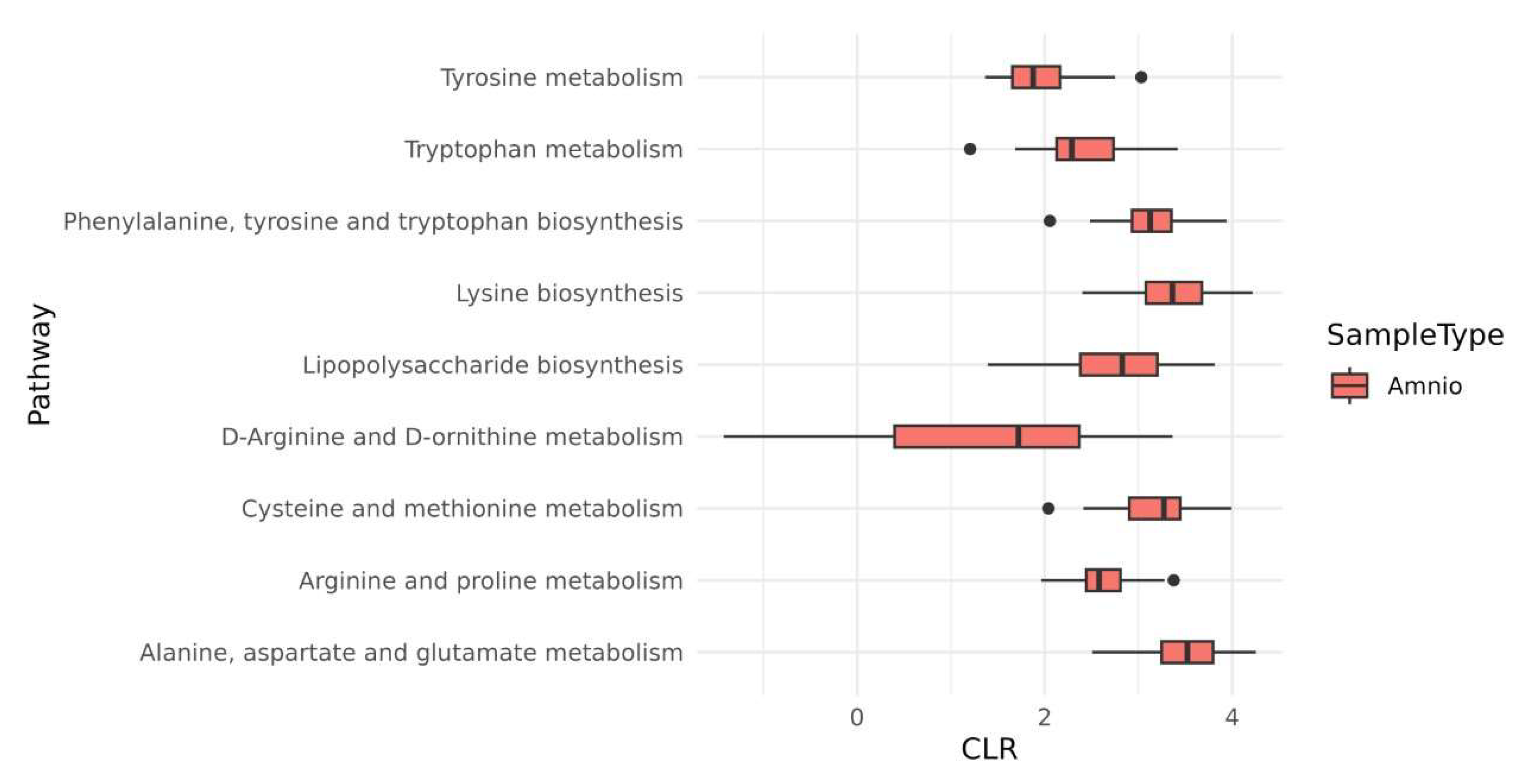
2.3. Microbiota Species Variants in HAF-EVs
3. Materials and Methods
3.1. EVs Derived from HAF Isolation
3.2. Nanoparticle Tracking Analysis (NTA)
3.3. Scanning Electron Mycroscopy (SEM) Analysis
3.4. Protein Content Analysis
3.4.1. SDS-PAGE and in-Gel Digestion
3.4.2. LC-MS/MS Analysis and Database Searching
3.4.3. Western Blot Analysis
3.4.4. THP-1 Cell Culture, Human Monocyte Purification and Treatments
3.5. Endotoxin Assay
3.6. Microbiome Analysis in Human Amniotic Fluid
3.7. Bioinformatics and Statistical Analyses
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Mao, Y.; Pierce, J.; Singh-Varma, A.; Boyer, M.; Kohn, J.; Reems, J.-A. Processed Human Amniotic Fluid Retains Its Antibacterial Activity. J. Transl. Med. 2019, 17, 68. [Google Scholar] [CrossRef] [PubMed]
- Underwood, M.A.; Gilbert, W.M.; Sherman, M.P. Amniotic Fluid: Not Just Fetal Urine Anymore. J. Perinatol. 2005, 25, 341–348. [Google Scholar] [CrossRef]
- Pierce, J.; Jacobson, P.; Benedetti, E.; Peterson, E.; Phibbs, J.; Preslar, A.; Reems, J.-A. Collection and Characterization of Amniotic Fluid from Scheduled C-Section Deliveries. Cell Tissue Bank. 2016, 17, 413–425. [Google Scholar] [CrossRef] [PubMed]
- Kim, Y.; Lee, K.-Y.; Lee, J.J.; Tak, H.; Park, S.-T.; Song, J.-E.; Son, G.-H. Expression of Antimicrobial Peptides in the Amniotic Fluid of Women with Cervical Insufficiency. Am. J. Reprod. Immunol. 2022, 88, e13577. [Google Scholar] [CrossRef] [PubMed]
- Pianta, S.; Bonassi Signoroni, P.; Muradore, I.; Rodrigues, M.F.; Rossi, D.; Silini, A.; Parolini, O. Amniotic membrane mesenchymal cells-derived factors skew T cell polarization toward Treg and downregulate Th1 and Th17 cells subsets. Stem Cell Rev. Rep. 2015, 11, 394–407. [Google Scholar] [CrossRef]
- Jankovičová, J.; Sečová, P.; Michalková, K.; Antalíková, J. Tetraspanins, more than Markers of Extracellular Vesicles in Reproduction. Int. J. Mol. Sci. 2020, 21, 7568. [Google Scholar] [CrossRef]
- Bebelman, M.P.; Smit, M.J.; Pegtel, D.M.; Baglio, S.R. Biogenesis and Function of Extracellular Vesicles in Cancer. Pharmacol. Ther. 2018, 188, 1–11. [Google Scholar] [CrossRef]
- Harrell, C.R.; Jovicic, N.; Djonov, V.; Arsenijevic, N.; Volarevic, V. Mesenchymal Stem Cell-Derived Exosomes and Other Extracellular Vesicles as New Remedies in the Therapy of Inflammatory Diseases. Cells 2019, 8, 1605. [Google Scholar] [CrossRef]
