Abstract
In this study, a new strain of Pantoea vagans, SRS89, was isolated from surface-sterilized stevia seeds. The isolate was evaluated using morphological, molecular, and biochemical methods. The bacterium was 1.5 μm long, yellowish in color, and classified as Gram-negative. Whole genome sequencing of our strain revealed the presence of a 4,610,019 bp chromosome, and genome annotation resulted in the detection of 4283 genes encoding 4204 putative coding sequences. Phylogenic analysis classified the genome of our strain close to the MP7 and LMG 24199 strains of P. vagans. Functional analysis showed that the highest number of genes within the analyzed bacterium genome were involved in transcription, amino acid transport and metabolism, and carbohydrate transport and metabolism. We also identified genes for enzymes involved in the biosynthesis of carotenoids and terpenoids. Furthermore, we showed the presence of growth regulators, with the highest amount noted for gibberellic acid A3, indole-3-acetic acid, and benzoic acid. However, the most promising property of this strain is its ability to synthesize rebaudioside A; the estimated amount quantified using reversed-phase (RP)-HPLC was 4.39 mg/g of the dry weight of the bacteria culture. The isolated endophytic bacterium may be an interesting new approach to the production of this valuable metabolite.
1. Introduction
Stevia rebaudiana Bertoni (Asteraceae) is an important plant due to its content of steviol glycosides (SGs), a kind of non-caloric and intensely sweet chemical compounds that were approved for human consumption by the Food and Drug Administration (US) in 2008 and by the European Union in 2011. The SG biosynthetic pathway in stevia has already been described [1,2,3]. The natural properties of SGs make them the subject of increasing interest as a natural sweetener whose consumption could exert beneficial effects on human health [4,5,6]. In stevia, more than 30 SGs have been identified, with stevioside (ST) and rebaudioside A (Reb A) being the most significant [7,8,9,10,11,12]. As reported by DuBois and Stephenson, the taste quality of Reb A is better than that of ST because it is sweeter and less bitter [13]. Reb A can be used in various foods or dietary supplements as a sweetener. In addition, studies have shown that Reb A attenuates aging by acting as an effective cellular antioxidant and lowering the ectopic accumulation of neutral lipids [14]. Recently, Reb A has also been demonstrated to have many physiological functions, such as antihypertension, anti-diabetes, and anticaries [15]. Research is also being conducted on the effect of Reb A and other SGs on intestinal microbiota [16,17]. The SGs are found in the leaves, where they are detected in the highest numbers and amounts, as well as in other organs with declined order: flowers, stems, seeds, and roots [18]. The concentrations of SGs in stevia leaves vary based on genotype, plant growth stage, fertilization level, and growing condition [19,20]. Stevia is naturally propagated by seeds; however, poor germination and the long time required for the development of seedlings suitable for planting in the field (which can last up to 60 days) means that it is not a widespread method in commercial stevia production. The possibility of improving stevia seed germination using physical factors or plant growth regulators has already been published [21,22,23,24,25,26]. As reported by Puente et al. [27], endophytic bacteria might also increase the rate of seed germination.
Endophytic bacteria are defined as organisms that live most of their lives inside plant tissues without eliciting any pathogenic symptoms [28]. As reported by Smith et al. [29], among the over 300,000 plant species found worldwide, each contains endophytes. Endophytic bacteria have been isolated from the leaves, stems, roots, flowers, fruits, and seeds of various plant species [30,31,32,33]. In light of this information, it is not surprising that endophytic bacteria have also been identified in stevia plants [34,35]. As reported by Yu et al. [36], in the seedling stage, the dominant genera of the endophytic bacteria of stevia are Enterobacterium and Erwinia, while Methylobacterium and Sphingomonas were found to be the principal endophytes in mature leaves. Interestingly, its concentration was positively correlated with stevioside content and with UGT74G1 and UGT76G1 gene expression [36]. These genes encode UDP-glycosyltransferases, which are engaged in the last stages of the SGs biosynthesis pathway. Additionally, in the research of Oviedo-Pereira et al. [35], reinoculation with the endophytic bacteria Enterobacter hormaechei increased SG synthesis, as well as flavonoid accumulation in the trichomes of S. rebaudiana plants. However, there is no information about the endophytic bacteria that occupy stevia seeds. Endophytic bacteria were isolated from the seeds and seedlings of eucalyptus species and hybrids [27,37]. In maize seeds, Gamma proteobacteria represent the most abundant class of endophytes, most of which are Pantoea and Enterobacter [38,39]. In rice seeds, one of the dominant genera of endophytic bacteria was also found to be Pantoea [40]. Scientists have suggested that endophytic bacteria from seeds are transferred vertically during subsequent germination inside the plant and further affect plant growth [41,42]. Various studies have shown that endophytes can regulate plant growth with nitrogen fixation, phosphate solubilization, siderophore, 1-aminocyclopropane-1-carboxylate (ACC) deaminase activity, and indole-3-acetic acid (IAA) synthesis [43,44,45,46]. It is also well known that endophytic bacteria can produce plant growth regulators and the same or similar secondary metabolites as their hosts [47,48]. Endophytic bacteria may also enhance plant tolerance against abiotic stresses, such as salinity, drought, cold, or heavy metal toxicity [49]. However, the interactions between plants and bacteria are often specific, and the endophytic bacteria of one plant species may be pathogenic to other plant species [50].
The aim of this study was to identify and characterize endophytic bacteria isolated from surface-sterilized seeds of Stevia rebaudiana Bertoni to elucidate their possible beneficial role in the host plant and to potentially use them as a source of valuable metabolites. To our knowledge, this is the first mention of endophytic bacteria from stevia seeds.
2. Results
2.1. Preliminary Identification of Endophytic Bacteria
The bacteria isolated from surface-sterilized stevia seeds have rod-shaped cells about 1.5 μm long and yellowish in color (Figure 1). Bacteria were classified as Gram-negative and were further identified via sequence analysis of the PCR product obtained with universal primers for the 16S rRNA coding gene. The basic local alignment search tool for nucleotides (BLASTn) search score analysis suggested that the bacteria strain isolated from stevia seeds belongs to the genus Pantoea. Our strain showed the greatest similarity to the strains of P. agglomerans and P. vagans (Table S1, Figure S1 in Supplementary File S1). We named the isolated strain SRS89.
