Abstract
Lung cancer is the deadliest cancer worldwide. Tissue biopsy is currently employed for the diagnosis and molecular stratification of lung cancer. Liquid biopsy is a minimally invasive approach to determine biomarkers from body fluids, such as blood, urine, sputum, and saliva. Tumor cells release cfDNA, ctDNA, exosomes, miRNAs, circRNAs, CTCs, and DNA methylated fragments, among others, which can be successfully used as biomarkers for diagnosis, prognosis, and prediction of treatment response. Predictive biomarkers are well-established for managing lung cancer, and liquid biopsy options have emerged in the last few years. Currently, detecting EGFR p.(Tyr790Met) mutation in plasma samples from lung cancer patients has been used for predicting response and monitoring tyrosine kinase inhibitors (TKi)-treated patients with lung cancer. In addition, many efforts continue to bring more sensitive technologies to improve the detection of clinically relevant biomarkers for lung cancer. Moreover, liquid biopsy can dramatically decrease the turnaround time for laboratory reports, accelerating the beginning of treatment and improving the overall survival of lung cancer patients. Herein, we summarized all available and emerging approaches of liquid biopsy—techniques, molecules, and sample type—for lung cancer.
1. Introduction
Lung cancer has the highest incidence rate for cancer type and the second-highest mortality rate in the world [1]. The high mortality rates associated with lung cancer are mostly due to its late detection and diagnosis, leading to a decrease in these overall survival rates [2,3]. However, even patients diagnosed in the early stages of the disease and still during cancer’s contained stage still show a poor five-years overall survival rate, which is lower than 60% compared to other cancer types. Despite the early detection, the poor prognosis is attributed to the disease progression [4].
Cigarette smoking and other tobacco exposure habits are the major risk factor for lung cancer, and it is associated with approximately 80% of all lung cancer cases [5,6]. However, additional risk factors associated with environmental and occupational exposures have also been linked to lung cancer development [7]. In addition to environmental risk factors, single nucleotide polymorphisms (SNPs) have also been associated with an increased risk for lung cancer development [8,9,10].
The biological processes behind lung cancer are complex, and the tumors are highly heterogeneous [11]. Histologically, lung cancer is divided into two major subtypes: small-cell lung cancer (SCLC) and non-small-cell lung cancer (NSCLC) [12]. The SCLC accounts for approximately 15–20% of all lung cancer cases, and SCLC patients show extremely low survival rates [13]. The NSCLC accounts for approximately 80–85% of all lung cancer patients, and it is further classified into three main histological subtypes: adenocarcinoma, squamous cell carcinoma (SCC), and large cell carcinoma [14]. NSCLC patients present heterogeneous survival rates depending on several features, including the stage of the disease at the time of diagnosis, the patient’s performance status, smoking habits, histological subtypes, and molecular characteristics [15,16].
In addition to the histological categorization, molecular characteristics are a critical additional component of the biological study of lung cancer due to the advances in precision medicine for both methodological procedural aspects and molecular biomarkers [17]. Currently, the number of FDA-approved drugs targeting molecular biomarkers has dramatically increased in the market, creating a more efficient route for treating NSCLC patients [18]. On the other hand, tumor tissue biopsy has continued to be mandatory for histological evaluation and diagnosis, and in the last decades, it has also been employed in the detection of molecular biomarkers [18,19]. However, the methods commonly used for tissue procurement are highly invasive and may have many side effects, which depending on the performance status of a patient, may not even be eligible for such an invasive procedure. Moreover, tumor tissue biopsy represents a small fragment of the tumor as a whole, which may result in unviable tissue biopsies for assessing tumor heterogeneity [20]. Thus, the development of new approaches is pivotal, given that the current approaches have some disadvantages, especially when it comes to offering a viable option for those who are not eligible for the currently existent methods of sample procurement [20,21,22,23].
In the last few years, a new approach has emerged, attracting various efforts for its implementation in the diagnostic routine of lung cancer [24,25]. Liquid biopsy is a minimally invasive approach for sample procurement, mostly body fluids, and it is used to detect molecular alterations, tumor cells, and metabolites. Furthermore, liquid biopsy, the emerging approach attracting substantial attention, has currently been employed for clinical management, specifically for guiding treatment and monitoring disease. In addition, to being a minimally invasive approach, the liquid biopsy also enables the serial collection of samples allowing early detection of residual disease and relapses and resistance to treatment [20,21,23,26,27]. For NSCLC, this approach is particularly valuable for patients who are not eligible for the conventional tissue biopsy, mainly due to patients’ individual conditions and the tumor location, as previously discussed. As a result, liquid biopsy has been employed for disease and treatment monitoring and for creating a targeted treatment for NSCLC patients within the field of precision medicine [21,28].
This review aims to gather the most up-to-date information for the procedure of liquid biopsy, given the context of NSCLC patients. This review will focus on information such as the various types of body fluids used for liquid biopsy, the currently available and promising biomarkers in the field, and a discussion of the major challenges involved in analyzing and securing samples for liquid biopsy.
2. Sample Types and Analytes for Liquid Biopsy from NSCLC
Body fluids such as plasma, sputum, saliva, urine, stool, cerebrospinal fluid, and pleural effusions, among others, are suitable sources for detecting diagnostic, prognostic, and predictive biomarkers for NSCLC [21,29,30,31,32,33]. As a result, liquid biopsy has been employed due to its minimally invasive sample procurement. However, it is important to point out that not all body fluids are collected by a minimally invasive sample procurement (e.g., cerebrospinal fluid). Thus, the type of body fluid selected for liquid biopsy must be carefully chosen based on the type of cancer, especially because some body fluids do not properly represent tumor origin (Table 1 and Figure 1). Moreover, the most important benefit of liquid biopsy is the minimally invasive sample procurement, and we should avail this advantage.

Table 1.
Liquid biopsy analytes for NSCLC: major clinical applications, biofluids, and methods.
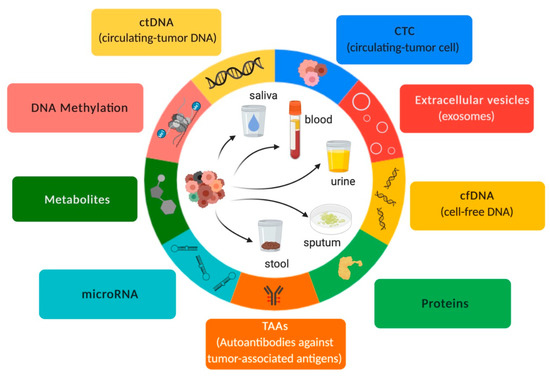
Figure 1.
Overview of liquid biopsy. Inspired by Hanahan & Weinberg (192).
Liquid biopsy samples are mostly composed of cell-free DNA (cfDNA), cell tumor DNA (ctDNA), circulating cell-free microRNAs (ccfmiRNAs), circulating tumor cells (CTCs), metabolites, and proteins as well as extracellular vesicles such as exosomes, which contain proteins and cell-free nucleic acids (cfNA) such as miRNAs (Figure 1). These components are released into body fluids through processes such as apoptosis, necrosis, and secretion [26]. Therefore, liquid biopsy has been an important “supporting actor” for guiding therapeutic strategies for NSCLC patients and has emerged as a “leading title-role actor” in the routine setting of precision medicine.
3. NSCLC Biomarkers for Detection in Liquid Biopsy Samples
Clinical biomarkers can be found in different biofluids; currently, the most frequently used in precision medicine are plasma and serum, both of which originate from the peripheral blood [49]. Both blood-derived biofluids can be used to analyze different biomarkers from distinctive sources, such as CTCs, ctDNA, cfDNA, ccfmiRNAs, and metabolites, among others [50]. The major applications of each type of molecule in the field of lung cancer are summarized hereafter.
3.1. Circulating-Tumor DNA (ctDNA) and Cell-Free DNA (cfDNA)
Fragments of cell-free DNA (cfDNA) are freely available throughout the blood. CfDNA is released into the bloodstream due to natural body mechanisms, such as apoptosis, necrosis, and active secretion [26,31]. CfDNA can be found in both healthy subjects and cancer patients, although cfDNA levels tend to be higher in cancer patients (Figure 2).

Figure 2.
Comparison of cfDNA and ctDNA. Individual aspects and similarities of both molecules are described.
Circulating tumor DNA (CtDNA), different from cfDNA, can harbor somatic mutations reflecting the tumor dynamics [51,52]. Tissue biopsy is a single “snapshot” of the tumor and, therefore, can mirror a unique subclone or a few subclones, while ctDNA can be more representative of the tumor’s entire tissue composition. Moreover, it is currently possible to trace the subclone’s origin of relapses and metastases through the phylogenetic profile of ctDNA. Tracing the clonal origin of the tumor can provide a collage of a tumor’s evolution, opening possibilities for the translation of such information into clinical practice [34].
Using liquid biopsy to detect ctDNA is extremely effective, as it is possible to collect a sample at any time during the disease and therapy course, assessing the progression of the disease in real-time [51]. However, liquid biopsy can become challenging when the concentration of ctDNA is measured to be extremely low (<1%) compared to the concentration of cfDNA in a patient’s bloodstream. Thence, highly sensitive and specific techniques are required for detecting mutations to track, detect, and monitor genomic alterations as cancer progresses [52,53,54]. The current standard methods for detecting somatic mutations, such as RT-PCR, Sanger sequencing, and next-generation sequencing (NGS), may not be sensitive enough for the detection of mutational ctDNA, specifically mutations presented with a low variant allele frequency (VAF) [51].
The main advantage of analyzing ctDNAs is the high specificity of the molecule, as it is proven that any molecular alterations present in these molecules are identical to those in the tumor tissue. In addition, advanced-stage cancer patients usually present with higher levels of ctDNA, which allows for easy monitoring of the disease’s course, tumor heterogeneity and dynamics, tumor evolution, and any tumor-acquired resistance to targeted treatments. Recently, ctDNA was associated with shorter survival, and actionable alterations in ctDNA were not detectable in time-matched tissue [55]. In addition, the turnaround time was decreased for the release of a biopsy report to guide therapeutic decisions that can be made earlier and safer [55].
Currently, highly sensitive PCR-based techniques, such as droplet digital PCR (ddPCR) and BEAMing, are considered sensitive enough for detecting mutations at low frequencies and have emerged as potential tools not only for advanced cancers but also for early detection [53]. NGS is also an extremely sensitive technique for detecting somatic mutations and identifying mutational frequencies as low as 0.02% VAF; however, the NGS technique produces an error rate of about 0.5–2.0% [56].
Therefore, the detection of rare variants by NGS remains challenging due to its limit of detection (LoD) and errors incorporated during sequencing. Furthermore, random errors can be incorporated into the DNA molecules during the library preparation or the sequencing processes, which could be mistaken for true variants. Strategies using molecular barcoding principles, such as hybrid capture and amplicon-based NGS, have also been used to detect low-frequency mutations in both liquid and conventional biopsy samples to decrease false negative results [51,53]. Each molecule in the sequencing library is marked with a small sequence of random nucleotides (8–16 N), which can be called a Unique Molecular Identifier (UMI), Tag Sequencing, or molecular barcodes and to decrease the chances of detecting false variants, the sequenced reads are grouped according to these UMI. As a result, sequencing artifacts can then be detected as they are not present in all reads with the same UMI, which increases the reliability of naming the true variants. Therefore, molecular barcoding technology allows for the correction of sequencing errors [57,58].
Due to the advances in sensitive technologies, ctDNA and mutational analysis are now possible for NSCLC patients. In addition, the detection rate of ctDNA can be higher than 80% in the plasma from NSCLC patients, suggesting that ctDNA analysis is an adequate alternative when sampling tissue biopsy is not an option [59]. EGFR, KRAS, ERBB2, and BRAF mutations, gene rearrangements (EML4—ALK, ROS1, NTRK1/2, and RET), exon skipping alterations, and gene amplifications (MET) are routinely evaluated in the management of care for NSCLC patients [51]. All these molecular alterations have been adapted into the clinical practice for guiding and monitoring patients’ treatment and disease state [53]. Currently, broader NGS panels have been employed in clinical practices, such as MSK-IMPACT (tissue) and MSK-ACCESS (plasma) [55]. A quarter of the patients presented alterations in ctDNA not detected in tissues [55], which suggests that plasma samples may present a higher specificity then previously reported, supporting the use of NGS as a sensitive, specific tool for plasma samples analysis [60].
Unfortunately, not all of the actionable alterations mentioned above have been translated for the clinical management of NSCLC patients through liquid biopsy sampling.
The FDA recently approved two IVD (in vitro diagnosis) tests (Cobas EGFR Mutation Test v2, Roche, and Idylla TM ctEGFR Mutation Assay) for NSCLC patients, which employs plasma samples for the detection of EGFR resistance mutation p.(Tyr790Met), exon 19 deletions and p/L858R mutations [61]. Although the IVD test shows a lower sensitivity when compared with other methods of variant detection (e.g., ddPCR and NGS), its approval was a great addition to the present tools used in guiding therapeutic decisions and monitoring treatment results for NSCLC patients [62].
3.2. cfDNA Methylation Biomarkers
Genetic mutations and epigenetic modifications, such as DNA methylation, have been detected in cfDNA and ctDNA. It has been considered a promising approach for translation into clinical applications to be used for diagnosis, prognosis, and predictive purposes.
Methylation is an incorporation of a methyl group (CH3) into a Cytosine in regions enriched with CG bases, also known as CpG islands [63]. Methylation occurs in CpG islands when found at the promoter regions of several genes, and it often results in gene silencing commonly found in tumor suppressor genes. On the other hand, the transcriptional activation of genes with the methylation present in the gene body is associated with various cancer types, including lung cancer [41,64]. Methylome analysis has yielded highly successful results on tumor tissues, especially when the analysis is focused on molecular subtyping and biomarkers discovery of several tumor types, including lung cancer [65,66]. In addition to plasma and serum, other body fluids deserve attention, such as sputum and bronchoalveolar lavage fluids, specifically due to their proximity to the tumor’s location [41].
