Utilizing the 1H-15N NMR Methods for the Characterization of Isomeric Human Milk Oligosaccharides
Abstract
1. Introduction
2. Results and Discussion
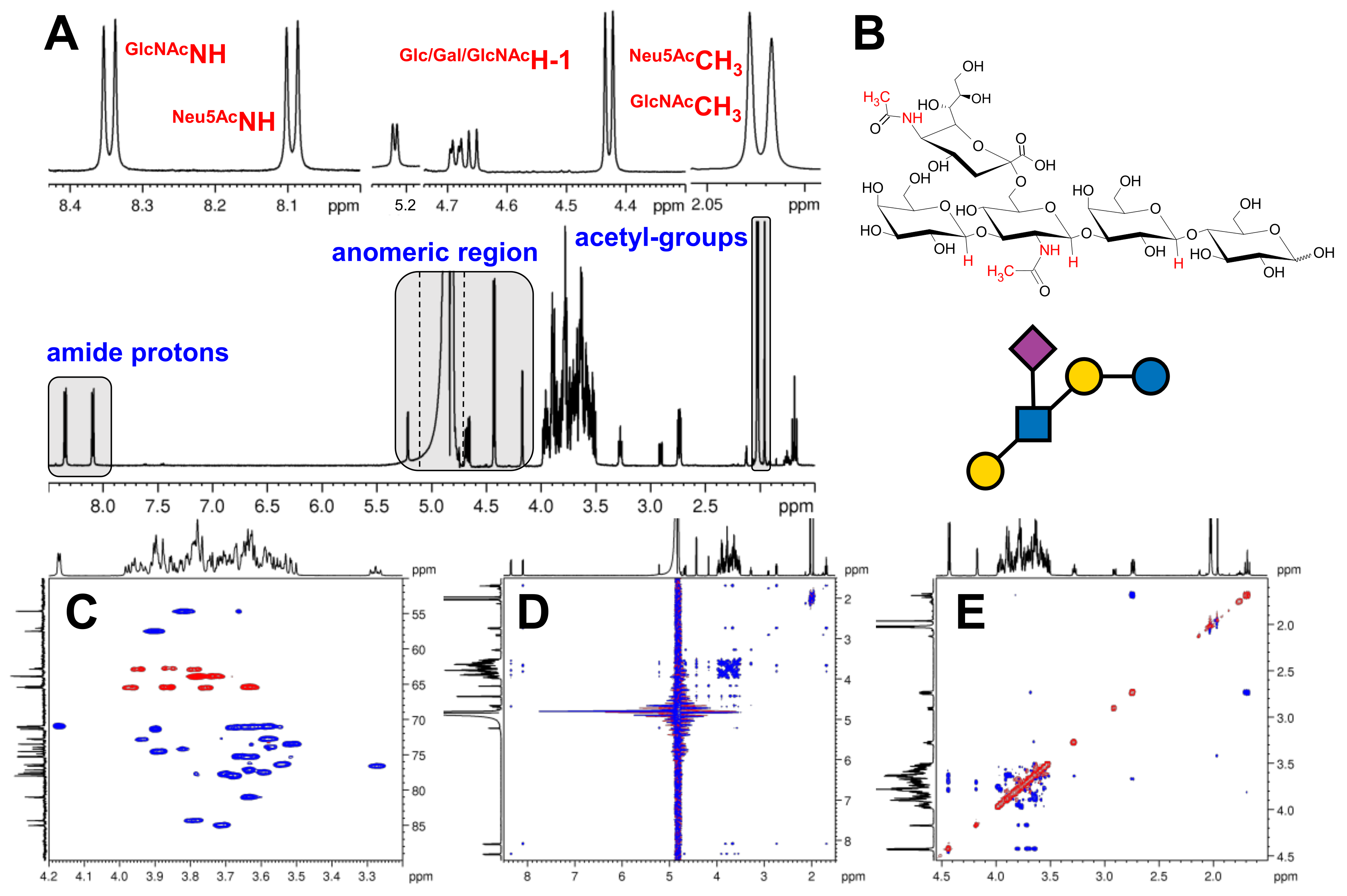
2.1. Optimization of Experimental Conditions
2.2. Complete 1H, 13C, and 15N Resonance Assignments of the HMO Samples
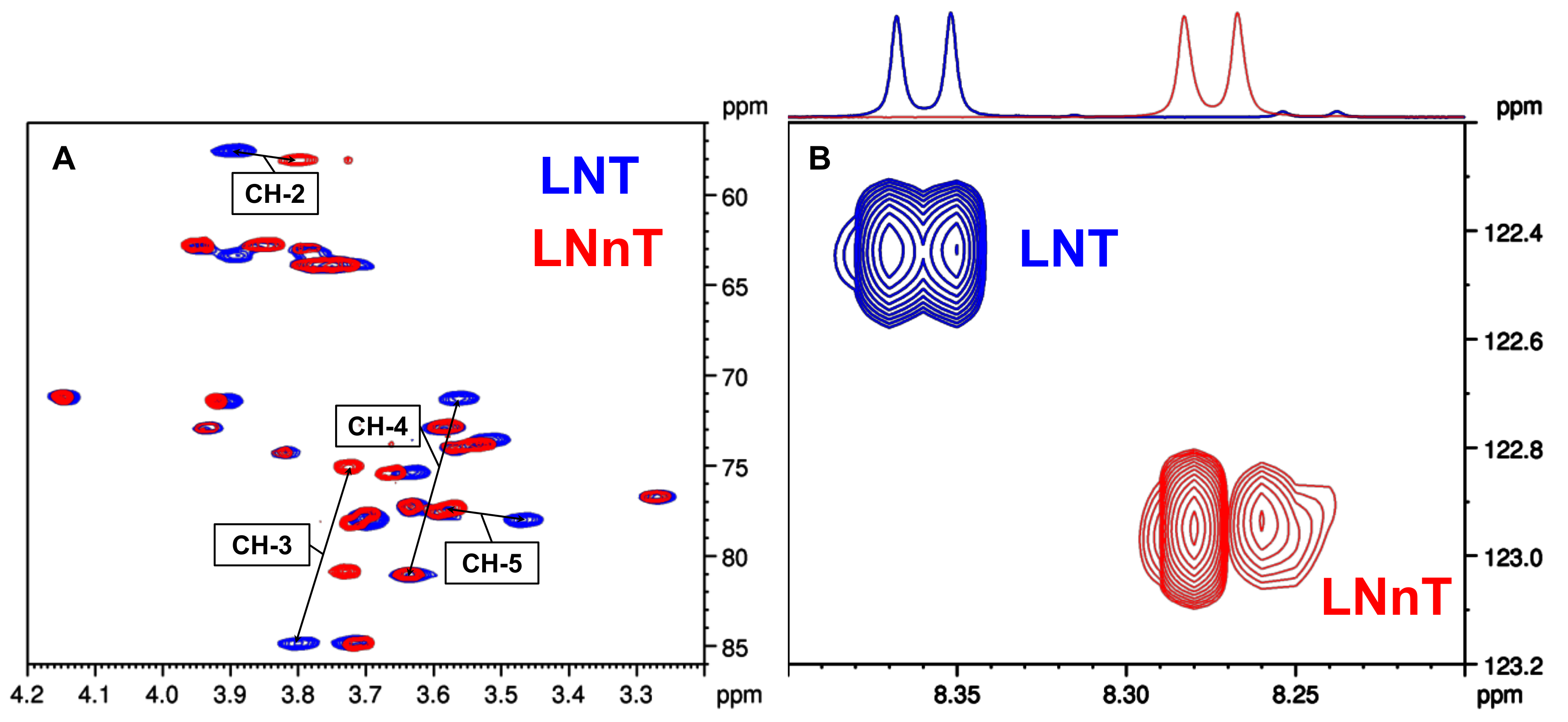
2.3. Comparison of the NMR Characteristics of the Isomeric HMOs
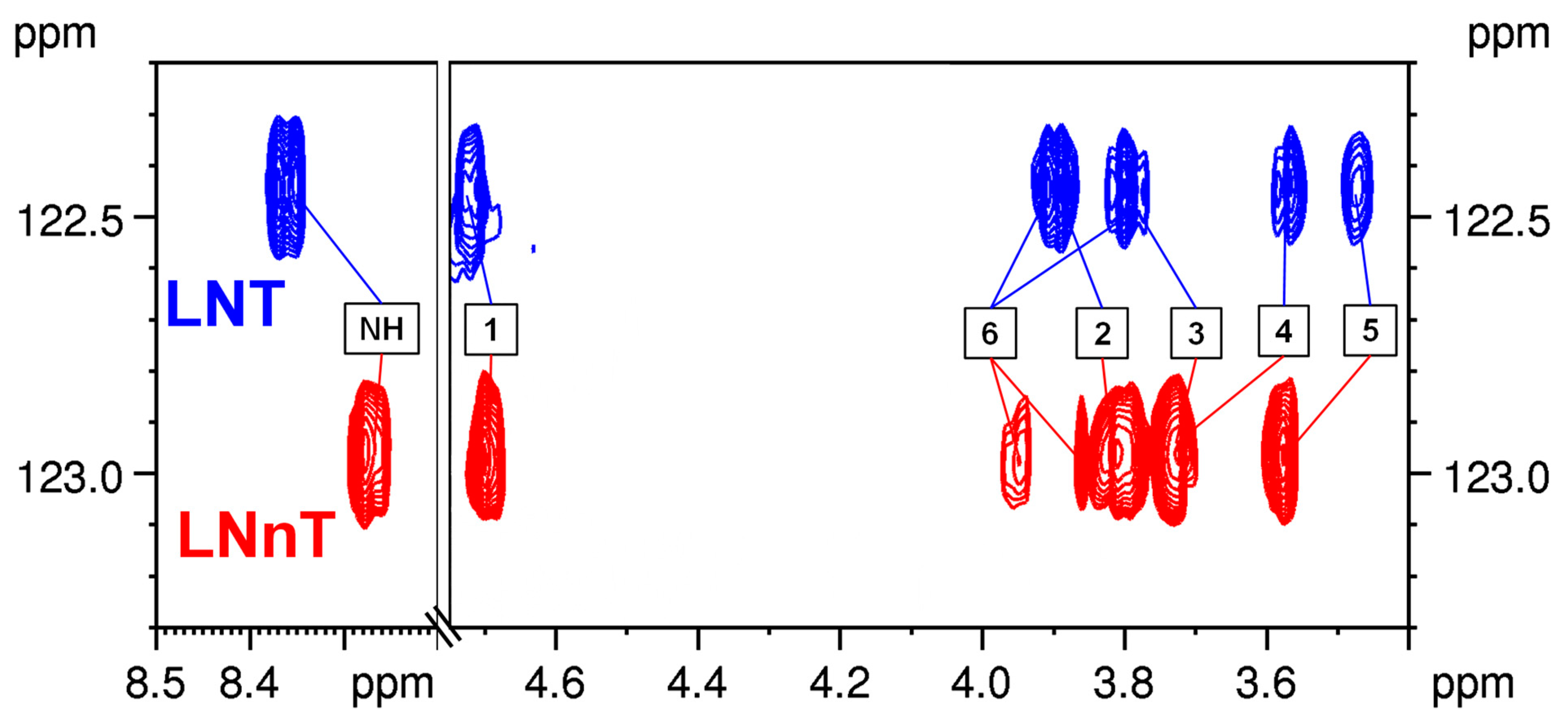
2.3.1. LNT–LNnT
2.3.2. LNFP II–LNFP III
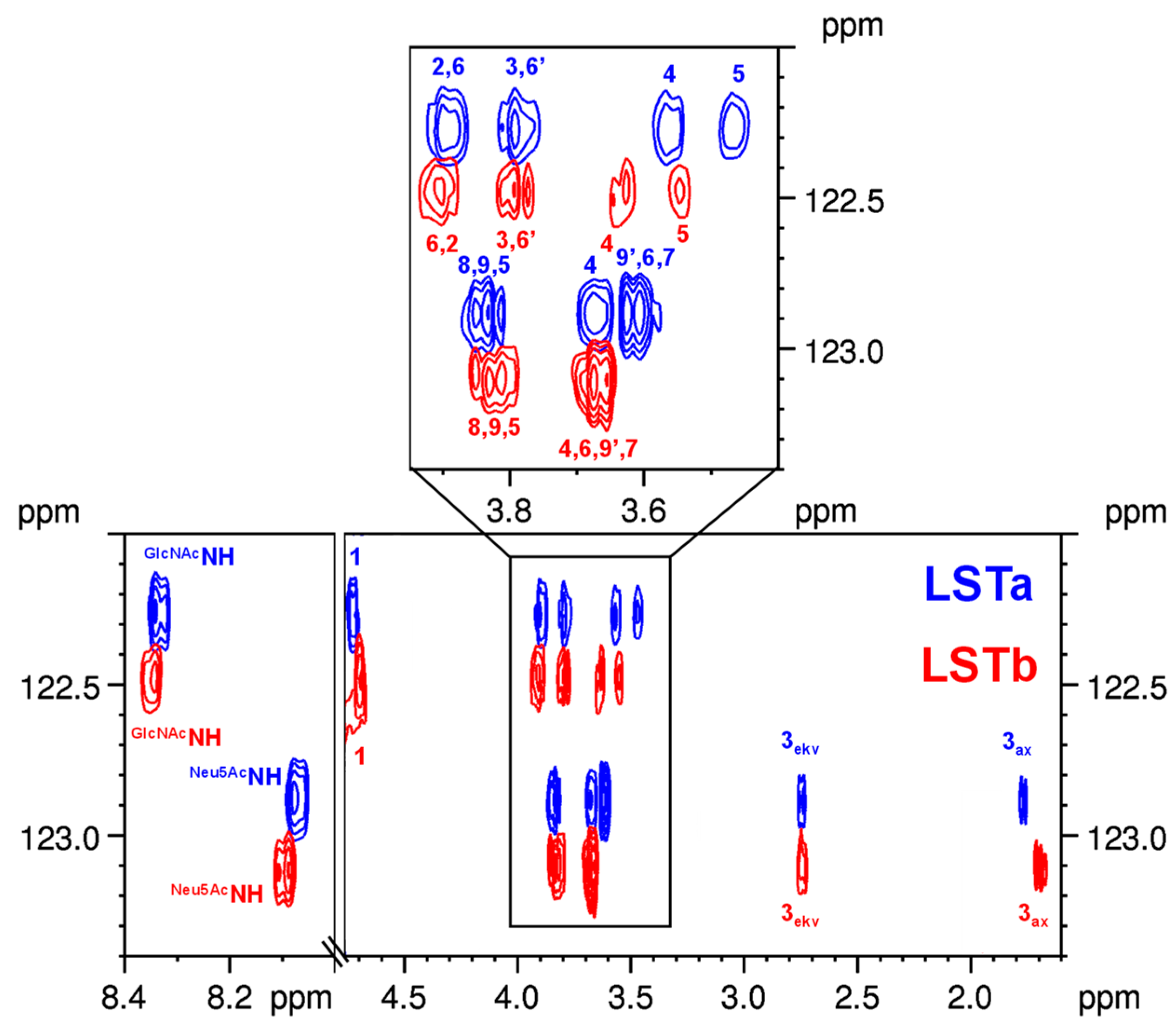
2.3.3. LSTa–LSTb
2.3.4. 3’SL–6’SL
2.4. The Use of 15N HSQC-TOCSY to Distinguish Isomers
3. Materials and Methods
3.1. Chemicals and Reagents
3.2. NMR Spectoscopic Measurements
NMR Experiments with Solvent Suppression
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Bode, L. Human Milk Oligosaccharides: Every Baby Needs a Sugar Mama. Glycobiology 2012, 22, 1147–1162. [Google Scholar] [CrossRef] [PubMed]
- Zhu, Y.; Zhang, W.; Mu, W. Human Milk Oligosaccharides: The New Gold Standard for Premium Infant Formula. J. Agric. Food Chem. 2022, 70, 2061–2063. [Google Scholar] [CrossRef] [PubMed]
- Bych, K.; Mikš, M.H.; Johanson, T.; Hederos, M.J.; Vigsnæs, L.K.; Becker, P. Production of HMOs Using Microbial Hosts—From Cell Engineering to Large Scale Production. Curr. Opin. Biotechnol. 2019, 56, 130–137. [Google Scholar] [CrossRef]
- Bode, L.; Contractor, N.; Barile, D.; Pohl, N.; Prudden, A.R.; Boons, G.-J.; Jin, Y.-S.; Jennewein, S. Overcoming the Limited Availability of Human Milk Oligosaccharides: Challenges and Opportunities for Research and Application. Nutr. Rev. 2016, 74, 635–644. [Google Scholar] [CrossRef] [PubMed]
- Ruhaak, L.R.; Lebrilla, C.B. The Glycobiology of Human Milk Oligosaccharides Advances in Analysis of Human Milk Oligosaccharides. Adv. Nutr. 2012, 3, 406S–414S. [Google Scholar] [CrossRef]