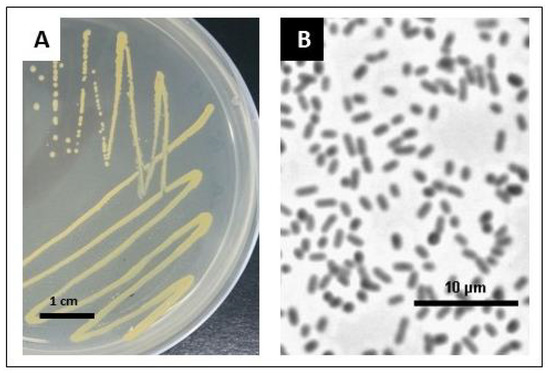
Figure 1.
Bacteria cultures grown overnight on the LB medium at 25 °C for single colony isolation. Bacteria cultures grown overnight on the LB solid medium (A). Single bacteria cells after 24 h of growth in the LB liquid medium (phase contrast microscopy was performed using a Nikon Eclipse E800 microscope at 40× magnification) (B).
2.2. New Strain SRS89 of Pantoea Vagans—Genome Features, Comparative Genome Analysis, and Gene Classification
Genomic DNA of the isolated strain was sequenced, and filtered reads were assembled into 158 contigs, 143 of which were longer than 1000 bp. The N50 for the assembly was 57,973, and the L50 was 23. The total assembly length was 4,610,019 bp, with an average GC content of 55.26% (Table 1). The average assembly coverage by the reads was 22.4×. Genome annotation with NCBI Prokaryotic Genome Annotation Pipeline (PGAP) resulted in the detection of 4283 genes encoding 4204 putative coding sequences (CDS) (NCBI WGS Project JAAALG01). Furthermore, 67 tRNA-coding genes, five rRNA coding genes, and one tmRNA coding gene were identified in the analyzed sequences. Analysis with Operon-mapper software resulted in the detection of 2312 operons (Supplementary File S2), and the genes belonged to 2272 unique clusters of orthologous groups of proteins (COGs).

Table 1.
Quality assessment for SRS89 strain assembly.
To phylogenetically classify the isolated strain and find the best reference genomes for comparative analysis, a genomic BLAST-based dendrogram was used. This analysis allowed for the selection of closely related P. vagans strains for comparative analysis. The strains included FDAARGOS_160, LMG24199, MP7, C9-1, and PV989 (Figure 2A). The visualization of our SRS89 strain genome with respect to the C9-1 strain revealed not only several highly homologous blocks but also the inversion of a genomic segment and several smaller rearrangements. Some of the other compared strain genomes showed more complex rearrangements (Figure 2B). Among the compared strains, SRS89 seemed to have a rather lower genome size (4.6 Mb), with a smaller genome observed only for the MP7 (4.59 Mb) strain (Table 2). The GC content of the analyzed genomes was comparable for all reference strains and oscillated within a range of 55.1–55.3%. The number of identified coding sequences (CDS) was the lowest in the MP7 strain (4005) and the highest in PV989 (4397), with an intermediate value for SRS89 (4204). The whole pan-genome for the compared bacteria involved 5562 genes, of which 3352 (60.2%) constituted a common core-genome (Figure 2C). The number of accessory genes ranged from 451 in SRS89 to 619 in C9-1. The number of strain-specific genes in the compared dataset of strains ranged from 102 to 349 and was 159 for the SRS89 strain (Supplementary File S3). Additionally, as many as 85 CDS were exclusively absent in this strain (Table 2).
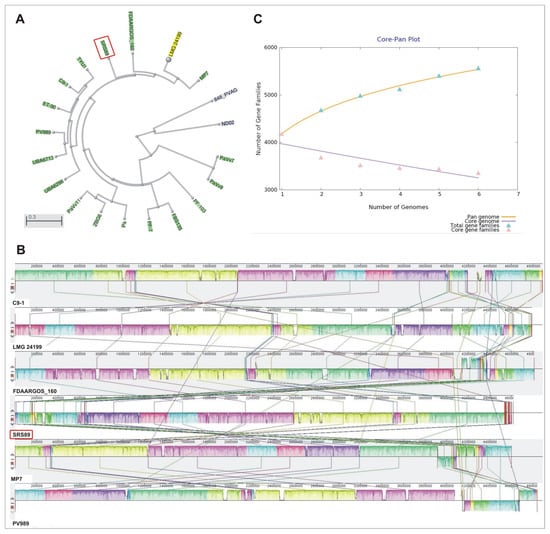
Figure 2.
Dendrogram created based on genomic BLAST analysis for all Pantoea vagans strains currently (December 2022) present in the NCBI database (A). Genome alignment of selected P. vagans strains using Mauve software (http://darlinglab.org/mauve/user-guide/viewer.html, accessed on 10 January 2023) (B). Pan-/Core-genome plot for the six compared P. vagans strains (C).

Table 2.
Genome and genes statistics for the analyzed strains of P. vagans.
Functional analysis of a pan- and core-genome showed that the highest number of genes within the analyzed genomes were engaged in transcription, amino acid transport and metabolism, and carbohydrate transport and metabolism (Figure 3A), with a significant number of genes related to COG involved in membrane transport (Figure 3B).
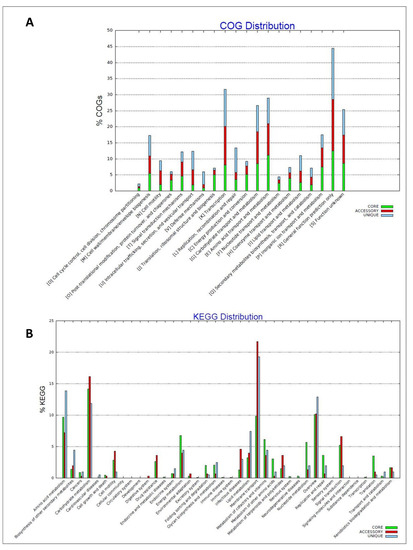
Figure 3.