- Keshtkar, S.; Azarpira, N.; Ghahremani, M.H. Mesenchymal Stem Cell-Derived Extracellular Vesicles: Novel Frontiers in Regenerative Medicine. Stem Cell Res. Ther. 2018, 9, 63. [Google Scholar] [CrossRef]
- Todorova, D.; Simoncini, S.; Lacroix, R.; Sabatier, F.; Dignat-George, F. Extracellular Vesicles in Angiogenesis. Circ. Res. 2017, 120, 1658–1673. [Google Scholar] [CrossRef]
- Veerman, R.E.; Akpinar, G.G.; Eldh, M.; Gabrielsson, S. Immune Cell-Derived Extracellular Vesicles–Functions and Therapeutic Applications. Trends Mol. Med. 2019, 25, 382–394. [Google Scholar] [CrossRef] [PubMed]
- Gebara, N.; Correia, Y.; Wang, K.; Bussolati, B. Angiogenic Properties of Placenta-Derived Extracellular Vesicles in Normal Pregnancy and in Preeclampsia. Int. J. Mol. Sci. 2021, 22, 5402. [Google Scholar] [CrossRef] [PubMed]
- Cargnoni, A.; Papait, A.; Masserdotti, A.; Pasotti, A.; Stefani, F.R.; Silini, A.R.; Parolini, O. Extracellular Vesicles From Perinatal Cells for Anti-Inflammatory Therapy. Front. Bioeng. Biotechnol. 2021, 9, 637737. [Google Scholar] [CrossRef] [PubMed]
- Magatti, M.; De Munari, S.; Vertua, E.; Gibelli, L.; Wengler, G.S.; Parolini, O. Human Amnion Mesenchyme Harbors Cells with Allogeneic T-Cell Suppression and Stimulation Capabilities. Stem Cells 2008, 26, 182–192. [Google Scholar] [CrossRef]
- Pianta, S.; Magatti, M.; Vertua, E.; Bonassi Signoroni, P.; Muradore, I.; Nuzzo, A.M.; Rolfo, A.; Silini, A.; Quaglia, F.; Todros, T.; et al. Amniotic Mesenchymal Cells from Pre-Eclamptic Placentae Maintain Immunomodulatory Features as Healthy Controls. J. Cell. Mol. Med. 2016, 20, 157–169. [Google Scholar] [CrossRef]
- Qiu, C.; Ge, Z.; Cui, W.; Yu, L.; Li, J. Human Amniotic Epithelial Stem Cells: A Promising Seed Cell for Clinical Applications. Int. J. Mol. Sci. 2020, 21, 7730. [Google Scholar] [CrossRef]
- Nair, S.; Salomon, C. Extracellular Vesicles and Their Immunomodulatory Functions in Pregnancy. Semin. Immunopathol. 2018, 40, 425–437. [Google Scholar] [CrossRef]
- Romani, R.; Pirisinu, I.; Calvitti, M.; Pallotta, M.T.; Gargaro, M.; Bistoni, G.; Vacca, C.; Di Michele, A.; Orabona, C.; Rosati, J.; et al. Stem Cells from Human Amniotic Fluid Exert Immunoregulatory Function via Secreted Indoleamine 2, 3-dioxygenase1. J. Cell. Mol. Med. 2015, 19, 1593–1605. [Google Scholar] [CrossRef]