Analysis of methylation-based biomarkers in cfDNA can also be used for diagnostic purposes for the management of lung cancer patients. Plasma cfDNA showed significant differences in DNA methylation level for early-stage NSCLC patients, including stage IA patients, indicating this type of biomarker could be a valuable tool for screening and early detection of NSCLC combined with imaging tests to improve the detection of early-stage pulmonary nodules [63,67,68,69,70]. Multi-cancer early detection (MCED) test may be a promising approach for cancer screening and early detection. MCED is a targeted methylation-based assay for complementary use in screening programs [71,72,73]. However, results for stage I lung cancer have not shown to be sensitive enough for being employed in a screening setting [72]. A recent study screened asymptomatic subjects from the NHS-Galleri trial (ISRCTN91431511) but results about the clinical utility of the MCED test for lung cancer still need to be addressed [73].
The combined methylation analysis of the CDO1 and HOXA9 was associated with unfavorable outcomes, while the combination of PTGDR and AJAP1 methylation was associated with favorable outcomes. A prognostic risk based on these methylation-based biomarkers may be useful to refine risk stratification [65,67]. In addition to prognostication, methylation-based biomarkers can also be useful as predictive biomarkers. Deregulation of methylation in cfDNA was also associated with EGFR-TKI resistance in early-stage NSCLC patients [74,75].
3.3. Circulating Tumor Cells (CTCs)
Circulating tumor cells (CTCs) are cells derived from primary tumors that were dissociated from tumor mass by either mechanical motion, the loss of adhesion molecules on the surface of cells entering the circulatory system, or a combination of both. CTCs are commonly detected in lower concentrations in the peripheral blood compared to other types of analytes (e.g., ctDNA), and it may be a challenge requiring the sampling of CTC-rich peripheral blood [38,52,54]. Although studies have shown that aggressive tumors can release thousands of these cells into the bloodstream every day, and it may be associated with the mechanism of distant metastasis, such a result would require completing a complex process [38,54]. Once these cells enter the bloodstream, they are capable of planting metastatic sites through active trans-endothelial migration while remaining inactive; however, no studies have yet been able to show how the biological process of transition from dormancy to active growth happens [52]. As a result, the mechanisms through which cancer spreads from one organ to another using CTCs are of great interest to the scientific community [54].
Furthermore, the methods of isolating CTCs include identifying these cells based on the presence of specific markers, such as the epithelial cell adhesion molecule (Ep-CAM), the cytokeratin of epithelial CTCs, and the N-Cadherin or vimentin of mesenchymal CTCs [76,77]. The FDA has confirmed an IVD method, based on anti-EpCAM ferromagnetic microbeads, called CellSearch CTC kit® (Veridex LLC, Raritan, NJ, USA) for prognostic assessment. This IVD method detects anti-EpCAM ferromagnetic microbeads on circulating tumor cells (CTC) in peripheral blood samples [78]. As previously mentioned, the characterization of CTCs is difficult since the detection method requires high sensitivity to detect these molecules’ low concentrations in extracorporeal fluids.
The presence of CTCs is considered a prognostic biomarker since it can help predict disease progression in cancer patients, including the progression of NSCLC [54]. One study showed that patients with metastatic lung cancer and a state of progressive disease presented a higher expression of the PIK3CA, AKT2, TWIST, and ALDH1 genes in CTCs compared to patients with non-metastatic disease. Thus, it is possible to correlate the presence of CTCs with the diagnostic scope of disease burden, including possible metastasis and disease progression [37]. The presence of CTCs was found to be independent of the tumor stage at diagnosis (49% stage I, 48% stage II, 48% stage III, and 52% stage IV patients) and histology (47% adenocarcinoma and 40% squamous cell carcinoma) when using a size-based filtration method for CTC detection [79]. Moreover, of the cells with the highest glucose uptake, hypermetabolic CTCs were isolated to analyze EGFR and KRAS mutations using ddPCR. The comparison between the primary tumor’s EGFR and KRAS mutations and that of CTCs showed a match in 70% of the cases [80].
3.4. Extracellular Vesicles
Extracellular vesicles (EVs) can be divided into microvesicles, vesicles, and exosomes. Exosomes are released by various methods and are detectable in several cell types and bodily fluids, including cancer cells. Moreover, exosomes are released during the process of exocytosis following the fusion of multivesicular bodies (MVBs) and cell membranes, and they can also be detected in body fluids such as blood (plasma and serum), urine, pleural effusions, saliva, cerebrospinal fluid, and semen [27,54]. In addition, exosomes have been shown to mediate intercellular communication between the tumor and the stroma, resulting in a rich source of molecular information that enables the location of origin cells for these exosomes [50,81].
Furthermore, ultracentrifugation or differential centrifugation (DC) and size-exclusion chromatography, based on size selection and chemical isolation during polymeric-based precipitation (PBP), are all commonly used for EV isolation [82,83,84].
Tumor cells can release more exosomes than non-tumor cells, making exosomes a potential biomarker in liquid biopsies for various types of tumors [27]. In lung cancer, exosomal RNA, DNA, and proteins can be used to detect molecular alterations, including actionable mutations [27,85]. Qu et al. (2019) identified exosomes harboring EGFR mutation, showcasing exosomes as a valuable component in the analysis’s tumor progression, pre-metastatic niche formation, and resistance to treatment [86]. The comparison between exosome-derived and tumor-derived mutations showed a sensitivity of 100% in detecting EGFR mutations and a specificity greater than 96% [86].
Furthermore, exosomal miRNAs (exo-miRNA) have also been generating interest from the scientific community as a potential biomarker in several types of cancer, including lung cancer. Exo-miRNA is protected from RNAse degradation in the bloodstream due to the lipid bilayer membrane, which results in their stability in body fluids and easily detectable form in body fluids [27]. As a result, exo-miRNA load was reported as a prognostic biomarker [87]. Additionally, exo-miRNAs miR-564 and miR-659 switched sensitive to resistant cells to gefitinib [88], which proved to be a predictive phenotype for exo-miRNA. On the other hand, the exo-miRNA miR-302-b is associated with the suppression of lung cancer cell proliferation and migration via the TGFβRII/ERK pathways, which has established the miR-302-b as a potential therapeutic target for lung cancer patients [89]. Thus, exosomal miRNAs may be employed in the process and measurements of diagnosis, prognosis, and monitoring of lung cancer patients, which reveals the need for further studies of exosomes and their clinical application in the context of precision medicine.
3.5. MicroRNA/CircRNA
MicroRNAs (miRNAs) are small RNAs (19–24 nucleotides) responsible for regulating gene expression. However, most miRNAs have unknown biological functions [90,91]. MiRNAs differ from mRNAs due to their stability, small size, and regulatory control in gene expression, rendering them a new generation of biomarkers [40]. Circulating-free miRNAs (cfmiRNAs) have already been reported in body fluids from cancer patients, including plasma, serum, urine, and saliva [92,93,94].
The detection of miRNAs in liquid biopsy samples was reported to distinguish NSCLC patients from healthy subjects paving the way for cfmiRNAs as promising early detection biomarkers [95]. Reis and collaborators (2020) recently reported differential expression of a set of miRNAs in plasma samples from early-stage lung cancer patients [96]. Similarly, plasma samples from early-stage NSCLC patients paired with non-cancer patients were screened for 754 circulating miRNAs, and a highly accurate 24-miRNAs panel was proposed for early detection of lung cancer in addition to the known risk factors [95]. In addition, dysregulation of miRNA expression can also be associated with exposure factors.
For example, the downregulation of let-7i-3p and miR-154-5p was found in serum from smokers, both non-cancer subjects and lung cancer patients. These miRNAs are involved in lung cancer development and progression, rendering these circulating miRNAs a potential role as a diagnostic and prognostic biomarker in lung cancer for tobacco-exposed patients [97,98]. Moreover, high expression of miR-34 and miR34-c in plasma samples from surgically resected NSCLC patients was associated with increased survival [98].
Recently, some publications have explored the potential of circular RNA (circRNA) as a biomarker in liquid biopsy [99]. This is because circRNA are resistant to RNases and other exonucleases due to the lack of final 5’ or 3’ regions [100,101], having a longer half-life than linear RNA [102]. Moreover, its specific tissue expression and stage of development increase this interest as a potential biomarker [98,103,104,105]. Studies show that circRNA expression varies in different tumor types, including lung cancer [104,106,107,108]. In adenocarcinoma patients, circRNA can be over-expressed, such as circ_0013958, which was upregulated [107], and circFARSA, a circRNA derived from exon 5–7 of the FARSA gene, in the plasma of patients with NSCLC compared to controls without cancer [109,110].
In addition, studies with circRNAs reported that they could detect specific signature profiles in tissue samples, capable of distinguishing histological groups in addition to NCLSC patients from healthy ones [105]. However, studies are being carried out to analyze the profile of circRNAs signatures in liquid biopsy samples, such as plasma, exosomes, and lymphocytes, to implement these molecules as diagnostic and predictive biomarkers.
Altogether, the detection of miRNAs/circRNAs in minimally invasive samples has emerged as a promising approach for the early detection of NSCLC to be incorporated in lung cancer screening programs and for prognostic purposes to be incorporated in the clinical management of these patients.
3.6. Metabolites and Proteins
In pathological conditions, including cancer, circulating metabolites can be detected in body fluids [45]. For detecting metabolites in biological fluids, Gas chromatography-mass spectrometry (GC-MS), nuclear magnetic resonance (NMR), and high-performance liquid chromatography UV detector (LC-UV) along with software (Metaboanalyst, Metabolite Set Enrichment Analysis [MSEA], Metlin, BioStatFlow and Human metabolome database [HMDB]) have been employed for detecting, processing, and analyzing metabolic data [111].
Lung cancer patients show changes in metabolic pathways compared to non-cancer patients; starch and sucrose metabolism, galactose metabolism, fructose, mannose degradation, purine metabolism, and tryptophan metabolism are among the unbalanced metabolic pathways [43]. In addition, some amino acids such as valine, leucine, and isoleucine are related to stress and energy production, regulating many signaling pathways, such as protein synthesis, lipid synthesis, cell growth, and autophagy, and can be found at higher levels in lung cancer patients compared with non-cancer patients [44]. However, these previously reported data must be clinically tested and reproduced in other series.
Although metabolomics has been a promising approach for managing and monitoring cancer patients, some limitations have still been experienced, such as identifying unknown compounds, detecting cancer-specific metabolites, and standardization of cutoffs [111].
In addition to metabolites, serum proteins have currently been used as cancer biomarkers, such as carcinoembryonic antigen (CEA), cytokeratin 19fragment (CYFRA 21-1), cancer antigen 125 (CA 125), neuron-specific enolase (NSE) and squamous cell carcinoma antigen (SCCA) [112,113]. Immunoassay (e.g., ELISA) and mass spectrometry can detect these serum proteins. In the last few years, considerable efforts have been made to find serum protein biomarkers for the early detection of lung cancer that will potentially be employed in screening programs soon.
Acute-phase reactant proteins (APRPs) are produced in response to inflammation caused by cancer and can also be used as potential biomarkers for the diagnosis of different types of cancer [114]. Lung cancer patients presented higher serum haptoglobin β (HP- β) chain levels than healthy controls. However, patients with other respiratory diseases also presented increased levels of Hp-β chain; thus, medical history, and radiological images should also be considered [115]. Lung cancer patients also presented higher serum amyloid A (SAA), another APRP, compared with healthy controls [116]. With higher levels of SAA1 and SAA2, lung adenocarcinoma patients also presented decreased serum levels of Apo A-1—a protein responsible for removing endogenous cholesterol from inflammatory sites. Thus, SAA1, SAA2, and Apo A-1 could also be considered potential biomarkers for the early detection of lung cancer [117].
In addition, to early detection applications, some proteins involved with metastasis and tumor progression are promising prognostic biomarkers. For example, plasma and pleural effusions from NSCLC patients with high levels of the S100A6 protein—a member of the S100 family with pro-apoptotic function—presented longer survival time compared with S100A6-negative cases [118,119]. On the other hand, NSCLC patients show high serum Cytokeratins (CKs) levels, such as CK 8, 18, and 19, which were associated with unfavorable prognoses [119,120]. In addition, the combined analysis of regulators of actin—calmodulin, thymosin β4, cofilin-1, and thymosin β10—was suitable for predicting patients’ outcomes [121].
The CancerSEEK reported forty-one potential protein biomarkers detectable at least in one out of the eight tumor types. The authors selected the best eight biomarkers for composing the final bead-based immunoassay test (CA-125, CA19-9, CEA, HGF, Myeloperoxidase, OPN, Prolactin, TIMP-1), which were highly effective in distinguishing cancer patients from healthy controls [122]. The positivity for the CancerSEEK test was about 70% for all cancer types. According to this study, combining protein biomarkers with cfDNA mutations increased sensitivity without significantly decreasing specificity [122].
Although proteomic approaches have been promising tools for precision medicine in the lung cancer field, there are some limitations regarding using proteins as biomarkers. Many of these proteins are associated with different tumor types. They present poor sensitivity, are not organ-specific, and can also be detected in non-malignant diseases [112,113,123]. Thus, clinical history and radiological exams should always be considered. Moreover, some proteins are presented with low serum levels, precluding their employment for early lung cancer diagnosis [123,124]. Finally, the analysis of these biomarkers can be influenced by the contamination of intracellular proteins caused by cell lysis during sample procurement and processing. Thus, pre-analytical steps are pivotal [118].
Scalable proteomics has also emerged as a tool for the identification of high-risk subjects for lung cancer. The most recent example is Olink Proteomics, which was created to detect circulating proteins (https://olink.com/, accessed on 16 November 2022). A 36-protein multiplexed assay was developed for risk assessment of lung cancer development, and this set of proteins includes growth factors, tumor necrosis factor receptors, and chemokines and cytokines [125]. Scalable proteomic approaches may not be affordable for low-middle-income countries, but when feasible, they should still be considered as a complementary tool along with LDCT in lung cancer screening programs. However, more studies should be conducted to prove these proteomic approaches are cost-effective for lung cancer screening, especially considering limited resources and minorities.
3.7. Autoantibodies against Tumor-Associated Antigens
Autoantibodies against tumor-associated antigens (TAAs) are circulating antibodies to autologous cellular antigens [126,127,128]. Tumor tissues can release cellular proteins, leading to the activation of the immune system and the production of autoantibodies [129,130]. Therefore, cancer patients produce autoantibodies against aberrant or overexpressed proteins produced by these cancer cells [131]. TAAs are stable in the serum and have been studied in several types of tumors, including lung cancer. TAAs can be detected through the immunoenzymatic assay (ELISA) [48,129,132]. Moreover, they can also provide information for early detection and disease monitoring [47,48,132,133,134,135].