Distribution of orthologous groups of proteins (COGs) across the analyzed Pantoea vagans strains pan- and core-genome (A). Distribution KEGG pathways across the analyzed P. vagans strains pan- and core-genome (B).
Regarding some details, in the SRS89 strain we detected genes involved in the early steps of the 2-C-methyl-D-erythritol-4 phosphate (MEP) pathway that is used, among others, for terpenoid biosynthesis. In stevia plants, that pathway is also used for SG biosynthesis. The detected genes were: dxs coding for geranylgeranyl diphosphate synthase type II [EC: 2.5.1.29] and 1-deoxy-D-xylulose-5-phosphate synthase [EC:2.2.1.7], dxr coding for 1-deoxy-D-xylulose-5-phosphate reductoisomerase [EC:1.1.1.267], and idi coding for isopentenyl-diphosphate delta-isomerase [EC:5.3.3.2]. The genes engaged in carotenoid biosynthesis were also identified: zep coding for zeaxanthin epoxidase [EC:1.14.13.90], crtB coding for phytoene synthase [EC:2.5.1.32] and crtZ coding for beta-carotene 3-hydroxylase [EC:1.14.13.129] (Supplementary File S4).
The phylogenic relationships among the compared strains were additionally visualized via pan- and core-genome analyses. Similarly, as in the initial phylogeny analysis results, the pan-genome analysis clustered SRS89 in a separate clade at a long distance to C9-1, while the core-genome-based analysis clustered SRS89 as genetically more like MP7 and LMG24199 (Figure 4). A relatively large pan-genome-based distance was observed between SRS89 and FDAARGOS_160, which were clustered in a direct vicinity in genome-based analysis.
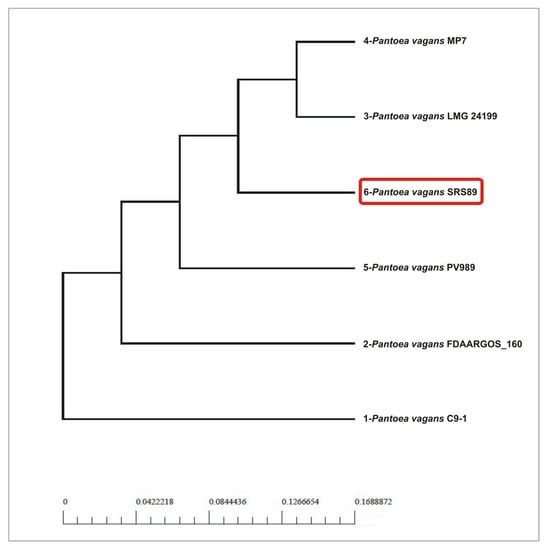
Figure 4.
Phylogenetic analysis the selected Pantoea vagans strains based on pan- and core-genome analysis.
2.3. Biochemical Properties of SRS89 Strain
The reversed-phase (RP)-HPLC analyses showed that the strain SRS89 of P. vagans contains Reb A. It was the only SG we identified in this bacterium, and its estimated amount quantified using RP-HPLC was 4.39 mg/g (±0.2) of the dry weight (DW) of the bacteria culture. We confirmed the presence of Reb A using Electrospray Ionization Mass Spectrometry (EMI-MS) analysis (Figure 5). Additionally, by using the RP-HPLC analyses, we demonstrated the presence of phenylalanine and tryptophan at concentrations of 0.75 mg/g DW (± 0.2) and 0.57 mg/g DW (±0.07), respectively (Table 3).
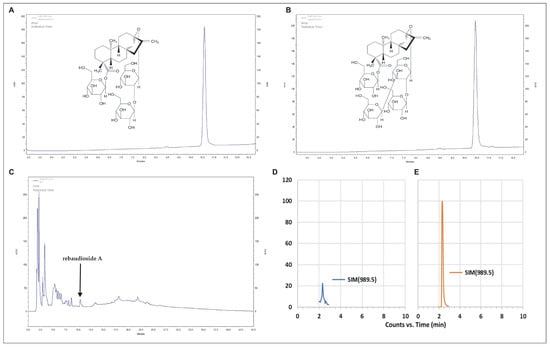
Figure 5.
RP-HPLC chromatograms of the stevioside (A), rebaudioside A (B) standards, and steviol glycosides fraction extract from the SRS89 strain of Pantoea vagans (C). Single ion [M+Na]+ monitoring (SIM) chromatogram of bacteria sample (D) and pure rebaudioside A standard (E) separated in HILIC mode.

Table 3.
Steviol glycosides, amino acids and growth regulars content in Pantoea vagans strain SRS89. The results are the means of three replications (n = 3) ±SD (nd—not detectable; ta—trace amount).
Ultrahigh-performance liquid chromatography–tandem mass spectrometry (UHPLC-MS/MS) analyses enabled us to identify growth regulators in bacteria extract such as indole-3-acetic acid (IAA, 831.7 pg/mg DW), indole-3-carboxylic acid (I3CA, 3716.6 pg/mg DW), indole-3-acetyl-aspartic acid (IAAsp, 112 pg/mg DW), gibberellic acid A3 (GA3, 17,402 pg/mg DW), gibberellic acid A6 (GA6, 318 pg/mg DW), benzoic acid (BeA, 10,770 pg/mg DW), salicylic acid (SA, 1830 pg/mg DW), and jasmonic acid (JA, 213 pg/mg DW) (Figure 6, Table 3). We were also able to detect trace amounts of abscisic acid (ABA) and auxins such as indole-3-acetyl-glutamic acid (IAGlu), indole-3-acetonitrile (IAN), indole-3-propionic acid (IPA), and some gibberellins: GA1, GA4, GA19, GA44, and GA53 (Table 3).