- Varki, A.; Cummings, R.D.; Aebi, M.; Packer, N.H.; Seeberger, P.H.; Esko, J.D.; Stanley, P.; Hart, G.; Darvill, A.; Kinoshita, T.; et al. Symbol Nomenclature for Graphical Representations of Glycans. Glycobiology 2015, 16, 1323–1324. [Google Scholar] [CrossRef]
- Van’t Land, B.; Joost Van Neerven, R.J.; Triantis, V.; Bode, L. Immunological Effects of Human Milk Oligosaccharides. Front. Pediatr. 2018, 1, 190. [Google Scholar] [CrossRef]
- Csernák, O.; Rácz, B.; Alberti, Á.; Béni, S. Quantitative Analysis of 3′- and 6′-Sialyllactose in Human Milk Samples by HPLC-MS/MS: A Validated Method for the Comparison of Two Consecutive Lactation Periods in the Same Woman. J. Pharm. Biomed. Anal. 2020, 184, 113184. [Google Scholar] [CrossRef]
- Grabarics, M.; Lettow, M.; Kirschbaum, C.; Greis, K.; Manz, C.; Pagel, K. Mass Spectrometry-Based Techniques to Elucidate the Sugar Code. Chem. Rev. 2021, 122, 7840–7908. [Google Scholar] [CrossRef]
- Huang, Y.; Dodds, E.D. Ion-Neutral Collisional Cross Sections of Carbohydrate Isomers as Divalent Cation Adducts and Their Electron Transfer Products. Analyst 2015, 140, 6912. [Google Scholar] [CrossRef]
- Huang, Y.; Dodds, E.D. Ion Mobility Studies of Carbohydrates as Group I Adducts: Isomer Specific Collisional Cross Section Dependence on Metal Ion Radius. Anal. Chem. 2013, 85, 9728–9735. [Google Scholar] [CrossRef]
- Han, L.; Costello, C.E. Electron Transfer Dissociation of Milk Oligosaccharides. J. Am. Soc. Mass Spectrom. 2011, 22, 997–1013. [Google Scholar] [CrossRef] [PubMed]
- Tang, Y.; Pu, Y.; Gao, J.; Hong, P.; Costello, C.E.; Lin, C. De Novo Glycan Sequencing by Electronic Excitation Dissociation and Fixed-Charge Derivatization. Anal. Chem. 2018, 90, 3793–3801. [Google Scholar] [CrossRef] [PubMed]
- van Leeuwen, S.S.; Schoemaker, R.J.W.; Gerwig, G.J.; van Leusen-Van Kan, E.J.M.; Dijkhuizen, L.; Kamerling, J.P. Rapid Milk Group Classification by 1H NMR Analysis of Le and H Epitopes in Human Milk Oligosaccharide Donor Samples. Glycobiology 2014, 24, 728–739. [Google Scholar] [CrossRef] [PubMed]