In the 90s, Lubin et al. (1995) analyzed the presence of p53 antibodies in the serum of patients with lung cancer [136]. These antibodies were 30% higher in cancer patients than in non-cancer patients, associated with TP53 mutations [136]. TAAs panels have also been developed, combining many different TAAs to improve test sensitivity [48,126,132,137,138]. The panels developed include TAAs for p53, NY-ESO-1, CAGE, GBU4–5, Annexin 1, SOX2, c-Myc, MDM2, NPM1, p16, cyclin B1, among others [48,132,139]. Although the levels of most biomarkers increase proportionally to tumor burden, TAAs levels do not differ among different stages of lung cancer, possibly due to the humoral immune response—present from the beginning of tumorigenesis [48,132,139]. This feature renders TAAs a potential tool for early detection [48,132,140,141]. However, studies including proper sample size and validation set are required for greater reliability of TAAs in the routine lung cancer setting.
4. Clinical Application of Liquid Biopsy for NSCLC
The absence of pathognomonic symptoms in lung cancer leads to late diagnosis—many patients may mistakenly receive another diagnosis, such as pneumonia, Chronic obstructive pulmonary disease (COPD), among others, especially from low-middle income countries, where resources are limited, and health systems are recurrently offscourings. The late diagnosis in limited therapeutic options and curative intent is no longer available, culminating in the highest lethality rate among all cancer types [3].
4.1. Prognosis
Lung cancer in the early stage has an excellent prognostic after surgery due to early screening but needs more information about the disease is needed to understand metastatic lung cancer [142]. After the liquid biopsy advent, the prediction of outcomes for NSCLC patients employing molecular biomarkers became more feasible for those who may not be eligible for conventional biopsy [21,27].
In NSCLC and SCLC, more CTCs were associated with adverse prognostic factors for survival [54,143,144]. A study compared the number of CTCs present in blood in patients with small-cell lung cancer. The CTC count was made at the beginning, before, and after treatment. The authors show that patients with eight or more CTCs, per 7.5 mL of blood had a worse survival than those with less than 8 CTCs at the pretreatment, and patients whose baseline CTC levels remained lower than 8 CTC at the posttreatment showed better survival [143].
Undetectable ctDNA blood levels may serve as a prognostic marker for targeted therapy and chemotherapy in patients with NSCLC [142,145]. CtDNA has been proposed as a non-invasive, real-time biomarker to provide prognostic information to monitor treatment since patients with the same mutation may differ in response to treatment [146]. Based on this, techniques can detect recurrent point mutations in controller genes. When using ddPCR, plasma G12/G13 status was associated with an unfavorable prognostic in progression-free survival and overall survival in NSCLC patients [147]. The prognostic value of TP53 in lung cancer is being debated. Several changes in this gene are found in patients with advanced NSCLC. Patients carrying pathogenic plasma mutations in TP53 have lower overall survival when compared to wild TP53. In addition, the risk of extrathoracic metastases in patients with TP53 ctDNA alteration is greater [148].
In addition to point mutations, the methylation status of critical genes can also be analyzed in minimally invasive samples, and the methylation status can serve as a prognostic biomarker [135,142]. For example, Wen et al. (2021) showed that circulating metHOXA9 is an adverse prognostic factor in patients with advanced NSCLC. In this study, the authors analyzed the levels of metHOXA9 in patients’ blood before starting treatment and before each cycle of chemotherapy. As a result, it was observed that the levels of metHOXA9 increased after the first treatment cycle; that is, the treatment changed the status of the biomarker, decreasing the overall survival of these patients [149].
The hypermethylation of some genes as p16, CDH1, FHIT, and APC, can be considered a prognostic factor. In addition to being classified by stages and histological type, genetic alterations among each stratified group may reveal additional and more accurate prognostic factors [150]. Such as, the methylation of genes p16, CDH13, RASSF1A, and APC in patients with early-stage NSCLC-treated surgery was associated with an early recurrence [151,152]. More studies are needed to analyze the best genes to use as prognostic biomarkers in lung cancer. By considering post-transcriptional mechanisms, miRNAs can also serve as prognostic biomarkers. For example, NSCLC patients with downregulated miR-590-5p had significantly lower median survival rates when compared to patients expressing high miR-590-5p, and it was associated as a potential prognostic marker for the progression of NSCLC [152].
High expression levels of miR-18a, miR-20a, miR- 92a, miR-126, miR-210, and miR-19a correlated with worse disease-free survival (DFS), in addition to a shorter overall survival (OS) than patients with low expression levels of these miRNAs. So, these results suggested that these miRNAs can be potential biomarkers for the prognosis of NSCLC patients [153]. In addition, elevated levels of miR-34a correlated with a prolonged DFS and OS compared to low levels of expression in 196 NSCLC patients, i.e., miR-34a has potential prognostic value for lung cancer patients [98].
Boeri et al. (2011) and Tian et al. (2016), with a short cohort, reported that miR-486-5p was downregulated in adenocarcinoma patients’ plasma, and miR-181b-5p was upregulated in squamous cell carcinoma patients’ plasma. In addition, the two miRNAs negatively regulate their targets—RASSF1 and PIK3R1 [148,149], while that elevated levels of miR-21 in plasma samples predicted poorer overall survival of NSCLC patients [154,155]. However, more studies with increased sample sizes must incorporate microRNAs into clinical practice as prognostic biomarkers [154].
4.2. Precision Medicine and Disease Monitoring
Positron Emission Tomography/Computed Tomography (PET/CT) and tissue biopsy are currently employed for NSCLC disease staging and monitoring and for guiding therapeutic strategies either based on disease staging or molecular alterations [43,156]. However, not all patients are eligible for tissue biopsy, depending on tumor location and the patient’s clinical performance [157]. Using liquid biopsy for clinical management, including treatment and disease monitoring, can better represent tumor heterogeneity, and predictive biomarkers can be successfully detected to guide therapeutic options for NSCLC. Unfortunately, the successful detection of predictive biomarkers in this sample depends on assay sensitivity and the variant allele frequency (VAF). Fortunately, susceptible technologies have emerged as suitable approaches for cfDNA analysis, making rare detecting alleles feasible [126,158,159].
There are two approved tests for the search for actionable mutations in liquid biopsy, the Idylla TM ctEGFR Mutation Assay and the Cobas® EGFR Mutation Test v2, equally real-time PCR-based [160,161]. Identifying EGFR mutations has become essential since EGFR-TKI therapy has become the standard treatment choice for EGFR mutant patients [162]. The acquisition of resistance to treatment is recurrent in up to 60% of patients, and disease recurrence undergoing EGFR-TKI therapy is generally mediated by the p.(Tyr790Met) mutation [163]. As Cabanero and Tsao (2018) suggest, in patients diagnosed with NSCLC, ctDNA analysis can provide tumor resistance responses acquired in real-time to TKIs for EGFR [164]. For them, the clinical application of these tests is about to reach reality because the current digital platforms approach greater sensitivity and precision to ctDNA [165]. Some patients presented with the EGFR p.(Tyr790Met) mutation early as 344 days before disease recurrence [160]. EGFR-mutated patients presented undetectable ctDNA after four weeks of TKi therapy, associated with a 12-week radiological response and progression-free and overall survival [66].
A prospective study was carried out in 2015 using available serial samples of blood—collected and follow-up for ten months—of 41 patients with lung cancer. All samples of blood were analyzed cfDNA observing EGFR mutation and p.(Tyr790Met) mutation, and the authors observed that the appearance or increase in a unit of the p.(Tyr790Met) allele frequency almost triples the risk of death and progression, and this information can be used for to estimate whether p.(Tyr790Met) positive patients should start second-line treatment based on molecular data rather than imaging data [166].
In addition to mutations, the decreased concentration of CTCs has been associated with the radiographic response of the tumor during the different treatments in patients with NSCLC (surgery, chemotherapy, radiotherapy, target therapy, immunotherapy), while the increased number of CTCs has been correlated with the disease progression [167].
Studies have also investigated the expression of PD-L1 in CTC and white blood cells (WBC) in NSCLC [68,168,169,170]. Expression of PD-L1 in CTC and WBC was highly correlated with the tumor tissue expression, pointing out the importance of this evaluation of the “liquid microenvironment” to assist in the immunotherapy stratification and the monitoring of disease [170]. Studies with tumor tissue from patients with NSCLC have shown a relationship between high tumor mutational burden (TMB) and a longer duration of response and survival in patients treated with anti-PD-1 or anti-PD-L1 therapy [171,172,173]. For example, Gandara et al. (2018) analyzed 797 plasma samples from NSCLC patients, and the authors observed that TMB in plasma (bTMB)≥ 16 had a higher benefit for progression-free survival with atezolizumab therapy [174]. However, few studies have been conducted with a blood-based assay to measure bTMB in lung cancer, and the effectiveness of bTMB remains unclear [175].
Gene fusions can also be detected in blood samples of NSCLC patients, such as anaplastic lymphoma kinase (ALK, ROS1, RET, and NTRK) [176]. The detection and permanence of this fusion were associated in this study with shorter progression-free survival to crizotinib [177].
4.3. Early Detection as an Emerging Application of Liquid Biopsy for NSCLC
The high mortality rate of lung cancer patients is related chiefly to late diagnosis when curative treatments are ineffective. In order to decrease lung cancer mortality, screening programs have been implemented worldwide [178,179].
There are currently cancer screening programs, such as breast cancer and lung cancer, which use X-ray and low-dose computed tomography (LDCT) emission, respectively, to increase the chances of early diagnosis and increase patient survival. However, these tests are performed only in the percentage of the population considered at high risk; that is, the young do not have the opportunity to undergo the screening procedure because they also have a chance of developing cancer. In addition, developing countries, associated with low income and the lack of easy access to public health programs, make screening programs difficult to occur due to the cost of equipment and the need for mobile units, which lead to late diagnosis of cancer [180]. For this reason, implementing public policies to aid research and development in the context of liquid biopsy is significant.
However, many challenges have been experienced in lung cancer screening programs, and additional strategies are required to increase the early detection of lung cancer [181]. Plasma levels of cfDNA from NSCLC patients are higher than controls with no cancer, rendering cfDNA plasma levels an exciting approach as a diagnostic biomarker [107,182]. CTCs can be eliminated by the primary tumor, even in the early stages of tumor development. However, they are available at low abundance, requiring precision and empathetic methodologies to detect them [183]. Improvements in the CTC isolation methods have emerged, making CTCs in clinical practice feasible to increase the patient’s possibility of care and quality of life [76]. Moreover, the use of biomarkers to distinguish benign from malignant lesions, and to identify molecules that can complement imaging tests during lung cancer screening, can reduce the number of false positives and false negatives [179,180].
MiRNAs released in plasma and serum may also be used for lung cancer screening [184]. The use of microRNAs for lung cancer screening and early detection should be considered to be used along with CT images in order to improve the accuracy of screening programs when CT images do not show clinically detectable disease. The Multicenter Italian Lung Detection (BioMILD) trial showed a miRNA signature classifier (MSC) combined with CT was more effective for risk stratification than only CT or only MSC [185]. Subjects with CT positive and MSC negative (CT+/MSC-) showed an HR of 13.73, and subjects with CT positive and MSC positive (CT+/MSC+) showed an HR of 30.71. However, validation and reproducibility studies are necessary for the implementation of this strategy proposed by the BioMILD trial. In addition, the employment of miRNAs as a minimally invasive tool together with CT in lung cancer screening programs still needs to be better addressed, especially due to conflict data about these biomarkers in blood samples [186].
Technologies are under development to improve the detection of rare and/or small molecules present in the human body. circRNAs-based signatures have emerged as a promising tool for the early detection of lung cancer [187,188].
The CancerSEEK improved the early detection of several types of tumors, including lung cancer, by combining several biomarkers in plasma [123]. The authors analyzed plasma samples from 1005 patients with eight different tumor types, and none of the patients received neoadjuvant chemotherapy nor harbored evident metastases before blood collection. CtDNA isolated from plasma for mutation analysis was initially submitted to PCR amplification using primers designed to amplify regions of interest in the 16 genes. Simultaneously, eight proteins were analyzed in the same samples by the Luminex bead-based immunoassays technique (Millipore, Bilerica, NY). The accuracy of the prediction of this approach was 39% for lung cancer. The sensitivity of CancerSEEK for lung cancer was approximately 60% [123].
The INTEGRAL Risk Biomarker and Nodule Malignancy project analyzes a proteomic panel, based on Olink assay, with 21 protein relevant for lung cancer using a minimum quantity of blood (<50 uL) to optimize LDCT screening [189]. This project was initiated with case-control cohorts—training and validation sets, totalizing more than four thousand people—of several screening programs, main in EUA, Australia, Singapura, and Canada. However, we expect results from this trial to prove the effectiveness of using proteomics panels in lung cancer screening scenarios.
On the other hand, a multiple cancer early detection test (MCED) developed a biomarker based on cfDNA methylation in the plasma of 4077 subjects. This biomarker showed an overall sensitivity and specificity of 51.5% and 99.5%, respectively [72]. For lung cancer, this same methylation panel has shown a sensitivity of 79.5% in cases with stage II (95% CI).
The DETECT-A study (Detecting cancers Earlier Through Elective mutation-based blood Collection and Testing) was performed with 10,000 women, 65 to 75 years old, with no personal history of cancer, and was analyze mutations of 16 genes in cfDNA and nine protein highly validated biomarkers in blood samples [190]. Twenty-six cancer patients were detected by blood testing, and nine were lung cancer. In addition, a PET-CT scan was performed for 15 patients to exclude distant metastasis [190]. Only 1.2% of the individuals tested in the blood test were submitted to PET-CT, decreasing the costs of imaging scans [190]. These data demonstrate the possible relevance of combining multi-cancer blood testing with PET-CT in the clinical routine, improving early detection of lung cancer in a non-invasive way [191]. In addition, lung cancer screening programs can shift the diagnosis scenario from metastatic to early-stage disease. Although overdiagnosis remains a concern, a recent study reported no increase in the overall incidence rate of lung cancer but a decrease in metastatic cases and an increased number of stage I cases compared with an unscreened population [192]. Furthermore, the development of alternative techniques for screening and early detection, such as liquid biopsy, can help the conventional methods already used, reducing the costs of public coffers and reducing exposure to radiation and patient discomfort.