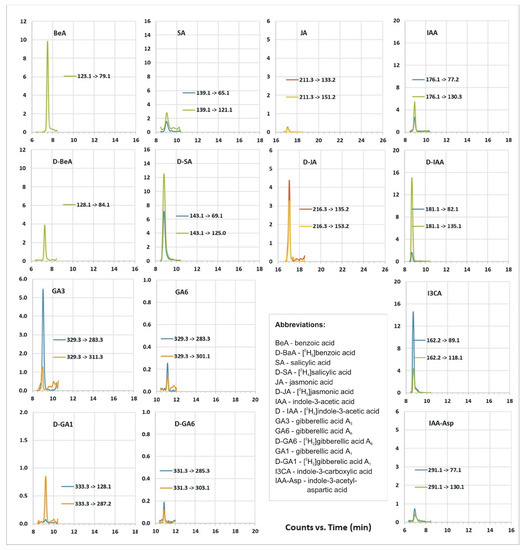
Figure 6.
UHPLC-MS/MS growth regulator profile of bacterial extract, chromatograms of multiple reaction monitoring (MRM) transitions for the analyzed plant growth regulators and related substances.
2.4. Seed Germination Promoting Potential of SRS89 Strain
The stevia seeds germinated properly after reinoculation and produced a well-developed root system with a large number of root hairs (Figure 7A–E). It can be seen that one hour of inoculation significantly promoted seed germination. It was visible from the second day of the experiment that the maximum value of germinated seeds reached 67.5% on the 9th day of germination compared to 51% of the non-reinoculated seeds. However, extending the inoculation time to 4 h decreased germination (Figure 7F). When the seeds were germinated on agar gel (AG) containing 50 mM NaCl, one-hour inoculation has no effect on germination when compared with the control, but four-hour inoculation significantly decreased the germination capacity (GC) (Figure 7G). In the experiment in which AG containing 150 mM NaCl was used, we observed a significant reduction in seed germination when compared to seed germination on other substrates. Here, however, the effect of inoculation on seed germination was not visible, and the GC was, on average, 19% (Figure 7H). We also observed that NaCl added to AG had a negative effect on the further growth of the seedlings, which was particularly evident when the concentration of NaCl was 150 mM. Here, the seedlings were small, poor in quality, and died shortly after germination.
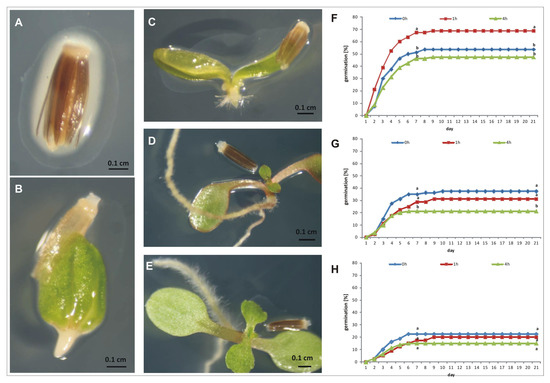
Figure 7.
The effect of SRS89 strain on stevia seed germination. Seeds were surface sterilized and inoculated with the isolated Pantoea vagans strain for one or four hours. Non inoculated seeds were used as a control. The following stages of stevia seed germination after inoculation with SRS89 strain (A–E). The stevia seed germination on agar gel (F), agar gel supplemented with 50 mM NaCl (G) or with 150 mM NaCl (H). The values of seed germination are means of four replications (n = 4). Different letters indicate a significant difference in the percentage of germinated seeds at the 7th (germination energy, GE) and 21st (germination capacity, GC) day of incubation at p < 0.05 according to Duncan’s test.
3. Discussion
Endophytic bacteria inhabit plant tissues without causing any harm to the host [51]. Numerous studies have revealed that these bacteria play a crucial role in the growth and development of a wide variety of host plant species. They also increase plant tolerance for environmental stresses and inhibit plant pathogen growth. These roles are mainly performed by secreting growth regulators and consequently helping to improve nutrition. Some previous papers have described the isolation of endophytic bacteria from different organs of stevia plants [34,35,36]; however, until now there has been no information about the endophytic bacteria inhabiting the stevia seeds. Here, for the first time, we report on bacteria isolated from Stevia rebaudiana seeds. Based on a sequence homology analysis of the 16S rRNA coding gene, we have shown that our bacterium shows the highest similarity to P. agglomerans and P. vagans. The verification carried out with the use of whole genome sequencing (WGS) confirmed that is definitely P. vagas strain. The genus Pantoea is a member of Enterobacteriales and represents several species that have been isolated from numerous different sources, mostly plants [52]. Anjum and Chandra [34] have already reported isolation from stevia plants of the bacteria belonging to the genus Pantoea, although species identification was not performed in that work. Originally, Pantoea was known as a plant pathogen; however, today, it is generally known that these bacteria have a beneficial effect on plants, especially in growth promotion and phytopathogen control [53,54,55,56]. P. vagans is very often isolated together with P. agglomerans since both species occupy the same ecological niches, and even some isolated P. vagans strains were previously identified as P. agglomerans [57]. As reported by Palmer et al. [58], P. vagans, in contrast to some strains of P. agglomerans, is not able to induce galls and is not a pathogenic bacterium. The bacteria belonging to the genus Pantoea were identified as the major endophytes of maize [39,59] and wheat [54] seeds. Additionally, P. agglomerans was isolated in the seeds of switchgrass [60]. One of the best-known P. vagans strains used in practice is the C9-1 strain isolated from apples (Malus x domestica ‘Jonathan’) (Michigan). This strain is the main component of the registered preparation BlightBan C9-1 (Nufarms America Inc., Burr Ridge, IL) for the biocontrol of fire blight caused by the related enterobacterium Erwinia amylovora [61]. As reported by Smits et al. [61], the P. vagans C9-1 genome does not contain known virulence determinants of enterobacteria, such as toxins, protolytic enzymes, or type II secretion system effectors. The strain of P. vagans isolated in our research—SRS89—was able to synthesize several types of growth regulators with great importance for plant growth, among which GA3 was the most abundant. Previous research has reported different Pantoea ssp. are able to produce growth regulators such as GA3 and ABA [62], IAA [54], cytokinins [63], ACC [64], and to phosphate solubilization or nitrogen fixation [54,65]. GAs have been recognized as regulators in numerous aspects of plant growth and development. The first reported bacterial strain with the ability to produce GA was Rhizobium phaseoli [66]. However, several other species with ability to biosynthesis active GAs have also been identified [67,68,69,70,71,72]. Here, for the first time, we indicated that strain SRS89 of P. vagans isolated from stevia seeds can synthesis GA3 and GA6. It has also been determined that the GA biosynthesis pathway in bacteria is identical to the 13-hydroxylation pathway found in plants, although the presence of ferredoxine (Fd) and short-chain alcohol dehydrogenase reductase (SDR) genes is specific to bacteria [73]. However, in plants, fungi, and bacteria, the biochemical route for GA synthesis starts from geranyl-geranyl diphosphate (GGPP) via isopentenyl-diphosphate (IPP), which is a building block for all terpenoid/isoprenoid compounds [74]. IPP can be generated via the MEP pathway, which in