- Spevacek, A.R.; Smilowitz, J.T.; Chin, E.L.; Underwood, M.A.; German, J.B.; Slupsky, C.M. Infant Maturity at Birth Reveals Minor Differences in the Maternal Milk Metabolome in the First Month of Lactation. J. Nutr. Genom. 2015, 148, 1698–1708. [Google Scholar] [CrossRef]
- Liu, H.; Zhang, Z.; Linhardt, R.J. Lessons Learned from the Contamination of Heparin. Nat. Prod. Rep. 2009, 26, 313–321. [Google Scholar] [CrossRef]
- Rudd, T.R.; MacChi, E.; Gardini, C.; Muzi, L.; Guerrini, M.; Yates, E.A.; Torri, G. How to Find a Needle (or Anything Else) in a Haystack: Two-Dimensional Correlation Spectroscopy-Filtering with Iterative Random Sampling Applied to Pharmaceutical Heparin. Anal. Chem. 2012, 84, 6841–6847. [Google Scholar] [CrossRef]
- Beni, S.; Limtiaco, J.F.K.; Larive, C.K. Analysis and Characterization of Heparin Impurities. Anal. Bioanal. Chem. 2011, 399, 527–539. [Google Scholar] [CrossRef]
- Limtiaco, J.F.K.; Langeslay, D.J.; Beni, S.; Larive, C.K. Getting to Know the Nitrogen next Door: HNMBC Measurements of Amino Sugars. J. Magn. Reson. 2011, 209, 323–331. [Google Scholar] [CrossRef]
- Pomin, V.H. Advances in Glycosaminoglycanomics by 15 N-NMR Spectroscopy. Anal. Bioanal. Chem. 2013, 405, 3035–3048. [Google Scholar] [CrossRef]
- Langeslay, D.J.; Beni, S.; Larive, C.K. Detection of the 1H and 15N NMR Resonances of Sulfamate Groups in Aqueous Solution: A New Tool for Heparin and Heparan Sulfate Characterization. Anal. Chem. 2011, 83, 8006–8010. [Google Scholar] [CrossRef]
- Langeslay, D.J.; Beecher, C.N.; Naggi, A.; Guerrini, M.; Torri, G.; Larive, C.K. Characterizing the Microstructure of Heparin and Heparan Sulfate Using N-Sulfoglucosamine 1 H and 15 N NMR Chemical Shift Analysis. Anal. Chem. 2013, 85, 1247–1255. [Google Scholar] [CrossRef]
- Blundell, C.D.; Reed, M.A.C.; Almond, A. Complete Assignment of Hyaluronan Oligosaccharides up to Hexasaccharides. Carbohydr. Res. 2006, 341, 2803–2815. [Google Scholar] [CrossRef]