5. Challenges and Limitations
Liquid biopsy has been a promising approach for detecting biomarkers in NSCLC patients. This minimally invasive approach better represents tumor heterogeneity and can also be effective for lung cancer screening.
The lack of standardization prevents liquid biopsy implementation in the clinical routine. For this reason, more studies involving protocol standardization and a more significant number of cases become necessary to obtain a more excellent population representation, generating accurate and applicable results. Another limitation is that some biomarkers are fragile, requiring great pre-analytical care. The interaction between genetics and environment is challenging to control. Additionally, specific and sensitive methodologies are necessary to isolate and analyze these biomarkers, mainly due to the low concentration of some molecules in the body fluid.
6. Open Issues e Future Perspectives
Although many studies and advances have been made over the last few years concerning liquid biopsy, little has been translated into clinical practice, probably due to the several challenges already mentioned in this review. In addition, we must consider that the vast majority of studies analyze populations of European and North American, opening a gap for the genomic analyses referring to more heterogeneous populations, such as the population of South America and Africa [193,194].
The development and improvement of specific techniques, both to isolate analytes from liquid biopsy and to analyze them, are necessary to increase the sensitivity and specificities of the tests so that they are safe to be used for both early detection, prognosis, and monitoring. Methodologies that increase confidence to detect rare variants or allow a small initial sample input is welcome in this area. Training professionals in this area is also critical for reliable results. In addition, improving new mathematical and computational methods based on machine learning can also improve liquid biopsy methods making this approach even closer to the routine setting.
In this review, we summarized promising biomarkers that can be used for lung cancer screening associated with gold standard methods—such as LDCT—improving detection rates of screening programs (Figure 3). In the near future, a liquid biopsy will hopefully increase the early detection of lung cancer.
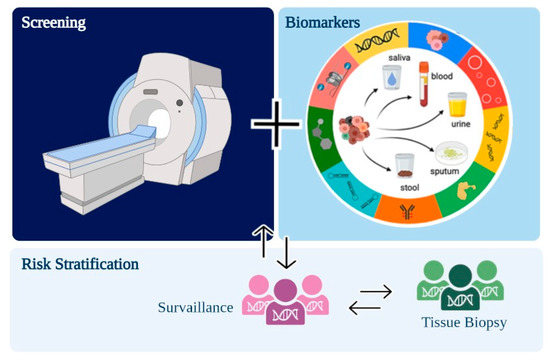
Figure 3.
Purposed workflow for lung cancer screening programs associating low-dose computed tomography (LDCT) and minimally invasive biomarkers.
Author Contributions
Wrote the manuscript: G.M.S.C. and M.d.O.S. (both equally contribute to this review). Review, Editing, and Supervising: L.F.L. and R.M.R. All authors have read and agreed to the published version of the manuscript.
Funding
GMSC is supported by Sao Paulo Research Foundation (FAPESP), and MOS is supported by the National Ministry of Health (Brazil). LFL was supported by the Public Ministry of Labor Campinas (Research, Prevention, and Education of Occupational Cancer). RMR is a recipient of a CNPq Productivity (Brazil) fellowship. This research was funded by PRONON—PRONON/MS (Abordagens móveis e de tecnologia para prevenção primária e secundária de câncer—NUP: 25000.015000/2019-53).
Institutional Review Board Statement
Review manuscripts do not require IRB approval.
Informed Consent Statement
Not applicable due to the nature of the article (Review article). We confirmed no patients data were collected for the current article.
Data Availability Statement
No original data were generated by the current manuscript. All information were gathered from open access and/or publicly available manuscripts.
Acknowledgments
Funding sources have no contribution to filling out authorship for the present study. We thank all members of the GTOP group (Translational Group of Pulmonary Oncology—Barretos Cancer Hospital, Brazil) for scientific discussion and suggestions and Nicole Mayara Dias de Souza for English editing.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Siegel, R.L.; Miller, K.D.; Jemal, A. Cancer statistics, 2019. CA Cancer J. Clin. 2019, 69, 7–34. [Google Scholar] [CrossRef]
- Henschke, C.I.; International Early Lung Cancer Action Program Investigators. Survival of patients with clinical stage I lung cancer diagnosed by computed tomography screening for lung cancer. Clin. Cancer Res. AACR 2007, 13, 4949–4950. [Google Scholar] [CrossRef] [PubMed]
- Warren, G.W.; Ostroff, J.S.; Goffin, J.R. Lung cancer screening, cancer treatment, and addressing the continuum of health risks caused by tobacco. Am. Soc. Clin. Oncol. Educ. Book 2016, 36, 223–229. [Google Scholar] [CrossRef] [PubMed]
- Howlader, N.N.; Noone, A.M.; Krapcho, M.; Miller, D.; Brest, A.; Yu, M.; Ruhl, J.; Tatalovich, Z.; Mariotto, A.; Lewis, D.R.; et al. SEER Cancer Statistics Review, 1975–2017; National Cancer Institute: Bethesda, MD, USA, 2020; Volume 4.
- Thun, M.J.; Henley, S.J.; Calle, E.E. Tobacco use and cancer: An epidemiologic perspective for geneticists. Oncogene 2002, 21, 7307–7325. [Google Scholar] [CrossRef]
- Ridge, C.A.; McErlean, A.M.; Ginsberg, M.S. (Eds.) Epidemiology of lung cancer. In Seminars in Interventional Radiology; Thieme Medical Publishers: New York, NY, USA, 2013. [Google Scholar]
- Markowitz, S.B.; Levin, S.M.; Miller, A.; Morabia, A. Asbestos, Asbestosis, Smoking, and Lung Cancer. New Findings from the North American Insulator Cohort. Am. J. Respir. Crit. Care Med. 2013, 188, 90–96. [Google Scholar] [CrossRef]
- Shankar, A.; Dubey, A.; Saini, D.; Singh, M.; Prasad, C.P.; Roy, S.; Bharati, S.J.; Rinki, M.; Singh, N.; Seth, T.; et al. Environmental and occupational determinants of lung cancer. Transl. Lung Cancer Res. 2019, 8, S31–S49. [Google Scholar] [CrossRef] [PubMed]
- McKay, J.D.; EPIC Study; Hung, R.J.; Gaborieau, V.; Boffetta, P.; Chabrier, A.; Byrnes, G.; Zaridze, D.; Mukeria, A.; Szeszenia-Dabrowska, N.; et al. Lung cancer susceptibility locus at 5p15.33. Nat. Genet. 2008, 40, 1404–1406. [Google Scholar] [CrossRef] [PubMed]
- McKay, J.D.; SpiroMeta Consortium; Hung, R.J.; Han, Y.; Zong, X.; Carreras-Torres, R.; Christiani, D.C.; Caporaso, N.E.; Johansson, M.; Xiao, X.; et al. Large-scale association analysis identifies new lung cancer susceptibility loci and heterogeneity in genetic susceptibility across histological subtypes. Nat. Genet. 2017, 49, 1126–1132. [Google Scholar] [CrossRef] [PubMed]
- Hanahan, D. Hallmarks of Cancer: New Dimensions. Cancer Discov. 2022, 12, 31–46. [Google Scholar] [CrossRef]
- Cruz, C.S.D.; Tanoue, L.T.; Matthay, R.A. Lung cancer: Epidemiology, etiology, and prevention. Clin. Chest Med. 2011, 32, 605–644. [Google Scholar] [CrossRef]
- Howlader, N.N.A.M.; Noone, A.M.; Krapcho, M.E.; Miller, D.; Brest, A.; Yu, M.E.; Ruhl, J.; Tatalovich, Z.; Mariotto, A.; Lewis, D.R.; et al. SEER Cancer Statistics Review, 1975–2016; National Cancer Institute: Bethesda, MD, USA, 2019; Volume 1.
- Planchard, D.; Smit, E.F.; Groen, H.J.M.; Mazieres, J.; Besse, B.; Helland, Å.; Giannone, V.; D’Amelio, A.M., Jr.; Zhang, P.; Mookerjee, B.; et al. Dabrafenib plus trametinib in patients with previously untreated BRAFV600E-mutant metastatic non-small-cell lung cancer: An open-label, phase 2 trial. Lancet Oncol. 2017, 18, 1307–1316. [Google Scholar] [CrossRef] [PubMed]
- Duma, N.; Santana-Davila, R.; Molina, J.R. (Eds.) Non–Small Cell Lung Cancer: Epidemiology, Screening, Diagnosis, and Treatment; Mayo Clinic Proceedings; Elsevier: Amsterdam, The Netherlands, 2019. [Google Scholar]
- Leal, L.F.; de Paula, F.E.; De Marchi, P.; Viana, L.D.S.; Pinto, G.D.J.; Carlos, C.D.; Berardinelli, G.N.; Miziara, J.E.; da Silva, C.M.; Silva, E.C.A.; et al. Mutational profile of Brazilian lung adenocarcinoma unveils association of EGFR mutations with high Asian ancestry and independent prognostic role of KRAS mutations. Sci. Rep. 2019, 9, 3209. [Google Scholar] [CrossRef] [PubMed]
- Kanwal, M.; Ding, X.-J.; Cao, Y. Familial risk for lung cancer. Oncol. Lett. 2017, 13, 535–542. [Google Scholar] [CrossRef]
- Tan, A.C.; Tan, D.S.W. Targeted Therapies for Lung Cancer Patients With Oncogenic Driver Molecular Alterations. J. Clin. Oncol. 2022, 40, 611–625. [Google Scholar] [CrossRef]
- Harrison, S.; Judd, J.; Chin, S.; Ragin, C. Disparities in Lung Cancer Treatment. Curr. Oncol. Rep. 2022, 24, 241–248. [Google Scholar] [CrossRef] [PubMed]
- Lim, M.; Kim, C.-J.; Sunkara, V.; Kim, M.-H.; Cho, Y.-K. Liquid Biopsy in Lung Cancer: Clinical Applications of Circulating Biomarkers (CTCs and ctDNA). Micromachines 2018, 9, 100. [Google Scholar] [CrossRef] [PubMed]
- Lu, J.; Han, B. Liquid Biopsy Promotes Non-Small Cell Lung Cancer Precision Therapy. Technol. Cancer Res. Treat. 2018, 17, 1533033818801809. [Google Scholar] [CrossRef] [PubMed]
- Pisapia, P.; Malapelle, U.; Troncone, G. Liquid Biopsy and Lung Cancer. Acta Cytol. 2019, 63, 489–496. [Google Scholar] [CrossRef]
- Hoseok, I.; Cho, J.-Y. Lung cancer biomarkers. Adv. Clin. Chem. 2015, 72, 107–170. [Google Scholar]
- Buder, A.; Tomuta, C.; Filipits, M. The potential of liquid biopsies. Curr. Opin. Oncol. 2016, 28, 130–134. [Google Scholar] [CrossRef]
- Castro-Giner, F.; Gkountela, S.; Donato, C.; Alborelli, I.; Quagliata, L.; Ng, C.K.Y.; Piscuoglio, S.; Aceto, N. Cancer Diagnosis Using a Liquid Biopsy: Challenges and Expectations. Diagnostics 2018, 8, 31. [Google Scholar] [CrossRef] [PubMed]
- Wan, J.C.M.; Massie, C.; Garcia-Corbacho, J.; Mouliere, F.; Brenton, J.D.; Caldas, C.; Pacey, S.; Baird, R.; Rosenfeld, N. Liquid biopsies come of age: Towards implementation of circulating tumour DNA. Nat. Rev. Cancer 2017, 17, 223–238. [Google Scholar] [CrossRef] [PubMed]
- Bracht, J.W.P.; Mayo-De-Las-Casas, C.; Berenguer, J.; Karachaliou, N.; Rosell, R. The Present and Future of Liquid Biopsies in Non-Small Cell Lung Cancer: Combining Four Biosources for Diagnosis, Prognosis, Prediction, and Disease Monitoring. Curr. Oncol. Rep. 2018, 20, 70. [Google Scholar] [CrossRef] [PubMed]
- Trombetta, D.; Sparaneo, A.; Fabrizio, F.P.; Muscarella, L.A. Liquid biopsy and NSCLC. Lung Cancer Manag. 2016, 5, 91–104. [Google Scholar] [CrossRef]
- Chan, M.H.M.; Chow, K.M.; Chan, A.T.C.; Leung, C.B.; Chan, L.Y.S.; Chow, K.C.K.; Lam, C.W.; Lo, Y.M.D. Quantitative Analysis of Pleural Fluid Cell-free DNA as a Tool for the Classification of Pleural Effusions. Clin. Chem. 2003, 49, 740–745. [Google Scholar] [CrossRef]
- Pu, D.; Liang, H.; Wei, F.; Akin, D.; Feng, Z.; Yan, Q.; Li, Y.; Zhen, Y.; Xu, L.; Dong, G.; et al. Evaluation of a novel saliva-based epidermal growth factor receptor mutation detection for lung cancer: A pilot study. Thorac. Cancer 2016, 7, 428–436. [Google Scholar] [CrossRef]