- Mezzasoma, L.; Bellezza, I.; Orvietani, P.; Manni, G.; Gargaro, M.; Sagini, K.; Llorente, A.; Scarpelli, P.; Pascucci, L.; Cellini, B.; et al. Amniotic Fluid Stem Cell-Derived Extracellular Vesicles Are Independent Metabolic Units Capable of Modulating Inflammasome Activation in THP-1 Cells. FASEB J. 2022, 36, e22218. [Google Scholar] [CrossRef]
- Del Rivero, T.; Milberg, J.; Bennett, C.; Mitrani, M.I.; Bellio, M.A. Human Amniotic Fluid Derived Extracellular Vesicles Attenuate T Cell Immune Response. Front. Immunol. 2022, 13, 977809. [Google Scholar] [CrossRef]
- Mitrani, M.I.; Bellio, M.A.; Sagel, A.; Saylor, M.; Kapp, W.; VanOsdol, K.; Haskell, G.; Stewart, D.; Abdullah, Z.; Santos, I.; et al. Case Report: Administration of Amniotic Fluid-Derived Nanoparticles in Three Severely Ill COVID-19 Patients. Front. Med. 2021, 8, 583842. [Google Scholar] [CrossRef] [PubMed]
- Bellio, M.A.; Bennett, C.; Arango, A.; Khan, A.; Xu, X.; Barrera, C.; Friedewald, V.; Mitrani, M.I. Proof-of-Concept Trial of an Amniotic Fluid-Derived Extracellular Vesicle Biologic for Treating High Risk Patients with Mild-to-Moderate Acute COVID-19 Infection. Biomater. Biosyst. 2021, 4, 100031. [Google Scholar] [CrossRef]
- Panaitescu, B.; Romero, R.; Gomez-Lopez, N.; Xu, Y.; Leng, Y.; Maymon, E.; Pacora, P.; Erez, O.; Yeo, L.; Hassan, S.S.; et al. In Vivo Evidence of Inflammasome Activation during Spontaneous Labor at Term. J. Matern. Fetal Neonatal Med. 2019, 32, 1978–1991. [Google Scholar] [CrossRef]
- Martinon, F.; Tschopp, J. Inflammatory Caspases: Linking an Intracellular Innate Immune System to Autoinflammatory Diseases. Cell 2004, 117, 561–574. [Google Scholar] [CrossRef] [PubMed]
- Kelley, N.; Jeltema, D.; Duan, Y.; He, Y. The NLRP3 Inflammasome: An Overview of Mechanisms of Activation and Regulation. Int. J. Mol. Sci. 2019, 20, 3328. [Google Scholar] [CrossRef] [PubMed]
- Mezzasoma, L.; Antognelli, C.; Talesa, V.N. Atrial Natriuretic Peptide Down-Regulates LPS/ATP-Mediated IL-1β Release by Inhibiting NF-KB, NLRP3 Inflammasome and Caspase-1 Activation in THP-1 Cells. Immunol. Res. 2016, 64, 303–312. [Google Scholar] [CrossRef] [PubMed]
- Gurung, P.; Li, B.; Subbarao Malireddi, R.K.; Lamkanfi, M.; Geiger, T.L.; Kanneganti, T.-D. Chronic TLR Stimulation Controls NLRP3 Inflammasome Activation through IL-10 Mediated Regulation of NLRP3 Expression and Caspase-8 Activation. Sci. Rep. 2015, 5, 14488. [Google Scholar] [CrossRef] [PubMed]
- Mosser, D.M.; Edwards, J.P. Exploring the Full Spectrum of Macrophage Activation. Nat. Rev. Immunol. 2008, 8, 958–969. [Google Scholar] [CrossRef] [PubMed]