- Hwang, T.L.; Shaka, A.J. Water Suppression That Works. Excitation Sculpting Using Arbitrary Wave-Forms and Pulsed-Field Gradients. J. Magn. Reason. A 1995, 112, 275–279. [Google Scholar] [CrossRef]

| LNT | LNnT | LNFP II | LNFP III | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1H | m, J [Hz] | 13C/15N | 1H | m, J [Hz] | 13C/15N | 1H | m, J [Hz] | 13C/15N | 1H | m, J [Hz] | 13C/15N | |
| α-Glc | ||||||||||||
| 1 | 5.22 | d, 3.7 | 94.6 | 5.22 | d, 3.7 | 94.6 | 5.22 | d, 3.7 | 94.6 | 5.22 | d, 3.7 | 94.6 |
| 2 | 3.57 | dd, 9.9, 3.8 | 74.0 | 3.57 | dd, 9.9, 3.8 | 74.0 | 3.57 | dd, 9.9, 3.8 | 74.0 | 3.57 | dd, 9.9, 3.8 | 74.0 |
| 3 | 3.82 | t, 9.5 | 74.2 | 3.82 | t, 9.3 | 74.2 | 3.82 | t, 9.4 | 74.2 | 3.82 | t, 9.4 | 74.2 |
| 4 | 3.63 | t, 9.5 | 81.0 | 3.63 | t, 9.3 | 81.0 | 3.63 | t, 9.4 | 80.9 | 3.63 | t, 9.4 | 81.0 |
| 5 | 3.94 | m | 72.8 | 3.94 | m | 72.8 | 3.94 | m | 72.8 | 3.94 | m | 72.8 |
| 6 | 3.86 | m | 62.7 | 3.86 | m | 62.7 | 3.85 | m | 62.7 | 3.85 | m | 62.7 |
| β-Glc | ||||||||||||
| 1 | 4.66 | d, 7.9 | 98.5 | 4.66 | d, 7.9 | 98.5 | 4.66 | d, 7.9 | 98.5 | 4.66 | d, 7.9 | 98.5 |
| 2 | 3.27 | m | 76.6 | 3.27 | m | 76.6 | 3.27 | m | 76.6 | 3.27 | m | 76.6 |
| 3 | 3.63 | m | 77.1 | 3.63 | m | 77.1 | 3.63 | m | 77.1 | 3.63 | m | 77.1 |
| 4 | 3.64 | m | 80.9 | 3.64 | m | 80.9 | 3.63 | m | 81.0 | 3.64 | m | 80.9 |
| 5 | n.a. | n.a. | n.a. | n.a. | n.a. | n.a. | n.a. | n.a. | ||||
| 6a | 3.95 | dd, 12.0, 2.0 | 62.9 | 3.95 | dd, 12.0, 2.0 | 62.9 | 3.95 | dd, 12.1, 2.0 | 62.8 | 3.95 | dd, 12.0, 2.0 | 62.8 |
| 6b | 3.79 | dd, 12.2, 5.0 | 3.79 | dd, 12.2, 5.0 | 3.79 | dd, 12.1, 5.0 | 3.79 | dd, 12.0, 5.0 | ||||
| β-Gal | ||||||||||||
| 1 | 4.43/4.43 | d, 7.7 | 105.60/57 | 4.43/4.43 | d, 7.9 | 105.61/58 | 4.42/4.42 | d, 7.9 | 105.62/58 | 4.42/4.42 | d, 7.9 | 105.61/58 |
| 2 | 3.60/3.58 | dd, 9.8, 7.8 | 72.80/77 | 3.60/3.58 | dd, 9.8, 7.9 | 72.76/73 | 3.59/3.57 | dd, 9.8, 7.9 | 72.75/73 | 3.59/3.57 | dd, 9.8, 7.9 | 72.76/73 |