- Wang, Y.; Springer, S.; Zhang, M.; McMahon, K.W.; Kinde, I.; Dobbyn, L.; Ptak, J.; Brem, H.; Chaichana, K.; Gallia, G.L.; et al. Detection of tumor-derived DNA in cerebrospinal fluid of patients with primary tumors of the brain and spinal cord. Proc. Natl. Acad. Sci. USA 2015, 112, 9704–9709. [Google Scholar] [CrossRef]
- Fujii, T.; Barzi, A.; Sartore-Bianchi, A.; Cassingena, A.; Siravegna, G.; Karp, D.D.; Piha-Paul, S.A.; Subbiah, V.; Tsimberidou, A.M.; Huang, H.J.; et al. Mutation-Enrichment Next-Generation Sequencing for Quantitative Detection of KRAS Mutations in Urine Cell-Free DNA from Patients with Advanced CancersKRAS Mutations in Urine cfDNA. Clin. Cancer Res. 2017, 23, 3657–3666. [Google Scholar] [CrossRef]
- Benlloch, S.; Martí-Ciriquián, J.L.; Galbis-Caravajal, J.M.; Martín, C.; Sánchez-Payá, J.; Rodríguez-Paniagua, J.M.; Romero, S.; Massutí, B. Cell-Free DNA Concentration in Pleural Fluid and Serum: Quantitative Approach and Potential Prognostic Factor in Patients with Cancer and Pleural Effusions. Clin. Lung Cancer 2006, 8, 140–145. [Google Scholar] [CrossRef]
- Abbosh, C.; Birkbak, N.J.; Wilson, G.A.; Jamal-Hanjani, M.; Constantin, T.; Salari, R.; Le Quesne, J.; Moore, D.A.; Veeriah, S.; Rosenthal, R.; et al. Phylogenetic ctDNA analysis depicts early-stage lung cancer evolution. Nature 2017, 545, 446–451. [Google Scholar] [CrossRef]
- Yamamoto, H.; Watanabe, Y.; Itoh, F. Cell-Free DNA. In Biomarkers in Cancer Therapy; Springer: Berlin/Heidelberg, Germany, 2019; pp. 11–24. [Google Scholar]
- Cescon, D.W.; Bratman, S.V.; Chan, S.M.; Siu, L.L. Circulating tumor DNA and liquid biopsy in oncology. Nat. Cancer 2020, 1, 276–290. [Google Scholar] [CrossRef] [PubMed]
- Hanssen, A.; Wagner, J.; Gorges, T.M.; Taenzer, A.; Uzunoglu, F.G.; Driemel, C.; Stoecklein, N.H.; Knoefel, W.T.; Angenendt, S.; Hauch, S.; et al. Characterization of different CTC subpopulations in non-small cell lung cancer. Sci. Rep. 2016, 6, 28010. [Google Scholar] [CrossRef] [PubMed]
- Shaffer, C. Circulating Tumor Cells and the Liquid Biopsy: Processed appropriately, liquid biopsy samples and circulating biomarkers may aid cancer diagnosis and predict treatment outcomes. Genet. Eng. Biotechnol. News 2019, 39, 42–44. [Google Scholar] [CrossRef]
- Jia, S.; Zhang, R.; Li, Z.; Li, J. Clinical and biological significance of circulating tumor cells, circulating tumor DNA, and exosomes as biomarkers in colorectal cancer. Oncotarget 2017, 8, 55632. [Google Scholar] [CrossRef]
- Iqbal, M.A.; Arora, S.; Prakasam, G.; Calin, G.A.; Syed, M.A. MicroRNA in lung cancer: Role, mechanisms, pathways and therapeutic relevance. Mol. Asp. Med. 2019, 70, 3–20. [Google Scholar] [CrossRef]
- Li, L.; Fu, K.; Zhou, W.; Snyder, M. Applying circulating tumor DNA methylation in the diagnosis of lung cancer. Precis. Clin. Med. 2019, 2, 45–56. [Google Scholar] [CrossRef]
- Moss, J.; Magenheim, J.; Neiman, D.; Zemmour, H.; Loyfer, N.; Korach, A.; Samet, Y.; Maoz, M.; Druid, H.; Arner, P.; et al. Comprehensive human cell-type methylation atlas reveals origins of circulating cell-free DNA in health and disease. Nat. Commun. 2018, 9, 5068. [Google Scholar] [CrossRef] [PubMed]
- Sun, Q.; Zhao, W.; Wang, L.; Guo, F.; Song, D.; Zhang, Q.; Zhang, D.; Fan, Y.; Wang, J. Integration of metabolomic and transcriptomic profiles to identify biomarkers in serum of lung cancer. J. Cell. Biochem. 2019, 120, 11981–11989. [Google Scholar] [CrossRef] [PubMed]
- Tang, Y.; Li, Z.; Lazar, L.; Fang, Z.; Tang, C.; Zhao, J. Metabolomics workflow for lung cancer: Discovery of biomarkers. Clin. Chim. Acta 2019, 495, 436–445. [Google Scholar] [CrossRef] [PubMed]
- Seow, W.J.; Shu, X.-O.; Nicholson, J.; Holmes, E.; Walker, D.I.; Hu, W.; Cai, Q.; Gao, Y.-T.; Xiang, Y.-B.; Moore, S.C.; et al. Association of Untargeted Urinary Metabolomics and Lung Cancer Risk Among Never-Smoking Women in China. JAMA Netw. Open 2019, 2, e1911970. [Google Scholar] [CrossRef]
- Yokota, H.; Guo, J.; Matoba, M.; Higashi, K.; Tonami, H.; Nagao, Y. Lactate, choline, and creatine levels measured by vitro 1H-MRS as prognostic parameters in patients with non-small-cell lung cancer. J. Magn. Reson. Imaging Off. J. Int. Soc. Magn. Reson. Med. 2007, 25, 992–999. [Google Scholar]
- Chapman, C.; Murray, A.; Chakrabarti, J.; Thorpe, A.; Woolston, C.; Sahin, U.; Barnes, A.; Robertson, J. Autoantibodies in breast cancer: Their use as an aid to early diagnosis. Ann. Oncol. 2007, 18, 868–873. [Google Scholar] [CrossRef] [PubMed]
- Du, Q.; Yu, R.; Wang, H.; Yan, N.; Yuan, Q.; Ma, Y.; Slamon, D.; Hou, D.; Wang, H.; Wang, Q. Significance of tumor-associated autoantibodies in the early diagnosis of lung cancer. Clin. Respir. J. 2018, 12, 2020–2028. [Google Scholar] [CrossRef] [PubMed]
- Luo, W.; Rao, M.; Qu, J.; Luo, D. Applications of liquid biopsy in lung cancer-diagnosis, prognosis prediction, and disease monitoring. Am. J. Transl. Res. 2018, 10, 3911. [Google Scholar] [PubMed]
- Miyanaga, A.; Masuda, M.; Yamada, T. Biomarkers of Lung Cancer: Liquid Biopsy Comes of Age. In Biomarkers in Cancer Therapy; Springer: Berlin/Heidelberg, Germany, 2019; pp. 105–113. [Google Scholar]
- Diaz, L.A., Jr.; Bardelli, A. Liquid biopsies: Genotyping circulating tumor DNA. J. Clin. Oncol. Off. J. Am. Soc. Clin. Oncol. 2014, 32, 579. [Google Scholar] [CrossRef]
- Davis, A.A.; Cristofanilli, M. Detection of Predictive Biomarkers Using Liquid Biopsies. In Predictive Biomarkers in Oncology; Springer: Berlin/Heidelberg, Germany, 2019; pp. 107–117. [Google Scholar]
- Gale, D.; Lawson, A.R.; Howarth, K.; Madi, M.; Durham, B.; Smalley, S.; Calaway, J.; Blais, S.; Jones, G.; Clark, J.; et al. Development of a highly sensitive liquid biopsy platform to detect clinically-relevant cancer mutations at low allele fractions in cell-free DNA. PLoS ONE 2018, 13, e0194630. [Google Scholar] [CrossRef]
- Revelo, A.E.; Martin, A.; Velasquez, R.; Kulandaisamy, P.C.; Bustamante, J.; Keshishyan, S.; Otterson, G. Liquid biopsy for lung cancers: An update on recent developments. Ann. Transl. Med. 2019, 7, 349. [Google Scholar] [CrossRef]
- Jee, J.; Lebow, E.S.; Murciano-Goroff, Y.R.; Jayakumaran, G.; Shen, R.; Brannon, A.R.; Benayed, R.; Namakydoust, A.; Offin, M.; Paik, P.K.; et al. Overall Survival with Circulating Tumor DNA-Guided Therapy in Advanced Non-Small Cell Lung Cancer; Wolters Kluwer Health: Philadelphia, PA, USA, 2021. [Google Scholar]
- Shendure, J.; Ji, H. Next-generation DNA sequencing. Nat. Biotechnol. 2008, 26, 1135–1145. [Google Scholar] [CrossRef]
- Wong, W.H.; Tong, R.S.; Young, A.L.; Druley, T.E. Rare Event Detection Using Error-corrected DNA and RNA Sequencing. J. Vis. Exp. 2018, e57509. [Google Scholar] [CrossRef]
- Teder, H.; Koel, M.; Paluoja, P.; Jatsenko, T.; Rekker, K.; Laisk-Podar, T.; Kukuškina, V.; Velthut-Meikas, A.; Fjodorova, O.; Peters, M.; et al. TAC-seq: Targeted DNA and RNA sequencing for precise biomarker molecule counting. NPJ Genom. Med. 2018, 3, 34. [Google Scholar] [CrossRef]
- Villaflor, V.; Won, B.; Nagy, R.; Banks, K.; Lanman, R.B.; Talasaz, A.; Salgia, R. Biopsy-free circulating tumor DNA assay identifies actionable mutations in lung cancer. Oncotarget 2016, 7, 66880. [Google Scholar] [PubMed]
- Sabari, J.K.; Offin, M.; Stephens, D.; Ni, A.; Lee, A.; Pavlakis, N.; Clarke, S.; Diakos, C.I.; Datta, S.; Tandon, N.; et al. A Prospective Study of Circulating Tumor DNA to Guide Matched Targeted Therapy in Lung Cancers. Gynecol. Oncol. 2019, 111, 575–583. [Google Scholar] [CrossRef] [PubMed]
- FuFaD, A. Aprovação Pré-Comercialização P150044, Teste de Mutação Cobas EGFR V2. 2017 FD. 2017. Available online: https://www.accessdata.fda.gov/cdrh_docs/pdf12/p120019s007c.pdf (accessed on 1 September 2022).
- Torres, S.; González, Á.; Tomás, A.J.C.; Fariñas, S.C.; Ferrero, M.; Mirda, D.; Sirera, R.; Jantus-Lewintre, E.; Camps, C. A profile on cobas® EGFR Mutation Test v2 as companion diagnostic for first-line treatment of patients with non-small cell lung cancer. Expert Rev. Mol. Diagn. 2020, 20, 575–582. [Google Scholar]
- Vrba, L.; Oshiro, M.M.; Kim, S.S.; Garland, L.L.; Placencia, C.; Mahadevan, D.; Nelson, M.A.; Futscher, B.W. DNA methylation biomarkers discovered in silico detect cancer in liquid biopsies from non-small cell lung cancer patients. Epigenetics 2020, 15, 419–430. [Google Scholar] [PubMed]
- Duruisseaux, M.; Esteller, M. (Eds.) Lung cancer epigenetics: From knowledge to applications. In Seminars in Cancer Biology; Elsevier: Amsterdam, The Netherlands, 2018. [Google Scholar]
- Bjaanaes, M.M.; Fleischer, T.; Halvorsen, A.R.; Daunay, A.; Busato, F.; Solberg, S.; Jørgensen, L.; Kure, E.; Edvardsen, H.; Børresen-Dale, A.-L.; et al. Genome-wide DNA methylation analyses in lung adenocarcinomas: Association with EGFR, KRAS and TP53 mutation status, gene expression and prognosis. Mol. Oncol. 2016, 10, 330–343. [Google Scholar] [PubMed]
- Wang, J.; Duan, Y.; Meng, Q.-H.; Gong, R.; Guo, C.; Zhao, Y.; Zhang, Y. Integrated analysis of DNA methylation profiling and gene expression profiling identifies novel markers in lung cancer in Xuanwei, China. PLoS ONE 2018, 13, e0203155. [Google Scholar]
- Ooki, A.; Maleki, Z.; Tsay, J.-C.J.; Goparaju, C.; Brait, M.; Turaga, N.; Nam, H.-S.; Rom, W.N.; Pass, H.I.; Sidransky, D.; et al. A Panel of Novel Detection and Prognostic Methylated DNA Markers in Primary Non–Small Cell Lung Cancer and Serum DNA. Clin. Cancer Res. 2017, 23, 7141–7152. [Google Scholar]
- Yang, Z.; Qi, W.; Sun, L.; Zhou, H.; Zhou, B.; Hu, Y. DNA methylation analysis of selected genes for the detection of early-stage lung cancer using circulating cell-free DNA. Adv. Clin. Exp. Med. Off. Organ Wroc. Med. Univ. 2019, 28, 355–360. [Google Scholar] [CrossRef]
- Pérez-Ramírez, C.; Cañadas-Garre, M.; Robles, A.I.; Molina, M.; Faus-Dáder, M.J.; Calleja-Hernández, M. Liquid biopsy in early stage lung cancer. Transl. Lung Cancer Res. 2016, 5, 517. [Google Scholar] [CrossRef]
- Hulbert, A.; Jusue-Torres, I.; Stark, A.; Chen, C.; Rodgers, K.; Lee, B.; Griffin, C.; Yang, A.; Huang, P.; Wrangle, J.; et al. Early Detection of Lung Cancer Using DNA Promoter Hypermethylation in Plasma and SputumEpigenetic Lung Cancer Screening. Clin. Cancer Res. 2017, 23, 1998–2005. [Google Scholar]
- Liu, M.C.; Oxnard, G.R.; Klein, E.A.; Swanton, C.; Seiden, M.V.; CCGA Consortium. Sensitive and specific multi-cancer detection and localization using methylation signatures in cell-free DNA. Ann. Oncol. 2020, 31, 745–759. [Google Scholar] [PubMed]
- Klein, E.; Richards, D.; Cohn, A.; Tummala, M.; Lapham, R.; Cosgrove, D.; Chung, G.; Clement, J.; Gao, J.; Hunkapiller, N.; et al. Clinical validation of a targeted methylation-based multi-cancer early detection test using an independent validation set. Ann. Oncol. 2021, 32, 1167–1177. [Google Scholar] [PubMed]