- Hotchkiss, R.S.; Monneret, G.; Payen, D. Sepsis-Induced Immunosuppression: From Cellular Dysfunctions to Immunotherapy. Nat. Rev. Immunol. 2013, 13, 862–874. [Google Scholar] [CrossRef]
- Foster, S.L.; Hargreaves, D.C.; Medzhitov, R. Gene-Specific Control of Inflammation by TLR-Induced Chromatin Modifications. Nature 2007, 447, 972–978. [Google Scholar] [CrossRef]
- Netea, M.G.; Domínguez-Andrés, J.; Barreiro, L.B.; Chavakis, T.; Divangahi, M.; Fuchs, E.; Joosten, L.A.B.; van der Meer, J.W.M.; Mhlanga, M.M.; Mulder, W.J.M.; et al. Defining Trained Immunity and Its Role in Health and Disease. Nat. Rev. Immunol. 2020, 20, 375–388. [Google Scholar] [CrossRef]
- Staude, B.; Oehmke, F.; Lauer, T.; Behnke, J.; Göpel, W.; Schloter, M.; Schulz, H.; Krauss-Etschmann, S.; Ehrhardt, H. The Microbiome and Preterm Birth: A Change in Paradigm with Profound Implications for Pathophysiologic Concepts and Novel Therapeutic Strategies. Biomed. Res. Int. 2018, 2018, 7218187. [Google Scholar] [CrossRef]
- Mishra, A.; Lai, G.C.; Yao, L.J.; Aung, T.T.; Shental, N.; Rotter-Maskowitz, A.; Shepherdson, E.; Singh, G.S.N.; Pai, R.; Shanti, A.; et al. Microbial Exposure during Early Human Development Primes Fetal Immune Cells. Cell 2021, 184, 3394–3409.e20. [Google Scholar] [CrossRef] [PubMed]
- Lechowicz, U.; Rudzinski, S.; Jezela-Stanek, A.; Janciauskiene, S.; Chorostowska-Wynimko, J. Post-Translational Modifications of Circulating Alpha-1-Antitrypsin Protein. Int. J. Mol. Sci. 2020, 21, 9187. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Bartlett, J.A.; Di, M.E.; Bomberger, J.M.; Chan, Y.R.; Gakhar, L.; Mallampalli, R.K.; McCray, P.B.; Di, Y.P. SPLUNC1/BPIFA1 Contributes to Pulmonary Host Defense against Klebsiella Pneumoniae Respiratory Infection. Am. J. Pathol. 2013, 182, 1519–1531. [Google Scholar] [CrossRef] [PubMed]
- Sayeed, S.; Nistico, L.; St Croix, C.; Di, Y.P. Multifunctional Role of Human SPLUNC1 in Pseudomonas Aeruginosa Infection. Infect. Immun. 2013, 81, 285–291. [Google Scholar] [CrossRef]
- Walton, W.G.; Ahmad, S.; Little, M.S.; Kim, C.S.K.; Tyrrell, J.; Lin, Q.; Di, Y.P.; Tarran, R.; Redinbo, M.R. Structural Features Essential to the Antimicrobial Functions of Human SPLUNC1. Biochemistry 2016, 55, 2979–2991. [Google Scholar] [CrossRef] [PubMed]