| 3 | 3.74/3.72 | t, 3.0 | 84.72/69 | 3.72/3.71 | t, 3.0 | 84.78/76 | 3.72/3.70 | t, 3.0 | 84.81/78 | 3.71/3.69 | t, 3.0 | 84.81/78 |
| 4 | 4.15 | d, 3.2 | 71.13/10 | 4.15 | d, 3.2 | 71.14/12 | 4.15 | d, 3.4 | 71.12/10 | 4.15 | d, 3.4 | 71.19/09 |
| 5 | n.a. | n.a. | n.a. | n.a. | n.a. | n.a. | n.a. | n.a. | ||||
| 6 | 3.76 | m | 63.78/77 | 3.76 | m | 63.78/77 | 3.76 | m | 63.79/78 | 3.76 | m | 63.79/77 |
| β-GlcNAc | ||||||||||||
| 1 | 4.72/4.72 | d, 8.4 | 105.3 | 4.69/4.69 | d, 8.3 | 105.46 | 4.69/4.69 | d, 8.5 | 105.4 | 4.70/4.70 | d, 8.5 | 105.3 |
| 2 | 3.90 | m | 57.5 | 3.80 | m | 58.0 | 3.95 | m | 58.6 | 3.96 | m | 58.7 |
| NH | 8.36 | d, 9.6 | 122.48 | 8.27 | d, 9.5 | 122.98 | 8.44 | d, 9.8 | 122.39 | 8.41 | d, 9.8 | 122.27 |
| CO | 117.7 | 177.7 | 177.5 | 177.5 | ||||||||
| CH3 | 2.02 | s | 25.0 | 2.03 | s | 25.0 | 2.03 | s | 25.1 | 2.02 | s | 25.0 |
| 3 | 3.80 | t, 9.6 | 84.8 | 3.73 | m | 75.0 | 4.07 | t, 9.6 | 78.6 | 3.87 | m | 77.5 |
| 4 | 3.57 | t, 9.6 | 71.2 | 3.73 | m | 80.8 | 3.75 | t, 9.6 | 74.8 | 3.94 | m | 75.7 |
| 5 | 3.47 | ddd, 10.0, 5.0, 2.3 | 77.9 | 3.58 | m | 77.3 | 3.53 | dt, 9.5, 3.0 | 77.9 | 3.57 | m | 77.8 |
| 6a | 3.90 | m | 63.3 | 3.95 | dd, 12.2, 2.0 | 62.6 | 3.94 | m | 62.4 | 3.97 | m | 62.4 |
| 6b | 3.78 | dd, 12.5, 5.3 | 3.85 | dd, 12.2, 4.7 | 3.86 | dd, 12.5, 3.2 | 3.87 | m | ||||
| β-Gal | ||||||||||||
| 1 | 4.43 | d, 8.4 | 106.2 | 4.47 | d, 7.8 | 105.53 | 4.50 | d, 7.7 | 105.55 | 4.45 | d, 7.8 | 104.46 |
| 2 | 3.52 | dd, 9.8, 7.8 | 73.5 | 3.54 | dd, 9.8, 7.8 | 73.8 | 3.48 | dd, 9.8, 7.7 | 73.3 | 3.49 | dd, 9.8, 7.9 | 73.8 |
| 3 | 3.64 | dd, 10.0, 3.0 | 75.3 | 3.67 | dd, 10.0, 3.3 | 75.3 | 3.62 | dd, 9.8, 3.4 | 75.1 | 3.65 | dd, 9.8, 3.3 | 75.3 |
| 4 | 3.90 | d, 3.4 | 71.3 | 3.92 | d, 3.5 | 71.3 | 3.88 | d, 3.5 | 71.2 | 3.90 | d, 3.4 | 71.2 |
| 5 | n.a. | n.a. | n.a. | n.a. | n.a. | n.a. | n.a. | n.a. | ||||
| 6 | 3.76 | m | 63.8 | 3.75 | m | 63.84 | 3.72 | m | 64.5 | 3.72 | m | 64.3 |
| α-Fuc | ||||||||||||