stevia plants is also used for SG biosynthesis [2]. We also detected some genes involved in the early steps of that pathway (dxs, dxr, and idi). It is also interesting that strain SRS89 isolated from stevia seeds seems to be able to perform Reb A biosynthesis. This is the first information about the capability of endophytic bacteria to produce a kind of SG. In the last few years, there have been reports of the possibility of synthesizing SGs by bacteria or fungi; however, this ability was obtained by genetic transformation of model species of microorganisms [75,76,77,78]. Therefore, further research is needed to understand the biosynthetic pathway of these compounds in the P. vagans SRS89 strain. According to all forecasts, the demand for SGs will increase, not only due to the projected increase in the number of diabetic patients but also due to changes in food preferences. However, in many countries, due to climatic conditions, the cultivation of stevia is either impossible or unprofitable. The use of endophytic bacteria for the production of SGs brings many benefits, including low cost, easily scaled-up production, shortening the time, and synthesis independent of environmental conditions. Recent studies have demonstrated that endophytes may be a rich source of natural products for medicinal, industrial, and agricultural use [79,80,81,82,83,84]. Numerous bioactive compounds produced by endophytes have already been commercialized and have been found useful in agriculture and pharmacology [85,86,87,88].
In the SRS89 strain of P. vagans, we also detected IAA and indole-derivative metabolite indole-3-carboxylic acid (I3CA). IAA is the most common auxin in plants and plays a key role in regulating numerous processes related to growth and development. IAA elicits seed growth by cell elongation. Depending on the type of organ and the developmental stage of the plant, IAA can be synthesized either independently or dependently of tryptophan. We also detected tryptophan in our bacteria strain, so we can conclude that it may be used for IAA synthesis. It should be noted that tryptophan is not detected in stevia plants [89]. It is worth mentioning that most auxins occur in plants as auxin conjugates. One of the major auxins, IAA, is inactivated to IAA-aspartate (IAAsp) via ATP-dependent conjugation catalyzed by amidosyntethases from the Gretchen Hagen 3 (GH3) acyl-adenylating enzyme family. Bacteria also have the ability to produce auxin. This ability concerns mainly plant growth-promoting rhizobacteria (PGPR) and endophytes. Our study showed that IAA production is also a feature of SRS89 strain isolated from stevia seeds. The ability to produce IAA was previously detected in different bacteria isolated from the rhizosphere of Stevia rebaudiana [90]. Some researchers have suggested that plants are colonized by high numbers of IAA-producing bacteria [40,49,91]. Some reports have also shown that inoculation of IAA-producing endophytic bacteria is a promising way to enhance plant biomass, root length, root tip number, and root surface area [47,92].
The other growth regulator that we identified in the SRS89 bacteria strain was jasmonic acid. Jasmonates (JAs) are lipid-derived signaling molecules produced by certain bacteria, fungi, and plants. Plants synthesize JAs in response to developmental cues or environmental stress. They take part in the regulation of many physiological processes, including seed germination, root growth, organ formation, flowering, fruit ripening, and leaf aging [93]. While much information exists on the biosynthesis and function of JA in plants, such knowledge regarding microorganisms is still scarce [94].
We also analyzed the effect of the identified strain on stevia seed germination. It is known that some endophytic bacteria have potential as biostimulators of crop species [40,49,95,96]. The surface-sterilized S. rebaudiana seeds were reinoculated with a bacterial culture before germination. Stevia belongs to a salt-sensitive species [26,97], and some evidence exists that endophytic bacteria promote plant growth in salt-stress conditions [98]. These beneficial functions are mainly performed by supplying nutrients, the detoxification of harmful compounds, and the production of bioactive compounds, such as secondary metabolites and hormones. The beneficial effect of bacteria on germination was observed only for control conditions. When there was no NaCl in the germination medium, inoculated stevia seeds germinated 15% better than non-reinoculated seeds. However, when NaCl was present in the medium, we did not observe a beneficial effect. This may indicate that the bacterium is sensitive to salinity and its lack of activity results in lower germination of stevia seeds.
4. Materials and Methods
4.1. Bacteria Isolation and Maintenance
The stevia seeds (POLAN Breeding and Seed Company, Krakow, Poland) were surface sterilized [23] and then placed in Petri dishes (7 cm in diameter) containing sterile solidified lysogeny broth (LB) medium (Sigma-Aldrich, Saint Louis, MO, USA). To confirm that the sterilization process was successful, 100 μL of the water used for the final washing of the surface-sterilized seeds was placed on the same medium and examined for microbial growth during incubation (25 °C for seven days). After sterilization, the seeds were incubated in sterile conditions at 25 °C for seven days. During this time, yellow bacteria appeared around some of the seeds. Bacterial colonies were picked up and further purified by repeated streaking on the same medium. A single bacterial colony was isolated and used to inoculate 5 mL of Tryptic Soy Broth culture medium (Oxoid, Basingstoke, UK). After 24 h of incubation, 4 mL of the bacteria culture was centrifuged at 4 °C with 38,903× g, for 15 min (Sigma 3–16 K), and the obtained pellet was used for genomic DNA extraction. To determine the DNA extraction method, 100 µL of bacteria culture was grown overnight on an LB solid medium at 25 °C. A single bacteria colony was isolated and stained using the standard Gram technique for microscopic observation. The remaining volume of bacterial culture was transferred to new tubes and stored at −80 °C in 20% (v/v) glycerol. DNA was extracted using the DNeasy Blood and Tissue Kit (Qiagen, Hilden, Germany) according to the manufacturer’s protocol. Bacteria DNA concentration was assessed spectrophotometrically using NanoDrop One (Thermo Scientific, Waltham, MA, USA).