- Mezzasoma, L.; Costanzi, E.; Scarpelli, P.; Talesa, V.N.; Bellezza, I. Extracellular Vesicles from Human Advanced-Stage Prostate Cancer Cells Modify the Inflammatory Response of Microenvironment-Residing Cells. Cancers 2019, 11, 1276. [Google Scholar] [CrossRef]
- Bohannon, J.K.; Hernandez, A.; Enkhbaatar, P.; Adams, W.L.; Sherwood, E.R. The Immunobiology of Toll-like Receptor 4 Agonists: From Endotoxin Tolerance to Immunoadjuvants. Shock 2013, 40, 451–462. [Google Scholar] [CrossRef] [PubMed]
- O’Carroll, C.; Fagan, A.; Shanahan, F.; Carmody, R.J. Identification of a Unique Hybrid Macrophage-Polarization State Following Recovery from Lipopolysaccharide Tolerance. J. Immunol. 2014, 192, 427–436. [Google Scholar] [CrossRef]
- Zheng, D.; Liwinski, T.; Elinav, E. Interaction between Microbiota and Immunity in Health and Disease. Cell Res. 2020, 30, 492–506. [Google Scholar] [CrossRef] [PubMed]
- Jiang, L.; Zhang, S.; Hu, H.; Yang, J.; Wang, X.; Ma, Y.; Jiang, J.; Wang, J.; Zhong, L.; Chen, M. Exosomes Derived from Human Umbilical Cord Mesenchymal Stem Cells Alleviate Acute Liver Failure by Reducing the Activity of the NLRP3 Inflammasome in Macrophages. Biochem. Biophys. Res. Commun. 2019, 508, 735–741. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Lou, G.; Li, A.; Zhang, T.; Qi, J.; Ye, D.; Zheng, M.; Chen, Z. AMSC-Derived Exosomes Alleviate Lipopolysaccharide/d-Galactosamine-Induced Acute Liver Failure by MiR-17-Mediated Reduction of TXNIP/NLRP3 Inflammasome Activation in Macrophages. EBioMedicine 2018, 36, 140–150. [Google Scholar] [CrossRef]
- Zeng, Q.; Zhou, Y.; Liang, D.; He, H.; Liu, X.; Zhu, R.; Zhang, M.; Luo, X.; Wang, Y.; Huang, G. Exosomes Secreted from Bone Marrow Mesenchymal Stem Cells Attenuate Oxygen-Glucose Deprivation/Reoxygenation-Induced Pyroptosis in PC12 Cells by Promoting AMPK-Dependent Autophagic Flux. Front. Cell. Neurosci. 2020, 14, 182. [Google Scholar] [CrossRef]
- Zhang, W.; Wang, Y.; Kong, Y. Exosomes Derived from Mesenchymal Stem Cells Modulate MiR-126 to Ameliorate Hyperglycemia-Induced Retinal Inflammation via Targeting HMGB1. Investig. Ophthalmol. Vis. Sci. 2019, 60, 294–303. [Google Scholar] [CrossRef]
- Soundara Rajan, T.; Giacoppo, S.; Diomede, F.; Bramanti, P.; Trubiani, O.; Mazzon, E. Human Periodontal Ligament Stem Cells Secretome from Multiple Sclerosis Patients Suppresses NALP3 Inflammasome Activation in Experimental Autoimmune Encephalomyelitis. Int. J. Immunopathol. Pharmacol. 2017, 30, 238–252. [Google Scholar] [CrossRef]