| 1 | 5.02 | d, 4.0 | 100.7 | 5.12 | d, 4.0 | 101.3 | ||||||
| 2 | 3.80 | dd, 10.5, 4.0 | 70.6 | 3.69 | dd, 10.4, 4.0 | 70.5 | ||||||
| 3 | 3.88 | dd, 10.3, 3.3 | 72.0 | 3.90 | dd, 10.4, 3.3 | 72.0 | ||||||
| 4 | 3.79 | d, 3.3 | 74.8 | 3.79 | d, 3.3 | 74.2 | ||||||
| 5 | 4.87 | o.l. | 69.6 | 4.83 | o.l. | 69.4 | ||||||
| CH3 | 1.17 | d, 6.7 | 18.1 | 1.17 | d, 6.6 | 18.0 | ||||||
| LSTa | LSTb | 3’SL | 6’SL | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1H | m, J [Hz] | 13C/15N | 1H | m, J [Hz] | 13C/15N | 1H | m, J [Hz] | 13C/15N | 1H | m, J [Hz] | 13C/15N | |
| α-Glc | ||||||||||||
| 1 | 5.22 | d, 3.7 | 94.6 | 5.22 | d, 3.8 | 94.6 | 5.22 | d, 3.8 | 94.6 | 5.22 | d, 3.7 | 94.6 |
| 2 | 3.57 | dd, 9.9, 3.8 | 74.0 | 3.57 | dd, 9.9, 3.8 | 74.0 | 3.57 | dd, 9.9, 3.8 | 74.0 | 3.60 | dd, 9.8, 3.8 | 73.9 |
| 3 | 3.82 | t, 9.4 | 74.2 | 3.82 | t, 9.4 | 74.2 | 3.83 | t, 9.4 | 74.2 | 3.84 | t, 9.3 | 74.4 |
| 4 | 3.63 | t, 9.4 | 81.1 | 3.63 | t, 9.4 | 81.1 | 3.66 | t, 9.4 | 80.9/80.8 | 3.61 | t, 9.3 | 82.4/82.3 |
| 5 | 3.94 | m | 72.8 | 3.94 | m | 72.8 | 3.94 | m | 72.8 | 3.95 | m | 72.7 |
| 6 | 3.86 | m | 62.7 | 3.86 | m | 62.7 | 3.88 | m | 62.7 | 3.88 | m | 62.9 |
| β-Glc | ||||||||||||
| 1 | 4.66 | d, 8.0 | 98.5 | 4.66 | d, 7.9 | 98.5 | 4.66 | d, 8.0 | 98.6 | 4.66 | d, 8.0 | 98.4 |
| 2 | 3.27 | m | 76.6 | 3.28 | m | 76.6 | 3.28 | dd, 9.0, 8.0 | 76.7 | 3.30 | t, 8.6 | 76.6 |
| 3 | 3.63 | m | 77.2 | 3.63 | m | 77.2 | 3.63 | t, 9.2 | 77.1 | 3.63 | m | 77.4 |
| 4 | 3.64 | m | 81.0 | 3.64 | m | 81.0 | 3.67 | dd, 9.5, 8.3 | 80.9/80.8 | 3.61 | m | 82.4/82.3 |
| 5 | n.a. | n.a. | n.a. | n.a. | 3.59 | ddd, 9.5, 5.0, 2.0 | 77.5 | 3.63 | m | 77.4 | ||
| 6a | 3.95 | dd, 12.0, 2.0 | 62.8 | 3.95 | dd, 12.0, 2.0 | 62.9 | 3.96 | dd, 12.3, 2.0 | 62.8 | 3.95 | dd, 12.3, 2.0 | 63.0 |
| 6b | 3.79 | dd, 12.2, 5.0 | 3.79 | dd, 12.2, 5.0 | 3.82 | dd, 12.3, 5.0 | 3.79 | dd, 12.3, 4.4 | ||||
| β-Gal | ||||||||||||