For bacteria identification, preliminary analysis using the 16S rRNA gene sequence similarity was performed, followed the detailed characteristics based on the whole genome sequence (WGS) data. Biochemical analysis of the isolated strain was also carried out and the ability to increase the germination capacity of stevia seeds was assessed.
4.2. Amplification and Sequencing of the 16S rRNA Gene
Bacteria species identification was performed according to Marchesi et al. [99]. For the amplification of the 16S rRNA gene, the specific par of primers 63F (5′-CAGGCCTAA CACATGCAAGTC-3′) and 1387R (5′-GGGCGGWGTGTACAAGGC-3′), was used. PCR was performed in 50 µL of a reaction mixture containing 1× GoTaq® Green Master mix, 1 µM of each primer, and <250 ng of bacteria genomic DNA. PCR was conducted as follows: Five min of initial denaturation at 95 °C followed by 30 cycles of denaturation for one min at 95 °C, annealing for one min at 55 °C, and elongation for 1.5 min at 72 °C, followed by a final five-min extension step at 72 °C (Biometra Tone; Analytik Jena, Jena, Germany). The PCR product was visualized via 1% agarose gel electrophoresis with GelRed (Biotium, Fremont, CA, USA) for DNA staining. The PCR product was excised from the gel and cleaned with a QIAquick PCR purification kit (Qiagen, Hilden, Germany). Its concentration was assessed spectrophotometrically (NanoDrop One; Thermo Scientific, Waltham, MA, USA) and adjusted to 10 ng/μL prior to sequencing (MWG, Birmingham, UK). The obtained sequence was analyzed with BLASTn using a database called “16S ribosomal RNA sequences (Bacteria and Archaea)” (NCBI, http://blast.ncbi.nlm.nih.gov/Blast.cgi (accessed on 3 January 2020)).
4.3. WGS Isolated Bacteria Strain and Data Analysis
Genomic DNA obtained from the endophytic bacteria was used for WGS. Sequence libraries were prepared using the Nextera XT v2 library preparation kit and sequenced on a MiSeq desktop sequencer (Illumina, San Diego, CA, USA) using MiSeq V3 reagent kits. The library was sequenced in a 300 bp paired-end run to obtain 404,752 reads. Raw reads were processed using Trimmomatic software [100] to remove adapter sequences (only keeping reads longer than 30 bp after timing) and filtered for quality to remove reads of a quality lower than Q20. Additionally, only reads with pairs after filtering were retained for further analysis. This resulted in 344,414 300-bp reads that were used for genome assembly. The initial assembly was created using Unicycler [101] software and the SPAdes algorithm [102] with an error correction step, in Pilon [103] for the polishing of the final assembly. Subsequently, Quast software [104] was used to evaluate assembly quality. The resulting contig annotation was done using NCBI PGAP. Additional annotation and comparative analysis were performed using the RAST server [105]. Operons were identified using the Operon-mapper software [106]. Taxonomic analysis of the studied strain was performed using a dendrogram created based on genomic BLAST available in the NCBI database in the Pantoea vagans genome data Table Using this approach, we identified closely related Pantoea vagans strains—FDAARGOS_160 (GCA_001558735.2), LMG24199 (GCA_004792415.1), MP7 (GCA_000757435.2), and slightly more distant C9-1 (GCA_000148935.1) and PV989 (GCA_003032455.1)—which were further used for comparative analysis. For visualization purposes using Mauve software [107] and for genome submission, the contigs of the analyzed strains were scaffolded against the C9-1 strain assembly (GCA_000148935.1) using MEDUSA software [108]. Pan- and core-genome analyses, as well as pan- and core-phylogeny, were performed for all the selected stains specified above using BPGA software [109]. The obtained contigs were deposited in the GenBank database under the assembly accession numbers: GCA_009905795.1, ASM990579v1, and the strain was labeled as SRS89.
4.4. HPLC Analysis of Isolated Bacteria
To prepare the bacteria for HPLC analysis, 100 µL of bacteria culture stored at −80 °C was used. Bacteria were grown overnight on an LB solid medium at 25 °C and then streaked onto an LB solid medium for overnight culturing at 25 °C. Single colonies were used to inoculate 100 mL of liquid LB medium and cultured for the next 24 h at 25 °C. The obtained bacterial cultures were centrifuged at 6000× g, and the resulting pellet was stored at −80 °C before lyophilization. To determine SGs and amino acids (phenylalanine and tryptophan) using RP-HPLC analysis, 250 mg of dry biomass was extracted according to Simlat et al. [26]. The analyses were performed in three replications.
4.5. Mass Spectrometry Confirmation of RebA
Samples in methanol were evaporated to dryness under N2 and redissolved in acetonitrile/water (3/1 v/v). Separation was achieved in hydrophilic interaction liquid chromatography (HILIC) mode on BlueOrchid NH2 (100 × 2 mm, 1.8 µm, Knauer, Berlin, Germany) in gradient mode of (A) 2 mM ammonium formate in water and (B) 5% 2 mM ammonium formate in acetonitrile, 0–4 min 100–75% B, 4–5 min 75–35% B, then returned to 100% B in 1 min. The system consisted of UHPLC (Agilent Infinity 1260, Agilent Technologies, Santa Clara, CA, USA) and a triple quadrupole mass spectrometer (Agilent 6410, Agilent Technologies, Santa Clara, CA, USA) with electrospray ionization (ESI). The measurements were performed in the positive mode in single ion monitoring (SIM), and the [M+Na]+ ion was monitored.