- Zhang, S.; Teo, K.Y.W.; Chuah, S.J.; Lai, R.C.; Lim, S.K.; Toh, W.S. MSC Exosomes Alleviate Temporomandibular Joint Osteoarthritis by Attenuating Inflammation and Restoring Matrix Homeostasis. Biomaterials 2019, 200, 35–47. [Google Scholar] [CrossRef] [PubMed]
- Bano, R.; Ahmad, F.; Mohsin, M. A Perspective on the Isolation and Characterization of Extracellular Vesicles from Different Biofluids. RSC Adv. 2021, 11, 19598–19615. [Google Scholar] [CrossRef] [PubMed]
- West, M.A.; Heagy, W. Endotoxin Tolerance: A Review. Crit. Care Med. 2002, 30, S64–S73. [Google Scholar] [CrossRef] [PubMed]
- Löb, S.; Amann, N.; Kuhn, C.; Schmoeckel, E.; Wöckel, A.; Zati Zehni, A.; Kaltofen, T.; Keckstein, S.; Mumm, J.-N.; Meister, S.; et al. Interleukin-1 Beta Is Significantly Upregulated in the Decidua of Spontaneous and Recurrent Miscarriage Placentas. J. Reprod. Immunol. 2021, 144, 103283. [Google Scholar] [CrossRef] [PubMed]
- Pena, O.M.; Pistolic, J.; Raj, D.; Fjell, C.D.; Hancock, R.E.W. Endotoxin Tolerance Represents a Distinctive State of Alternative Polarization (M2) in Human Mononuclear Cells. J. Immunol. 2011, 186, 7243–7254. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.; Yang, G.X.; Hu, Y.; Lam, P.; Sangha, K.; Siciliano, D.; Swenerton, A.; Miller, R.; Tilley, P.; Von Dadelszen, P.; et al. Comprehensive Human Amniotic Fluid Metagenomics Supports the Sterile Womb Hypothesis. Sci. Rep. 2022, 12, 6875. [Google Scholar] [CrossRef] [PubMed]
- Winters, A.D.; Romero, R.; Greenberg, J.M.; Galaz, J.; Shaffer, Z.D.; Garcia-Flores, V.; Kracht, D.J.; Gomez-Lopez, N.; Theis, K.R. Does the Amniotic Fluid of Mice Contain a Viable Microbiota? Front. Immunol. 2022, 13, 820366. [Google Scholar] [CrossRef] [PubMed]
- Aagaard, K.; Ma, J.; Antony, K.M.; Ganu, R.; Petrosino, J.; Versalovic, J. The Placenta Harbors a Unique Microbiome. Sci. Transl. Med. 2014, 6, 237ra65. [Google Scholar] [CrossRef] [PubMed]
- Cao, B.; Stout, M.J.; Lee, I.; Mysorekar, I.U. Placental Microbiome and Its Role in Preterm Birth. Neoreviews 2014, 15, e537–e545. [Google Scholar] [CrossRef] [PubMed]
- McGovern, N.; Shin, A.; Low, G.; Low, D.; Duan, K.; Yao, L.J.; Msallam, R.; Low, I.; Shadan, N.B.; Sumatoh, H.R.; et al. Human Fetal Dendritic Cells Promote Prenatal T-Cell Immune Suppression through Arginase-2. Nature 2017, 546, 662–666. [Google Scholar] [CrossRef]