| 1 | 4.43/4.43 | d, 7.9 | 105.60/58 | 4.43/4.43 | d, 7.7 | 105.60/57 | 4.52 | d, 7.7 | 105.3 | 4.42 | d, 7.9 | 105.93/90 |
| 2 | 3.60/3.58 | dd, 9.8, 7.9 | 72.80/78 | 3.59/3.57 | dd, 9.8, 7.9 | 72.78/74 | 3.57 | dd, 9.9, 7.9 | 72.2 | 3.53 | dd, 9.8, 7.9 | 73.6 |
| 3 | 3.74/3.72 | t, 2.9 | 84.65/63 | 3.72/3.70 | t, 3.3 | 85.04/84.99 | 4.11/4.10 | t, 3.0 | 78.21/20 | 3.67/3.65 | t, 3.0 | 75.2 |
| 4 | 4.15 | d, 3.2 | 71.12/10 | 4.17 | d, 3.3 | 70.97/94 | 3.95 | d, 3.0 | 70.2 | 3.93 | d, 3.3 | 71.33/31 |
| 5 | n.a. | n.a. | n.a. | n.a. | n.a. | n.a. | n.a. | n.a. | ||||
| 6 | 3.78 | m | 63.79/78 | 3.78 | m | 63.92/91 | 3.74 | m | 63.85/84 | 3.96 | m | 66.3 |
| 3.59 | ||||||||||||
| β-GlcNAc | ||||||||||||
| 1 | 4.73/4.72 | d, 8.4 | 105.2 | 4.69/4.68 | d, 8.54 | 105.4 | ||||||
| 2 | 3.90 | m | 57.4 | 3.98 | m | 57.5 | ||||||
| NH | 8.34 | d, 9.6 | 122.30 | 8.35 | d, 9.6 | 122.50 | ||||||
| CO | 177.68 | 177.67 | ||||||||||
| CH3 | 2.02 | s | 25.1/24.8 | 2.02 | s | 25.0 | ||||||
| 3 | 3.80 | t, 9.5 | 84.9 | 3.79 | t, 9.5 | 84.4 | ||||||
| 4 | 3.57 | t, 9.5 | 71.2 | 3.63 | t, 9.5 | 71.1 | ||||||
| 5 | 3.47 | ddd, 10.0, 5.3, 2.2 | 77.9 | 3.54 | ddd, 9.8, 5.2, 2.2 | 76.4 | ||||||
| 6a | 3.90 | m | 63.3 | 3.97 | dd, 10.8, 5.7 | 65.5 | ||||||
| 6b | 3.78 | dd, 12.0, 5.3 | 3.76 | d, 10.8 | ||||||||
| β-Gal | ||||||||||||
| 1 | 4.50 | d, 7.8 | 106.1 | 4.43 | d, 7.7 | 106.1 | ||||||
| 2 | 3.54 | dd, 9.7, 8.0 | 71.9 | 3.52 | dd 10.0, 7.8 | 73.5 | ||||||
| 3 | 4.08 | dd, 9.7, 3.0 | 78.3 | 3.63 | dd, 10.0, 3.3 | 75.3 | ||||||
| 4 | 3.93 | d, 3.0 | 70.0 | 3.90 | d, 3.4 | 71.4 | ||||||
| 5 | n.a. | n.a. | n.a. | n.a. | ||||||||
| 6 | 3.72 | m | 63.8 | 3.78/3.74 | m | 63.9 | ||||||
| α-Neu5Ac | ||||||||||||
| 1 | 176.5 | 176.1 | 176.5 | 176.10/09 | ||||||||
| 2 | 102.3 | 102.9 | 102.4 | 102.9 | ||||||||
| 3α | 1.78 | t, 12.1 | 42.5 | 1.68 | t, 12.1 | 42.8 | 1.80 | t, 12.2 dd, 12.4, 4.7 | 42.3 | 1.74 | t, 12.2 | 42.8 |
| 3β | 2.75 | dd, 12.4, 4.7 | 2.74 | dd, 12.3, 4.7 | 2.75 | 2.70 | dd, 12.4, 4.6 | |||||