4.6. Profiling of Plant Hormones
Ultrahigh-performance liquid chromatography coupled with tandem mass spectrometry (UHPLC-MS/MS) was used for the analysis of phytohormones, according to Hura et al. [110] and Dziurka et al. [111]. A stable isotope-labeled internal standard mixture was added to lyophilized bacteria samples of about 20 mg. Samples were extracted (methanol/H2O/formic acid, 15:4:1 (v/v/v)) and then evaporated under an N2 stream (TurboVap LV, Caliper, Hopkinton, MA, USA). After dissolution in 3% (v/v) methanol in 1 M HCOOH, samples were cleaned up on a hybrid solid-phase extraction (SPE; Bond Elut Plexa PCX; Agilent Technologies, Santa Clara, CA, USA) column. Targeted profiling of phytohormones and related compounds was conducted in multiple reaction monitoring (MRM) mode on an Agilent Infinity 1260 UHPLC system (Agilent Technologies), coupled with 6410 QQQ LC/MS with an electrospray interface (ESI) ion source (Agilent Technologies). The separation was achieved on an Ascentis Express RP-amide analytical column (2.7 μm, 2.1 mm × 150 mm; Supelco, Bellefonte, PA, USA) in a linear gradient of H2O vs. acetonitrile with 0.01% (v/v) of HCOOH. Further technical details are given in Table 4. As internal standard, [2H2]GA1 (D-GA1), [2H2]GA6 (D-GA6) [2H5]indoleacetic acid (D-IAA), [2H4]salicylic acid (D-SA), and [2H5]benzoic acid (D-BeA) (OlChemim, Olomouc, Czech Republic), and [2H5]jasmonic acid (D-JA) (CND Isotopes, Quebec, Canada) were used.

Table 4.
MRM parameters at positive ion mode (+ESI), capillary voltage 4 kV, gas temperature 350 °C, gas flow 12 L/min and nebulizer pressure 35 psi. MassHunter software was used to control the LC–MS/MS system and in data analysis. For MRM parameters optimization MassHunter Optimizer was used. Monitored compounds: [2H5]benzoic acid (D-BeA), benzoic acid (BeA), [2H4]salicylic acid (D-SA), salicylic acid (SA), [2H5]jasmonic acid (D-JA), jasmonic acid (JA), [2H5]indole-3-acetic acid (D-IAA) indole-3-acetic acid (IAA), indole-3-carboxylic acid (I3CA), indole-3-acetyl-aspartic acid (IAA-Asp), gibberellic acid A3 (GA3), [2H2]gibberellic acid A1 (D-GA1), [2H2]gibberellic acid A6 (D-GA6), gibberellic acid A6 (GA6).
4.7. Effect of Endophytic Bacteria on Stevia Seed Germination
Before inoculation, the stevia seeds were surface sterilized according to Simlat et al. [23]. The efficiency of sterilization was confirmed by plating 100 µL of the final rinse water onto LB plates and incubating them for seven days at 25 °C. After disinfection, the seeds were dried on filter paper (to a moisture content of approximately 12%) and then reinoculated by dipping them into the bacterial culture. To prepare the inoculum, 100 µL of the bacteria culture stored at −80 °C was used. Bacteria culture was grown overnight on an LB solid medium at 25 °C and then streaked onto an LB solid medium for overnight culturing at 25 °C. A single colony was used to inoculate 100 mL of the liquid LB medium and cultured for 24 h at 25 °C. After incubation, the bacteria culture was adjusted to 0.5 OD using LB liquid medium and used for seed treatment. Seed inoculation lasted either one or four hours in darkness at 25 °C, during which the seeds were mixed (130 rpm) (Innova, Fredericton, NB, Canada). After inoculation, the seeds were dried and placed on Petri dishes containing AG (dH2O solidified with 0.7% Difco Bacto Agar), AG with 50 mM NaCl, or AG with 150 mM NaCl. Disinfected, non-reinoculated seeds were used as controls for each germination substrate. The germination conditions in the controlled plant growth chamber were as follows: 25 °C, white fluorescent light with an intensity expressed as a photosynthetic photon flux density (PPFD) of 60 μmol m−2s−1 (which we previously tested: [23]) for 12 h/day, and 70 ± 5% of relative humidity (Adaptis-A1000TC, Conviron, Winnipeg, MB, Canada). The experiment was performed in four replications, each of which consisted of 20 seeds. The number of germinated seeds was recorded each day, starting from the first day after the seeds had been placed in Petri dishes. The percentage of germinated seeds after seven days of incubation was expressed as the germination energy (GE), and after 21 days as the germination capacity (GC).
4.8. Statistical Analysis
The obtained data (Reb A, amino acids, growth regulators content) were reported as mean ± standard deviation (SD). To determine the significant differences between inoculation time, the results for GE and GC were analyzed using STATISTICA software (version 13.1, www.statsoft.com, accessed on 10 January 2023) by one-way ANOVA, followed by Duncan’s multiple test (p < 0.05).
5. Conclusions
In summary, the present study reports on the isolation, identification, and characteristics of endophytic bacteria from Stevia rebaudiana Bertoni seeds. Based on the results of phylogenetic analysis based on sequencing data, the isolated strain SRS89 is considered to represent a novel strain of Pantoea vagans. We detected the ability of the isolated strain to promote host seed germination, and to synthesize growth regulators, among which GA3 was the most present. However, in our opinion, the most promising property of isolated strain SRS89 is its ability to synthesize rebaudioside A—a kind of SGs typical of stevia plants. We also found some genes involved in SG biosynthesis. The discovery of endophytic bacteria with the capacity to synthesize Reb A fits with the increasing interest in the use of microorganisms for producing valuable metabolites with health benefits as alternatives to chemical synthesis or plant-derived metabolites. Further research on the SRS89 strain should focus on optimizing the industrial-scale production of Reb A using a fermenter. Additionally, the detailed elucidation of how Reb A is synthesized in bacterial cells is valuable and should be investigated for the possibility of synthesizing other types of rebaudioside.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/ijms24032174/s1.