- Neal, R.D.; Johnson, P.; Clarke, C.A.; Hamilton, S.A.; Zhang, N.; Kumar, H.; Swanton, C.; Sasieni, P. Cell-Free DNA–Based Multi-Cancer Early Detection Test in an Asymptomatic Screening Population (NHS-Galleri): Design of a Pragmatic, Prospective Randomised Controlled Trial. Cancers 2022, 14, 4818. [Google Scholar] [PubMed]
- Poggiana, C.; Rossi, E.; Zamarchi, R. Possible role of circulating tumor cells in early detection of lung cancer. J. Thorac. Dis. 2020, 12, 3821. [Google Scholar] [CrossRef]
- Santarpia, M.; Liguori, A.; Karachaliou, N.; Gonzalez-Cao, M.; Daffinà, M.G.; D’Aveni, A.; Marabello, G.; Altavilla, G.; Rosell, R. Osimertinib in the treatment of non-small-cell lung cancer: Design, development and place in therapy. Lung Cancer Targets Ther. 2017, 8, 109. [Google Scholar] [CrossRef]
- He, Y.; Shi, J.; Schmidt, B.; Liu, Q.; Shi, G.; Xu, X.; Liu, C.; Gao, Z.; Guo, T.; Shan, B. Circulating Tumor Cells as a Biomarker to Assist Molecular Diagnosis for Early Stage Non-Small Cell Lung Cancer. Cancer Manag. Res. 2020, 12, 841. [Google Scholar] [CrossRef]
- Correnti, M.; Raggi, C. Stem-like plasticity and heterogeneity of circulating tumor cells: Current status and prospect challenges in liver cancer. Oncotarget 2017, 8, 7094. [Google Scholar]
- Zhang, Y.; Wang, Y.; Du, Z.; Wu, M.; Zhang, G. Detection of micrometastases in lung cancer with magnetic nanoparticles and quantum dots. Int. J. Nanomed. 2012, 7, 2315. [Google Scholar] [CrossRef]
- Tu, Q.; Wu, X.; Le Rhun, E.; Blonski, M.; Wittwer, B.; Taillandier, L.; Bittencourt, M.D.C.; Faure, G.C. CellSearch® technology applied to the detection and quantification of tumor cells in CSF of patients with lung cancer leptomeningeal metastasis. Lung Cancer 2015, 90, 352–357. [Google Scholar]
- Hofman, V.; Long, E.; Ilie, M.; Bonnetaud, C.; Vignaud, J.M.; Fléjou, J.F.; Lantuejoul, S.; Piaton, E.; Mourad, N.; Butori, C.; et al. Morphological analysis of circulating tumour cells in patients undergoing surgery for non-small cell lung carcinoma using the isolation by size of epithelial tumour cell (ISET) method. Cytopathology 2012, 23, 30–38. [Google Scholar] [CrossRef]
- Turetta, M.; Bulfoni, M.; Brisotto, G.; Fasola, G.; Zanello, A.; Biscontin, E.; Mariuzzi, L.; Steffan, A.; Di Loreto, C.; Cesselli, D.; et al. Assessment of the Mutational Status of NSCLC Using Hypermetabolic Circulating Tumor Cells. Cancers 2018, 10, 270. [Google Scholar] [PubMed]
- Reclusa, P.; Taverna, S.; Pucci, M.; Durendez, E.; Calabuig, S.; Manca, P.; Serrano, M.J.; Sober, L.; Pauwels, P.; Russo, A.; et al. Exosomes as diagnostic and predictive biomarkers in lung cancer. J. Thorac. Dis. 2017, 9 (Suppl. S13), S1373–S1382. [Google Scholar] [CrossRef] [PubMed]
- Vanni, I.; Alama, A.; Grossi, F.; Dal Bello, M.G.; Coco, S. Exosomes: A new horizon in lung cancer. Drug Discov. Today 2017, 22, 927–936. [Google Scholar] [PubMed]
- Momen-Heravi, F.; Balaj, L.; Alian, S.; Mantel, P.-Y.; Halleck, A.E.; Trachtenberg, A.J.; Soria, C.E.; Oquin, S.; Bonebreak, C.M.; Saracoglu, E.; et al. Current methods for the isolation of extracellular vesicles. Biol. Chem. 2013, 394, 1253–1262. [Google Scholar]
- Böing, A.N.; van der Pol, E.; Grootemaat, A.E.; Coumans, F.A.W.; Sturk, A.; Nieuwland, R. Single-step isolation of extracellular vesicles by size-exclusion chromatography. J. Extracell. Vesicles 2014, 3, 23430. [Google Scholar]
- Qu, X.; Li, Q.; Yang, J.; Zhao, H.; Wang, F.; Zhang, F.; Zhang, S.; Zhang, H.; Wang, R.; Wang, Q.; et al. Double-Stranded DNA in Exosomes of Malignant Pleural Effusions as a Novel DNA Source for EGFR Mutation Detection in Lung Adenocarcinoma. Front. Oncol. 2019, 9, 931. [Google Scholar]
- Siravegna, G.; Marsoni, S.; Siena, S.; Bardelli, A. Integrating liquid biopsies into the management of cancer. J. Clin. Oncol. Off. J. Am. Soc. Clin. Oncol. 2017, 14, 531–548. [Google Scholar]
- Rodríguez, M.; Silva, J.; López-Alfonso, A.; López-Muñiz, M.B.; Peña, C.; Domínguez, G.; García, J.M.; López-Gónzalez, A.; Méndez, M.; Provencio, M.; et al. Different exosome cargo from plasma/bronchoalveolar lavage in non-small-cell lung cancer. Genes Chromosom. Cancer 2014, 53, 713–724. [Google Scholar]
- Rabinowits, G.; Gerçel-Taylor, C.; Day, J.M.; Taylor, D.D.; Kloecker, G.H. Exosomal MicroRNA: A Diagnostic Marker for Lung Cancer. Clin. Lung Cancer 2009, 10, 42–46. [Google Scholar] [CrossRef]
- Tomasetti, M.; Lee, W.; Santarelli, L.; Neuzil, J. Exosome-derived microRNAs in cancer metabolism: Possible implications in cancer diagnostics and therapy. Exp. Mol. Med. 2017, 49, e285. [Google Scholar]
- Galka-Marciniak, P.; Urbanek-Trzeciak, M.O.; Nawrocka, P.M.; Dutkiewicz, A.; Giefing, M.; Lewandowska, M.A.; Kozlowski, P. Somatic Mutations in miRNA Genes in Lung Cancer—Potential Functional Consequences of Non-Coding Sequence Variants. Cancers 2019, 11, 793. [Google Scholar] [PubMed]
- Almeida, M.I.; Reis, R.M.; Calin, G.A. MicroRNA history: Discovery, recent applications, and next frontiers. Mutat. Res./Fundam. Mol. Mech. Mutagen. 2011, 717, 1–8. [Google Scholar]
- Mitchell, P.S.; Parkin, R.K.; Kroh, E.M.; Fritz, B.R.; Wyman, S.K.; Pogosova-Agadjanyan, E.L.; Peterson, A.; Noteboom, J.; O’Briant, K.C.; Allen, A.; et al. Circulating microRNAs as stable blood-based markers for cancer detection. Proc. Natl. Acad. Sci. USA 2008, 105, 10513–10518. [Google Scholar] [CrossRef] [PubMed]
- Puerta-Gil, P.; García-Baquero, R.; Jia, A.Y.; Ocaña, S.; Alvarez-Múgica, M.; Alvarez-Ossorio, J.L.; Cordon-Cardo, C.; Cava, F.; Sánchez-Carbayo, M. miR-143, miR-222, and miR-452 Are Useful as Tumor Stratification and Noninvasive Diagnostic Biomarkers for Bladder Cancer. Am. J. Pathol. 2012, 180, 1808–1815. [Google Scholar] [CrossRef]
- Zhi, F.; Cao, X.; Xie, X.; Wang, B.; Dong, W.; Gu, W.; Ling, Y.; Wang, R.; Yang, Y.; Liu, Y. Identification of Circulating MicroRNAs as Potential Biomarkers for Detecting Acute Myeloid Leukemia. PLoS ONE 2013, 8, e56718. [Google Scholar]
- Reis, P.P.; Drigo, S.A.; Carvalho, R.F.; Lapa, R.M.L.; Felix, T.F.; Patel, D.; Cheng, D.; Pintilie, M.; Liu, G.; Tsao, M.-S. Circulating miR-16-5p, miR-92a-3p, and miR-451a in Plasma from Lung Cancer Patients: Potential Application in Early Detection and a Regulatory Role in Tumorigenesis Pathways. Cancers 2020, 12, 2071. [Google Scholar] [CrossRef]
- Wozniak, M.B.; Scelo, G.; Muller, D.C.; Mukeria, A.; Zaridze, D.; Brennan, P. Circulating MicroRNAs as Non-Invasive Biomarkers for Early Detection of Non-Small-Cell Lung Cancer. PLoS ONE 2015, 10, e0125026. [Google Scholar]
- Huang, J.; Wu, J.; Li, Y.; Li, X.; Yang, T.; Yang, Q.; Jiang, Y. Deregulation of Serum MicroRNA Expression Is Associated with Cigarette Smoking and Lung Cancer. BioMed Res. Int. 2014, 2014, 364316. [Google Scholar] [CrossRef]
- Zhao, K.; Cheng, J.; Chen, B.; Liu, Q.; Xu, D.; Zhang, Y. Circulating microRNA-34 family low expression correlates with poor prognosis in patients with non-small cell lung cancer. J. Thorac. Dis. 2017, 9, 3735. [Google Scholar] [CrossRef]
- Arnaiz, E.; Sole, C.; Manterola, L.; Iparraguirre, L.; Otaegui, D.; Lawrie, C.H. (Eds.) CircRNAs and cancer: Biomarkers and master regulators. In Seminars in Cancer Biology; Elsevier: Amsterdam, The Netherlands, 2019. [Google Scholar]
- Suzuki, H.; Zuo, Y.; Wang, J.; Zhang, M.Q.; Malhotra, A.; Mayeda, A. Characterization of RNase R-digested cellular RNA source that consists of lariat and circular RNAs from pre-mRNA splicing. Nucleic Acids Res. 2006, 34, e63. [Google Scholar] [CrossRef]
- Vincent, H.A.; Deutscher, M.P. Substrate Recognition and Catalysis by the Exoribonuclease RNase R. J. Biol. Chem. 2006, 281, 29769–29775. [Google Scholar] [CrossRef] [PubMed]
- Salzman, J.; Chen, R.E.; Olsen, M.N.; Wang, P.L.; Brown, P.O. Cell-type specific features of circular RNA expression. PLoS Genet. 2013, 9, e1003777. [Google Scholar] [CrossRef]
- Jeck, W.R.; Sharpless, N.E. Detecting and characterizing circular RNAs. Nat. Biotechnol. 2014, 32, 453–461. [Google Scholar] [CrossRef]
- De Fraipont, F.; Gazzeri, S.; Cho, W.C.; Eymin, B. Circular RNAs and RNA splice variants as biomarkers for prognosis and therapeutic response in the liquid biopsies of lung cancer patients. Front. Genet. 2019, 10, 390. [Google Scholar] [PubMed]
- Pedraz-Valdunciel, C.; Giannoukakos, S.; Potie, N.; Giménez-Capitán, A.; Huang, C.; Hackenberg, M.; Fernandez-Hilario, A.; Bracht, J.; Filipska, M.; Aldeguer, E.; et al. Digital multiplexed analysis of circular RNAs in FFPE and fresh non-small cell lung cancer specimens. Mol. Oncol. 2022, 16, 2367–2383. [Google Scholar] [CrossRef]
- Chen, K.-Z.; Lou, F.; Yang, F.; Zhang, J.-B.; Ye, H.; Chen, W.; Guan, T.; Zhao, M.-Y.; Su, X.-X.; Shi, R.; et al. Circulating Tumor DNA Detection in Early-Stage Non-Small Cell Lung Cancer Patients by Targeted Sequencing. Sci. Rep. 2016, 6, 31985. [Google Scholar] [CrossRef]
- Zhu, X.; Wang, X.; Wei, S.; Chen, Y.; Chen, Y.; Fan, X.; Han, S.; Wu, G. hsa_circ_0013958: A circular RNA and potential novel biomarker for lung adenocarcinoma. FEBS J. 2017, 284, 2170–2182. [Google Scholar] [CrossRef]
- Zhang, X.; Zhou, H.; Jing, W.; Luo, P.; Qiu, S.; Liu, X.; Zhu, M.; Liang, C.; Yu, M.; Tu, J. The Circular RNA hsa_circ_0001445 Regulates the Proliferation and Migration of Hepatocellular Carcinoma and May Serve as a Diagnostic Biomarker. Dis. Markers 2018, 2018, 3073467. [Google Scholar] [CrossRef]
- Hang, D.; Zhou, J.; Qin, N.; Zhou, W.; Ma, H.; Jin, G.; Hu, Z.; Dai, J.; Shen, H. A novel plasma circular RNA circFARSA is a potential biomarker for non-small cell lung cancer. Cancer Med. 2018, 7, 2783–2791. [Google Scholar] [CrossRef]
- Yu, L.; Li, K.; Zhang, X. Next-generation metabolomics in lung cancer diagnosis, treatment and precision medicine: Mini review. Oncotarget 2017, 8, 115774. [Google Scholar] [CrossRef]
- Kumar, A.; Misra, B.B. Challenges and Opportunities in Cancer Metabolomics. Proteomics 2019, 19, e1900042. [Google Scholar] [CrossRef] [PubMed]
- Molina, R.; Filella, X.; Augé, J.; Fuentes, R.; Bover, I.; Rifa, J.; Moreno, V.; Canals, E.; Viñolas, N.; Marquez, A.; et al. Tumor Markers (CEA, CA 125, CYFRA 21-1, SCC and NSE) in Patients with Non-Small Cell Lung Cancer as an Aid in Histological Diagnosis and Prognosis. Tumor Biol. 2003, 24, 209–218. [Google Scholar] [CrossRef] [PubMed]
- Barlési, F.; Gimenez, C.; Torre, J.-P.; Doddoli, C.; Mancini, J.; Greillier, L.; Roux, F.; Kleisbauer, J.-P. Prognostic value of combination of Cyfra 21-1, CEA and NSE in patients with advanced non-small cell lung cancer. Respir. Med. 2004, 98, 357–362. [Google Scholar] [CrossRef]