| 4 | 3.68 | ddd, 12.4, 4.7, 2.2 | 71.2 | 3.68 | m | 71.1 | 3.68 | ddd, 11.7, 9.7, 4.7 | 71.2 | 3.65 | ddd, 11.7, 9.7, 4.6 | 71.2 |
| 5 | 3.84 | m | 54.4 | 3.82 | m | 54.7 | 3.85 | m | 54.5 | 3.85 | m | 54.6 |
| CH3 | 2.02 | s | 25.1/24.8 | 2.03 | s | 24.8 | 2.03 | s | 24.8 | 2.02 | s | 24.8 |
| NH | 8.07 | d, 9.3 | 122.91 | 8.09 | d, 9.3 | 123.14 | 8.08 | d, 9.3 | 122.98 | 8.06 | d, 9.4 | 123.01 |
| CO | 177.72 | 177.79 | 177.8 | |||||||||
| 6 | 3.62 | dd, 10.4, 1.8 | 75.5 | 3.66 | dd, 10.4, 1.8 | 75.2 | 3.62 | dd, 10.4, 1.8 | 75.6 | 3.72 | dd, 10.4, 1.7 | 75.2 |
| 7 | 3.59 | d, 9.0 | 70.8 | 3.58 | d, 9.0 | 71.0 | 3.59 | 70.9 | 3.56 | d, 9.0 | 71.2 | |
| 8 | 3.87 | m | 74.7 | 3.88 | m | 74.5 | 3.88 | m | 74.6 | 3.89 | m | 74.6 |
| 9a | 3.85 | m | 65.3 | 3.87 | m | 65.4 | 3.87 | m | 65.4 | 3.88 | m | 65.5 |
| 9b | 3.64 | m | 3.63 | m | 3.64 | m | 3.64 | m | ||||
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Garádi, Z.; Tóth, A.; Gáti, T.; Dancsó, A.; Béni, S. Utilizing the 1H-15N NMR Methods for the Characterization of Isomeric Human Milk Oligosaccharides. Int. J. Mol. Sci. 2023, 24, 2180. https://doi.org/10.3390/ijms24032180
Garádi Z, Tóth A, Gáti T, Dancsó A, Béni S. Utilizing the 1H-15N NMR Methods for the Characterization of Isomeric Human Milk Oligosaccharides. International Journal of Molecular Sciences. 2023; 24(3):2180. https://doi.org/10.3390/ijms24032180
Chicago/Turabian StyleGarádi, Zsófia, András Tóth, Tamás Gáti, András Dancsó, and Szabolcs Béni. 2023. "Utilizing the 1H-15N NMR Methods for the Characterization of Isomeric Human Milk Oligosaccharides" International Journal of Molecular Sciences 24, no. 3: 2180. https://doi.org/10.3390/ijms24032180
APA StyleGarádi, Z., Tóth, A., Gáti, T., Dancsó, A., & Béni, S. (2023). Utilizing the 1H-15N NMR Methods for the Characterization of Isomeric Human Milk Oligosaccharides. International Journal of Molecular Sciences, 24(3), 2180. https://doi.org/10.3390/ijms24032180