- Rackaityte, E.; Halkias, J.; Fukui, E.M.; Mendoza, V.F.; Hayzelden, C.; Crawford, E.D.; Fujimura, K.E.; Burt, T.D.; Lynch, S.V. Viable Bacterial Colonization Is Highly Limited in the Human Intestine in Utero. Nat. Med. 2020, 26, 599–607. [Google Scholar] [CrossRef]
- Schreurs, R.R.C.E.; Baumdick, M.E.; Sagebiel, A.F.; Kaufmann, M.; Mokry, M.; Klarenbeek, P.L.; Schaltenberg, N.; Steinert, F.L.; van Rijn, J.M.; Drewniak, A.; et al. Human Fetal TNF-α-Cytokine-Producing CD4+ Effector Memory T Cells Promote Intestinal Development and Mediate Inflammation Early in Life. Immunity 2019, 50, 462–476.e8. [Google Scholar] [CrossRef]
- Rackaityte, E.; Halkias, J. Mechanisms of Fetal T Cell Tolerance and Immune Regulation. Front. Immunol. 2020, 11, 588. [Google Scholar] [CrossRef]
- Stras, S.F.; Werner, L.; Toothaker, J.M.; Olaloye, O.O.; Oldham, A.L.; McCourt, C.C.; Lee, Y.N.; Rechavi, E.; Shouval, D.S.; Konnikova, L. Maturation of the Human Intestinal Immune System Occurs Early in Fetal Development. Dev. Cell 2019, 51, 357–373.e5. [Google Scholar] [CrossRef]
- Théry, C.; Zitvogel, L.; Amigorena, S. Exosomes: Composition, Biogenesis and Function. Nat. Rev. Immunol. 2002, 2, 569–579. [Google Scholar] [CrossRef]
- Chiasserini, D.; Davidescu, M.; Orvietani, P.L.; Susta, F.; Macchioni, L.; Petricciuolo, M.; Castigli, E.; Roberti, R.; Binaglia, L.; Corazzi, L. 3-Bromopyruvate Treatment Induces Alterations of Metabolic and Stress-Related Pathways in Glioblastoma Cells. J. Proteom. 2017, 152, 329–338. [Google Scholar] [CrossRef] [PubMed]
- Wingett, S.W.; Andrews, S. FastQ Screen: A Tool for Multi-Genome Mapping and Quality Control. F1000 Res. 2018, 7, 1338. [Google Scholar] [CrossRef]
- Bolyen, E.; Rideout, J.R.; Dillon, M.R.; Bokulich, N.A.; Abnet, C.C.; Al-Ghalith, G.A.; Alexander, H.; Alm, E.J.; Arumugam, M.; Asnicar, F.; et al. Reproducible, Interactive, Scalable and Extensible Microbiome Data Science Using QIIME 2. Nat. Biotechnol. 2019, 37, 852–857. [Google Scholar] [CrossRef] [PubMed]
- Felicetti, L.; Femminella, M.; Reali, G.; Gresele, P.; Malvestiti, M.; Daigle, J.N. Modeling CD40-Based Molecular Communications in Blood Vessels. IEEE Trans. Nanobiosci. 2014, 13, 230–243. [Google Scholar] [CrossRef]
- Callahan, B.J.; McMurdie, P.J.; Rosen, M.J.; Han, A.W.; Johnson, A.J.A.; Holmes, S.P. DADA2: High-Resolution Sample Inference from Illumina Amplicon Data. Nat. Methods 2016, 13, 581–583. [Google Scholar] [CrossRef]
- Katoh, K.; Misawa, K.; Kuma, K.; Miyata, T. MAFFT: A Novel Method for Rapid Multiple Sequence Alignment Based on Fast Fourier Transform. Nucleic Acids Res. 2002, 30, 3059–3066. [Google Scholar] [CrossRef]