- Pang, W.W.; Abdul-Rahman, P.S.; Wan-Ibrahim, W.I.; Hashim, O.H. Can the acute-phase reactant proteins be used as cancer biomarkers? Int. J. Biol. Markers 2010, 25, 1–11. [Google Scholar] [CrossRef]
- Kang, S.-M.; Sung, H.-J.; Ahn, J.-M.; Park, J.-Y.; Lee, S.-Y.; Park, C.-S.; Cho, J.-Y. The Haptoglobin β chain as a supportive biomarker for human lung cancers. Mol. Biosyst. 2011, 7, 1167–1175. [Google Scholar] [CrossRef] [PubMed]
- Sung, H.-J.; Ahn, J.-M.; Yoon, Y.-H.; Rhim, T.-Y.; Park, C.-S.; Park, J.-Y.; Lee, S.-Y.; Kim, J.-W.; Cho, J.-Y. Identification and Validation of SAA as a Potential Lung Cancer Biomarker and its Involvement in Metastatic Pathogenesis of Lung Cancer. J. Proteome Res. 2011, 10, 1383–1395. [Google Scholar] [CrossRef] [PubMed]
- Jensen, L.; Whitehead, A.S. Regulation of serum amyloid A protein expression during the acute-phase response. Biochem. J. 1998, 334, 489–503. [Google Scholar] [CrossRef]
- De Petris, L.; Orre, L.M.; Kanter, L.; Pernemalm, M.; Koyi, H.; Lewensohn, R.; Lehtiö, J. Tumor expression of S100A6 correlates with survival of patients with stage I non-small-cell lung cancer. Lung Cancer 2009, 63, 410–417. [Google Scholar] [CrossRef] [PubMed]
- Pujol, J.L.; Grenier, J.; Daurès, J.P.; Daver, A.; Pujol, H.; Michel, F.B. Serum fragment of cytokeratin subunit 19 measured by CYFRA 21-1 immunoradiometric assay as a marker of lung cancer. Cancer Res. 1993, 53, 61–66. [Google Scholar]
- Bergqvist, M.; Brattström, D.; Hesselius, P.; Wiklund, B.; Silen, A.; Wagenius, G.; Brodin, O. Cytokeratin 8 and 18 fragments measured in serum and their relation to survival in patients with non-small cell lung cancer. Anticancer Res. 1999, 19, 1833–1836. [Google Scholar]
- Xu, B.J.; Gonzalez, A.L.; Kikuchi, T.; Yanagisawa, K.; Massion, P.P.; Wu, H.; Mason, S.E.; Olson, S.J.; Shyr, Y.; Carbone, D.P.; et al. MALDI-MS derived prognostic protein markers for resected non-small cell lung cancer. Proteom. Clin. Appl. 2008, 2, 1508–1517. [Google Scholar] [CrossRef] [PubMed]
- Cohen, J.D.; Li, L.; Wang, Y.; Thoburn, C.; Afsari, B.; Danilova, L.; Douville, C.; Javed, A.A.; Wong, F.; Mattox, A.; et al. Detection and localization of surgically resectable cancers with a multi-analyte blood test. Science 2018, 359, 926–930. [Google Scholar] [CrossRef] [PubMed]
- Zamay, T.N.; Zamay, G.S.; Kolovskaya, O.S.; Zukov, R.A.; Petrova, M.M.; Gargaun, A.; Berezovski, M.V.; Kichkailo, A.S. Current and Prospective Protein Biomarkers of Lung Cancer. Cancers 2017, 9, 155. [Google Scholar] [CrossRef]
- Alcala, K.; Guida, F.; Johansson, M.; Johansson, M.; Robbins, H.A.; Smith-Byrne, K.; Stevens, V.; Zahed, H. Lung Cancer Cohort Consortium The Blood Proteome of Imminent Lung Cancer Diagnosis. medRxiv 2022. [Google Scholar] [CrossRef]
- Mehan, M.R.; Williams, S.A.; Siegfried, J.M.; Bigbee, W.L.; Weissfeld, J.L.; Wilson, D.O.; Pass, H.I.; Rom, W.N.; Muley, T.; Meister, M.; et al. Validation of a blood protein signature for non-small cell lung cancer. Clin. Proteom. 2014, 11, 32. [Google Scholar] [CrossRef] [PubMed]
- Boyle, P.; Chapman, C.J.; Holdenrieder, S.; Murray, A.; Robertson, C.; Wood, W.C.; Maddison, P.; Healey, G.; Fairley, G.H.; Barnes, A.C.; et al. Clinical validation of an autoantibody test for lung cancer. Ann. Oncol. 2011, 22, 383–389. [Google Scholar] [CrossRef]
- Tan, E.M. Autoantibodies as reporters identifying aberrant cellular mechanisms in tumorigenesis. J. Clin. Investig. 2001, 108, 1411–1415. [Google Scholar] [CrossRef]
- Houghton, A.N. Cancer antigens: Immune recognition of self and altered self. J. Exp. Med. 1994, 180, 1–4. [Google Scholar] [CrossRef]
- Tan, E.M.; Zhang, J. Autoantibodies to tumor-associated antigens: Reporters from the immune system. Immunol. Rev. 2008, 222, 328–340. [Google Scholar] [CrossRef]
- Zhang, J.-Y.; Tan, E.M. Autoantibodies to tumor-associated antigens as diagnostic biomarkers in hepatocellular carcinoma and other solid tumors. Expert Rev. Mol. Diagn. 2010, 10, 321–328. [Google Scholar] [CrossRef]
- Veronesi, G.; Bianchi, F.; Infante, M.; Alloisio, M. The challenge of small lung nodules identified in CT screening: Can biomarkers assist diagnosis? Biomark. Med. 2016, 10, 137–143. [Google Scholar] [CrossRef] [PubMed]
- Dai, L.; Tsay, J.-C.J.; Li, J.; Yie, T.-A.; Munger, J.S.; Pass, H.; Rom, W.N.; Zhang, Y.; Tan, E.M.; Zhang, J.-Y. Autoantibodies against tumor-associated antigens in the early detection of lung cancer. Lung Cancer 2016, 99, 172–179. [Google Scholar] [CrossRef] [PubMed]
- Burotto, M.; Thomas, A.; Subramaniam, D.; Giaccone, G.; Rajan, A. Biomarkers in Early-Stage Non–Small-Cell Lung Cancer: Current Concepts and Future Directions. J. Thorac. Oncol. 2014, 9, 1609–1617. [Google Scholar] [CrossRef] [PubMed]
- Chen, H.; Werner, S.; Tao, S.; Zörnig, I.; Brenner, H. Blood autoantibodies against tumor-associated antigens as biomarkers in early detection of colorectal cancer. Cancer Lett. 2014, 346, 178–187. [Google Scholar] [CrossRef]
- Lubin, R.; Zalcman, G.; Bouchet, L.; Trédaniel, J.; Legros, Y.; Cazals, D.; Hirsch, A.; Soussi, T. Serum p53 antibodies as early markers of lung cancer. Nat. Med. 1995, 1, 701–702. [Google Scholar] [CrossRef] [PubMed]
- Murray, A.; Chapman, C.; Healey, G.; Peek, L.; Parsons, G.; Baldwin, D.; Barnes, A.; Sewell, H.; Fritsche, H.; Robertson, J. Technical validation of an autoantibody test for lung cancer. Ann. Oncol. 2010, 21, 1687–1693. [Google Scholar] [CrossRef]
- Farlow, E.C.; Patel, K.; Basu, S.; Lee, B.S.; Kim, A.W.; Coon, J.S.; Faber, L.P.; Bonomi, P.; Liptay, M.J.; Borgia, J.A. Development of a Multiplexed Tumor-Associated Autoantibody-Based Blood Test for the Detection of Non–Small Cell Lung CancerEarly Detection Serum Test for NSCLC. Clin. Cancer Res. 2010, 16, 3452–3462. [Google Scholar] [CrossRef]
- Wu, L.; Chang, W.; Zhao, J.; Yu, Y.; Tan, X.; Su, T.; Zhao, L.; Huang, S.; Liu, S.; Cao, G. Development of Autoantibody Signatures as Novel Diagnostic Biomarkers of Non–Small Cell Lung CancerNew Serum Diagnostic Biomarkers for NSCLC. Clin. Cancer Res. 2010, 16, 3760–3768. [Google Scholar] [CrossRef]
- Lam, S.; Boyle, P.; Healey, G.F.; Maddison, P.; Peek, L.; Murray, A.; Chapman, C.J.; Allen, J.; Wood, W.C.; Sewell, H.F.; et al. EarlyCDT-Lung: An Immunobiomarker Test as an Aid to Early Detection of Lung Cancer. Cancer Prev. Res. 2011, 4, 1126–1134. [Google Scholar] [CrossRef]
- Wang, T.; Liu, H.; Pei, L.; Wang, K.; Song, C.; Wang, P.; Ye, H.; Zhang, J.; Ji, Z.; Ouyang, S.; et al. Screening of tumor-associated antigens based on Oncomine database and evaluation of diagnostic value of autoantibodies in lung cancer. Clin. Immunol. 2020, 210, 108262. [Google Scholar] [CrossRef]
- Jiang, D.; Wang, Y.; Liu, M.; Si, Q.; Wang, T.; Pei, L.; Wang, P.; Ye, H.; Shi, J.; Wang, X.; et al. A panel of autoantibodies against tumor-associated antigens in the early immunodiagnosis of lung cancer. Immunobiology 2020, 225, 151848. [Google Scholar] [CrossRef]
- Li, R.-Y.; Liang, Z.-Y. Circulating tumor DNA in lung cancer: Real-time monitoring of disease evolution and treatment response. Chin. Med. J. 2020, 133, 2476–2485. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Z.; Xiao, Y.; Zhao, J.; Chen, M.; Xu, Y.; Zhong, W.; Xing, J.; Wang, M. Relationship between circulating tumour cell count and prognosis following chemotherapy in patients with advanced non-small-cell lung cancer. Respirology 2016, 21, 519–525. [Google Scholar] [CrossRef] [PubMed]
- Hou, J.-M.; Krebs, M.G.; Lancashire, L.; Sloane, R.; Backen, A.; Swain, R.K.; Priest, L.J.C.; Greystoke, A.; Zhou, C.; Morris, K.; et al. Clinical Significance and Molecular Characteristics of Circulating Tumor Cells and Circulating Tumor Microemboli in Patients With Small-Cell Lung Cancer. J. Clin. Oncol. 2012, 30, 525–532. [Google Scholar] [CrossRef] [PubMed]
- Kim, T.; Kim, E.Y.; Lee, S.H.; Kwon, D.S.; Kim, A.; Chang, Y.S. Presence of mEGFR ctDNA predicts a poor clinical outcome in lung adenocarcinoma. Thorac. Cancer 2019, 10, 2267–2273. [Google Scholar] [CrossRef] [PubMed]
- Hur, J.Y.; Lee, J.S.; Kim, I.A.; Kim, H.J.; Kim, W.S.; Lee, K.Y. Extracellular vesicle-based EGFR genotyping in bronchoalveolar lavage fluid from treatment-naive non-small cell lung cancer patients. Transl. Lung Cancer Res. 2019, 8, 1051. [Google Scholar] [CrossRef] [PubMed]
- Michaelidou, K.; Koutoulaki, C.; Mavridis, K.; Vorrias, E.; Papadaki, M.A.; Koutsopoulos, A.V.; Mavroudis, D.; Agelaki, S. Detection of KRAS G12/G13 Mutations in Cell Free-DNA by Droplet Digital PCR, Offers Prognostic Information for Patients with Advanced Non-Small Cell Lung Cancer. Cells 2020, 9, 2514. [Google Scholar] [CrossRef]
- Wen, S.W.C.; Andersen, R.F.; Hansen, T.F.; Nyhus, C.H.; Hager, H.; Hilberg, O.; Jakobsen, A. The prognostic impact of circulating homeobox A9 methylated DNA in advanced non-small cell lung cancer. Transl. Lung Cancer Res. 2021, 10, 855. [Google Scholar] [CrossRef]
- Pavan, A.; Bragadin, A.B.; Calvetti, L.; Ferro, A.; Zulato, E.; Attili, I.; Nardo, G.; Maso, A.D.; Frega, S.; Menin, A.G.; et al. Role of next generation sequencing-based liquid biopsy in advanced non-small cell lung cancer patients treated with immune checkpoint inhibitors: Impact of STK11, KRAS and TP53 mutations and co-mutations on outcome. Transl. Lung Cancer Res. 2021, 10, 202. [Google Scholar] [CrossRef]
- Suzuki, M.; Yoshino, I. Aberrant methylation in non-small cell lung cancer. Surg. Today 2010, 40, 602–607. [Google Scholar] [CrossRef]
- Brock, M.V.; Hooker, C.M.; Ota-Machida, E.; Han, Y.; Guo, M.; Ames, S.; Glöckner, S.; Piantadosi, S.; Gabrielson, E.; Pridham, G.; et al. DNA Methylation Markers and Early Recurrence in Stage I Lung Cancer. N. Engl. J. Med. 2008, 358, 1118–1128. [Google Scholar] [CrossRef] [PubMed]
- Khandelwal, A.; Seam, R.K.; Gupta, M.; Rana, M.K.; Prakash, H.; Vasquez, K.M.; Jain, A. Circulating micro RNA-590-5p functions as a liquid biopsy marker in non-small cell lung cancer. Cancer Sci. 2020, 111, 826–839. [Google Scholar] [CrossRef] [PubMed]
- Boeri, M.; Verri, C.; Conte, D.; Roz, L.; Modena, P.; Facchinetti, F.; Calabrò, E.; Croce, C.M.; Pastorino, U.; Sozzi, G. MicroRNA signatures in tissues and plasma predict development and prognosis of computed tomography detected lung cancer. Proc. Natl. Acad. Sci. USA 2011, 108, 3713–3718. [Google Scholar] [CrossRef]
- Tian, F.; Shen, Y.; Chen, Z.; Li, R.; Lu, J.; Ge, Q. Aberrant miR-181b-5p and miR-486-5p expression in serum and tissue of non-small cell lung cancer. Gene 2016, 591, 338–343. [Google Scholar] [CrossRef]
- Wang, Z.X.; Bian, H.B.; Wang, J.R.; Cheng, Z.X.; Wang, K.M.; De, W. Prognostic significance of serum miRNA-21 expression in human non-small cell lung cancer. J. Surg. Oncol. 2011, 104, 847–851. [Google Scholar] [CrossRef]
- Bianconi, F.; Palumbo, I.; Spanu, A.; Nuvoli, S.; Fravolini, M.L.; Palumbo, B. PET/CT Radiomics in Lung Cancer: An Overview. Appl. Sci. 2020, 10, 1718. [Google Scholar] [CrossRef]
- Yoneda, K.; Imanishi, N.; Ichiki, Y.; Tanaka, F. A liquid biopsy in primary lung cancer. Surg. Today 2019, 49, 1–14. [Google Scholar] [CrossRef]
- Kessler, M.D.; Pawar, N.R.; Martin, S.S.; Antalis, T.M.; O’Connor, T.D. Improving Cancer Detection and Treatment with Liquid Biopsies and ptDNA. Trends Cancer 2018, 4, 643–654. [Google Scholar] [CrossRef]
- Sorensen, B.S.; Wu, L.; Wei, W.; Tsai, J.; Weber, B.; Nexo, E.; Meldgaard, P. Monitoring of epidermal growth factor receptor tyrosine kinase inhibitor-sensitizing and resistance mutations in the plasma DNA of patients with advanced non–small cell lung cancer during treatment with erlotinib. Cancer 2014, 120, 3896–3901. [Google Scholar] [CrossRef]
- Reijans, M.; Gestel, S.; Haes, E.; Vandesteene, C.; Gomez, J.C.; Gouedard, C.; Patera, S.; Murray, S.; Maertens, G. Feasibility study of a ctEGFR prototype assay on the fully automated Idylla™ platform. Ann. Oncol. 2019, 30, v577. [Google Scholar] [CrossRef]
- Heeke, S.; Benzaquen, J.; Hofman, V.; Ilié, M.; Allegra, M.; Long-Mira, E.; Lassalle, S.; Tanga, V.; Salacroup, C.; Bonnetaud, C.; et al. Critical Assessment in Routine Clinical Practice of Liquid Biopsy for EGFR Status Testing in Non–Small-Cell Lung Cancer: A Single-Laboratory Experience (LPCE, Nice, France). Clin. Lung Cancer 2020, 21, 56–65.e8. [Google Scholar] [CrossRef] [PubMed]
- Lee, D.H. Treatments for EGFR-mutant non-small cell lung cancer (NSCLC): The road to a success, paved with failures. Pharmacol. Ther. 2017, 174, 1–21. [Google Scholar] [CrossRef] [PubMed]
- Cabanero, M.; Tsao, M.S. Circulating Tumour DNA in EGFR-Mutant Non-Small-Cell Lung Cancer. Curr. Oncol. 2018, 25, 38–44. [Google Scholar] [CrossRef]
- Ding, P.; Becker, T.; Bray, V.; Chua, W.; Ma, Y.; Lynch, D.; Po, J.; Luk, A.; Caixeiro, N.; de Souza, P.; et al. The predictive and prognostic significance of liquid biopsy in advanced epidermal growth factor receptor-mutated non-small cell lung cancer: A prospective study. Lung Cancer 2019, 134, 187–193. [Google Scholar] [CrossRef] [PubMed]
- Provencio, M.; Torrente, M.; Calvo, V.; Pérez-Callejo, D.; Gutiérrez, L.; Franco, F.; Pérez-Barrios, C.; Barquín, M.; Royuela, A.; García-García, F.; et al. Prognostic value of quantitative ctDNA levels in non small cell lung cancer patients. Oncotarget 2018, 9, 488. [Google Scholar] [CrossRef]
- Pérez-Callejo, D.; Torrente, M.; Romero, A.; Provencio, M. Clinical and Investigational Applications of Liquid Biopsy in Non-Small Cell Lung Cancer. J. Tumor 2016, 4, 461–468. [Google Scholar] [CrossRef]
- Nicolazzo, C.; Raimondi, C.; Mancini, M.; Caponnetto, S.; Gradilone, A.; Gandini, O.; Mastromartino, M.; Del Bene, G.; Prete, A.A.; Longo, F.; et al. Monitoring PD-L1 positive circulating tumor cells in non-small cell lung cancer patients treated with the PD-1 inhibitor Nivolumab. Sci. Rep. 2016, 6, 31726. [Google Scholar] [CrossRef]
- Boffa, D.J.; Graf, R.P.; Salazar, M.C.; Hoag, J.; Lu, D.; Krupa, R.; Louw, J.; Dugan, L.; Wang, Y.; Landers, M.; et al. Cellular Expression of PD-L1 in the Peripheral Blood of Lung Cancer Patients is Associated with Worse Survival. Cancer Epidemiol. Biomark. Prev. 2017, 26, 1139–1145. [Google Scholar] [CrossRef]
- Ilié, M.; Szafer-Glusman, E.; Hofman, V.; Chamorey, E.; Lalvée, S.; Selva, E.; Leroy, S.; Marquette, C.-H.; Kowanetz, M.; Hedge, P.; et al. Detection of PD-L1 in circulating tumor cells and white blood cells from patients with advanced non-small-cell lung cancer. Ann. Oncol. 2018, 29, 193–199. [Google Scholar] [CrossRef]
- Rizvi, N.A.; Hellmann, M.D.; Snyder, A.; Kvistborg, P.; Makarov, V.; Havel, J.J.; Lee, W.; Yuan, J.; Wong, P.; Ho, T.S.; et al. Cancer immunology. Mutational landscape determines sensitivity to PD-1 blockade in non–small cell lung cancer. Science 2015, 348, 124–128. [Google Scholar] [CrossRef]
- Goodman, A.M.; Kato, S.; Bazhenova, L.; Patel, S.P.; Frampton, G.M.; Miller, V.; Stephens, P.J.; Daniels, G.A.; Kurzrock, R. Tumor Mutational Burden as an Independent Predictor of Response to Immunotherapy in Diverse CancersTMB Predicts Response to Immunotherapy in Diverse Cancers. Mol. Cancer Ther. 2017, 16, 2598–2608. [Google Scholar] [CrossRef] [PubMed]
- Singal, G.; Miller, P.; Agarwala, V.; Li, G.; Kaushik, G.; Backenroth, D.; Gossai, A.; Frampton, G.M.; Torres, A.Z.; Lehnert, E.M.; et al. Association of Patient Characteristics and Tumor Genomics With Clinical Outcomes Among Patients With Non–Small Cell Lung Cancer Using a Clinicogenomic Database. JAMA 2019, 321, 1391–1399. [Google Scholar] [CrossRef] [PubMed]
- Gandara, D.R.; Paul, S.M.; Kowanetz, M.; Schleifman, E.; Zou, W.; Li, Y.; Rittmeyer, A.; Fehrenbacher, L.; Otto, G.; Malboeuf, C.; et al. Blood-based tumor mutational burden as a predictor of clinical benefit in non-small-cell lung cancer patients treated with atezolizumab. Nat. Med. 2018, 24, 1441–1448. [Google Scholar] [CrossRef]
- Zill, O.A.; Banks, K.C.; Fairclough, S.R.; Mortimer, S.A.; Vowles, J.V.; Mokhtari, R.; Gandara, D.R.; Mack, P.C.; Odegaard, J.I.; Nagy, R.J.; et al. The Landscape of Actionable Genomic Alterations in Cell-Free Circulating Tumor DNA from 21,807 Advanced Cancer PatientsSomatic Genomic Landscape of Circulating Tumor DNA. Clin. Cancer Res. 2018, 24, 3528–3538. [Google Scholar] [CrossRef]
- Shaw, A.T.; Yeap, B.Y.; Mino-Kenudson, M.; Digumarthy, S.R.; Costa, D.B.; Heist, R.S.; Solomon, B.; Stubbs, H.; Admane, S.; McDermott, U.; et al. Clinical Features and Outcome of Patients With Non–Small-Cell Lung Cancer Who Harbor EML4-ALK. J. Clin. Oncol. 2009, 27, 4247. [Google Scholar] [CrossRef] [PubMed]
- Nilsson, R.J.A.; Karachaliou, N.; Berenguer, J.; Gimenez-Capitan, A.; Schellen, P.; Teixido, C.; Tannous, J.; Kuiper, J.L.; Drees, E.; Grabowska, M.; et al. Rearranged EML4-ALK fusion transcripts sequester in circulating blood platelets and enable blood-based crizotinib response monitoring in non-small-cell lung cancer. Oncotarget 2016, 7, 1066. [Google Scholar] [CrossRef]
- de Koning, H.J.; van der Aalst, C.M.; de Jong, P.A.; Scholten, E.T.; Nackaerts, K.; Heuvelmans, M.A.; Lammers, J.W.J.; Weenink, C.; Yousaf-Khan, U.; Horeweg, N.; et al. Reduced lung-cancer mortality with volume CT screening in a randomized trial. N. Engl. J. Med. 2020, 382, 503–513. [Google Scholar] [CrossRef]
- Hochhegger, B.; Camargo, S.; Teles, G.B.D.S.; Chate, R.C.; Szarf, G.; Guimarães, M.D.; Gross, J.L.; Barbosa, P.N.V.P.; Chiarantano, R.S.; Reis, R.M.; et al. Challenges of Implementing Lung Cancer Screening in a Developing Country: Results of the Second Brazilian Early Lung Cancer Screening Trial (BRELT2). JCO Glob. Oncol. 2022, 8, e2100257. [Google Scholar] [CrossRef]
- Chiarantano, R.S.; Vazquez, F.L.; Franco, A.; Ferreira, L.C.; da Costa, M.C.; Talarico, T.; Oliveira, N.; Miziara, J.E.; Mauad, E.C.; da Silva, E.C.; et al. Implementation of an Integrated Lung Cancer Prevention and Screening Program Using a Mobile Computed Tomography (CT) Unit in Brazil. Cancer Control 2022, 29, 10732748221121385. [Google Scholar] [CrossRef]
- Blandin Knight, S.; Crosbie, P.A.; Balata, H.; Chudziak, J.; Hussell, T.; Dive, C. Progress and prospects of early detection in lung cancer. Open Biol. 2017, 7, 170070. [Google Scholar] [CrossRef] [PubMed]
- Phallen, J.; Sausen, M.; Adleff, V.; Leal, A.; Hruban, C.; White, J.; Anagnostou, V.; Fiksel, J.; Cristiano, S.; Papp, E.; et al. Direct detection of early-stage cancers using circulating tumor DNA. Sci. Transl. Med. 2017, 9, eaan2415. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Z.; Ramnath, N.; Nagrath, S. Current status of CTCs as liquid biopsy in lung cancer and future directions. Front. Oncol. 2015, 5, 209. [Google Scholar] [CrossRef] [PubMed]
- Foss, K.M.; Sima, C.; Ugolini, D.; Neri, M.; Allen, K.E.; Weiss, G.J. miR-1254 and miR-574-5p: Serum-Based microRNA Biomarkers for Early-Stage Non-small Cell Lung Cancer. J. Thorac. Oncol. 2011, 6, 482–488. [Google Scholar] [CrossRef]
- Pastorino, U.; Boeri, M.; Sestini, S.; Sabia, F.; Milanese, G.; Silva, M.; Suatoni, P.; Verri, C.; Cantarutti, A.; Sverzellati, N.; et al. Baseline computed tomography screening and blood microRNA predict lung cancer risk and define adequate intervals in the BioMILD trial. Ann. Oncol. 2022, 33, 395–405. [Google Scholar] [CrossRef] [PubMed]
- Kumar, S.; Gaur, V.; Mir, I.A.; Saikia, J.; Kumar, S. MicroRNA-3692-3p is overexpressed in lung tumors but may not serve as a prognostic biomarker in lung cancer patients. Mol. Biol. Rep. 2022, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Lian, X.; Cao, D.; Hu, X.; Mo, W.; Yao, X.; Mo, J.; Wang, H. Circular RNAs Hsa_circ_101555 and Hsa_circ_008068 as Diagnostic Biomarkers for Early-Stage Lung Adenocarcinoma. Int. J. Gen. Med. 2022, 15, 5579. [Google Scholar] [CrossRef]
- Zhang, Q.; Qin, S.; Peng, C.; Liu, Y.; Huang, Y.; Ju, S. Circulating circular RNA hsa_circ_0023179 acts as a diagnostic biomarker for non-small-cell lung cancer detection. J. Cancer Res. Clin. Oncol. 2022, 1–12. [Google Scholar] [CrossRef]
- Robbins, H.A.; Alcala, K.; Moez, E.K.; Guida, F.; Thomas, S.; Zahed, H.; Warkentin, M.T.; Smith-Byrne, K.; Brhane, Y.; Muller, D.; et al. Design and methodological considerations for biomarker discovery and validation in the Integrative Analysis of Lung Cancer Etiology and Risk (INTEGRAL) Program. Ann. Epidemiol. 2023, 77, 1–12. [Google Scholar] [CrossRef]
- Lennon, A.M.; Buchanan, A.H.; Kinde, I.; Warren, A.; Honushefsky, A.; Cohain, A.T.; Ledbetter, D.H.; Sanfilippo, F.; Sheridan, K.; Rosica, D.; et al. Feasibility of blood testing combined with PET-CT to screen for cancer and guide intervention. Science 2020, 369, eabb9601. [Google Scholar] [CrossRef]
- LaRose, T.L.; Meheus, F.; Brennan, P.; Johansson, M.; Robbins, H.A. Assessment of Biomarker Testing for Lung Cancer Screening Eligibility. JAMA Netw. Open 2020, 3, e200409. [Google Scholar] [CrossRef]
- Vachani, A.; Carroll, N.M.; Simoff, M.J.; Neslund-Dudas, C.; Honda, S.; Greenlee, R.T.; Rendle, K.A.; Burnett-Hartman, A.; Ritzwoller, D.P. Stage Migration and Lung Cancer Incidence After Initiation of Low-Dose Computed Tomography Screening. J. Thorac. Oncol. 2022, 17, 1355–1364. [Google Scholar] [CrossRef] [PubMed]
- Fatumo, S.; Chikowore, T.; Choudhury, A.; Ayub, M.; Martin, A.R.; Kuchenbaecker, K. A roadmap to increase diversity in genomic studies. Nat. Med. 2022, 28, 243–250. [Google Scholar] [CrossRef] [PubMed]
- Saulsberry, L.; Olopade, O.I. Precision oncology: Directing genomics and pharmacogenomics toward reducing cancer inequities. Cancer Cell 2021, 39, 730–733. [Google Scholar] [CrossRef] [PubMed]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).