- Price, M.N.; Dehal, P.S.; Arkin, A.P. FastTree: Computing Large Minimum Evolution Trees with Profiles Instead of a Distance Matrix. Mol. Biol. Evol. 2009, 26, 1641–1650. [Google Scholar] [CrossRef]
- Bokulich, N.A.; Subramanian, S.; Faith, J.J.; Gevers, D.; Gordon, J.I.; Knight, R.; Mills, D.A.; Caporaso, J.G. Quality-Filtering Vastly Improves Diversity Estimates from Illumina Amplicon Sequencing. Nat. Methods 2013, 10, 57–59. [Google Scholar] [CrossRef]
- Konopka, A. What Is Microbial Community Ecology? ISME J. 2009, 3, 1223–1230. [Google Scholar] [CrossRef]
- Douglas, G.M.; Maffei, V.J.; Zaneveld, J.R.; Yurgel, S.N.; Brown, J.R.; Taylor, C.M.; Huttenhower, C.; Langille, M.G.I. PICRUSt2 for Prediction of Metagenome Functions. Nat. Biotechnol. 2020, 38, 685–688. [Google Scholar] [CrossRef] [PubMed]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Nunzi, E.; Mezzasoma, L.; Bellezza, I.; Zelante, T.; Orvietani, P.; Coata, G.; Giardina, I.; Sagini, K.; Manni, G.; Di Michele, A.; et al. Microbiota-Associated HAF-EVs Regulate Monocytes by Triggering or Inhibiting Inflammasome Activation. Int. J. Mol. Sci. 2023, 24, 2527. https://doi.org/10.3390/ijms24032527
Nunzi E, Mezzasoma L, Bellezza I, Zelante T, Orvietani P, Coata G, Giardina I, Sagini K, Manni G, Di Michele A, et al. Microbiota-Associated HAF-EVs Regulate Monocytes by Triggering or Inhibiting Inflammasome Activation. International Journal of Molecular Sciences. 2023; 24(3):2527. https://doi.org/10.3390/ijms24032527
Chicago/Turabian StyleNunzi, Emilia, Letizia Mezzasoma, Ilaria Bellezza, Teresa Zelante, Pierluigi Orvietani, Giuliana Coata, Irene Giardina, Krizia Sagini, Giorgia Manni, Alessandro Di Michele, and et al. 2023. "Microbiota-Associated HAF-EVs Regulate Monocytes by Triggering or Inhibiting Inflammasome Activation" International Journal of Molecular Sciences 24, no. 3: 2527. https://doi.org/10.3390/ijms24032527
APA StyleNunzi, E., Mezzasoma, L., Bellezza, I., Zelante, T., Orvietani, P., Coata, G., Giardina, I., Sagini, K., Manni, G., Di Michele, A., Gargaro, M., Talesa, V. N., Di Renzo, G. C., Fallarino, F., & Romani, R. (2023). Microbiota-Associated HAF-EVs Regulate Monocytes by Triggering or Inhibiting Inflammasome Activation. International Journal of Molecular Sciences, 24(3), 2527. https://doi.org/10.3390/ijms24032527