Author Contributions
Conceptualization, M.S. and A.P.; performed the experiments and analyses, M.S., A.P., A.J., A.S., M.D. and A.G.; writing-original draft preparation, M.S., M.D., A.S. and A.G.; writing—review and editing, M.S., A.P., A.J., A.S., M.D. and A.G. All authors have read and agreed to the published version of the manuscript.
Funding
This research received no external funding.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
The data presented in this study are available on request from the corresponding author.
Acknowledgments
The authors would like to thank Ewa Piotrowska (Department of Plant Breeding, Physiology and Seed Science, University of Agriculture in Krakow) for her help in the seed germination experiments. The research was financed from the subsidy of the Ministry of Science and Higher Education of the Republic of Poland awarded to the University of Agriculture in Krakow, Poland.
Conflicts of Interest
The authors declare no conflict of interest. The funders had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript; or in the decision to publish the results.
References
- Richman, A.; Swanson, A.; Humphrey, T.; Chapman, R.; McGarvey, B.; Pocs, R.; Brandle, J. Functional genomics uncovers three glucosyltransferases involved in the synthesis of the major sweet glucosides of Stevia rebaudiana. Plant J. 2005, 41, 56–67. [Google Scholar] [CrossRef] [PubMed]
- Brandle, J.E.; Telmer, P.G. Steviol glycoside biosynthesis. Phytochemistry 2007, 68, 1855–1863. [Google Scholar] [CrossRef] [PubMed]
- Guleria, P.; Yadav, S.K. Agrobacterium mediated transient gene silencing (AMTS) in Stevia rebaudiana: Insights into steviol glycoside biosynthesis pathway. PLoS ONE 2013, 8, e74731. [Google Scholar] [CrossRef]
- Yadav, A.K.; Singh, S.; Dhyani, D.; Ahuja, P.S. A review on the improvement of stevia [Stevia rebaudiana (Bertoni)]. Can. J. Plant Sci. 2011, 91, 1–27. [Google Scholar] [CrossRef]
- Momtazi-Borojeni, A.A.; Esmaeili, S.A.; Abdollahi, E.; Sahebkar, A. A Review on the pharmacology and toxicology of steviol glycosides extracted from Stevia rebaudiana. Curr. Pharm. Des. 2017, 23, 1616–1622. [Google Scholar] [CrossRef]
- Peteliuk, V.; Rybchuk, L.; Bayliak, M.; Storey, K.B.; Lushchak, O. Natural sweetener Stevia rebaudiana: Functionalities, health benefits and potential risks. Excli. J. 2021, 20, 14121430. [Google Scholar] [CrossRef]
- Ohta, M.; Sasa, S.; Inoue, A.; Tamai, T.; Fujita, I.; Morita, K.; Matsuura, F. Characterization of novel steviol glycosides from leaves of Stevia rebaudiana Morita. J. Appl. Glycosci. 2010, 57, 199–209. [Google Scholar] [CrossRef]
- Chaturvedula, V.S.; Prakash, I. Structures of the novel diterpene glycosides from Stevia rebaudiana. Carbohydr. Res. 2011, 346, 1057–1060. [Google Scholar] [CrossRef]
- Chaturvedula, V.S.; Prakash, I. Additional minor diterpene glycosides from Stevia rebaudiana. Nat. Prod. Commun. 2011, 6, 1059–1062. [Google Scholar] [CrossRef]
- Chaturvedula, V.S.; Rhea, J.; Milanowski, D.; Mocek, U.; Prakash, I. Two minor diterpene glycosides from the leaves of Stevia rebaudiana. Nat. Prod. Commun. 2011, 6, 175–178. [Google Scholar] [CrossRef]
- Ceunen, S.; Geuns, J.M.C. Steviol glycosides: Chemical diversity, metabolism, and function. J. Nat. Prod. 2013, 76, 1201–1228. [Google Scholar] [CrossRef] [PubMed]
- Prakash, I.; Chaturvedula, V.S.P. Steviol glycosides: Natural noncaloric sweeteners. In Sweeteners: Pharmacology, Biotechnology, and Applications; Mérillon, J.-M., Ramawat, K.G., Eds.; Springer: Cham, Switzerland, 2018; Volume 5, pp. 101–128. [Google Scholar]
- DuBois, G.E.; Stephenson, R.A. Diterpenoid sweeteners. Synthesis and sensory evaluation of stevioside analogs with improved organoleptic properties. J. Med. Chem. 1985, 28, 93–98. [Google Scholar] [CrossRef] [PubMed]
- Li, P.; Wang, Z.; Lam, S.M.; Shui, G. Rebaudioside A enhances pesistance to oxidative stress and extends lifespan and healthspan in Caenorhabditis elegans. Antioxidants 2021, 10, 262. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Luo, X.; Chen, L.; Mustapha, A.T.; Yu, X.; Zhou, C.; Okonkwo, C.E. Natural and low-caloric rebaudioside A as a substitute for dietary sugars: A comprehensive review. Compr. Rev. Food Sci. Food Saf. 2023, 22, 615–642. [Google Scholar] [CrossRef] [PubMed]
- Nettleton, J.E.; Klancic, T.; Schick, A.; Choo, A.C.; Shearer, J.; Borgland, S.L.; Chleilat, F.; Mayengbam, S.; Reimer, R.A. Low-dose stevia (rebaudioside A) consumption perturbs gut microbiota and the mesolimbic dopamine reward system. Nutrients 2019, 11, 1248. [Google Scholar] [CrossRef]
- Kasti, A.N.; Nikolaki, M.D.; Synodinou, K.D.; Katsas, K.N.; Petsis, K.; Lambrinou, S.; Pyrousis, I.A.; Triantafyllou, K. The effects of stevia consumption on gut bacteria: Friend or foe? Microorganisms 2022, 10, 744. [Google Scholar] [CrossRef]
- Bondarev, N.; Sukhanova, M.; Reshetnyak, O.; Nosov, A.M. Steviol glycoside content in different organs of Stevia rebaudiana and its dynamics during ontogeny. Biol. Plant 2003, 47, 261–264. [Google Scholar] [CrossRef]
- Kang, K.H.; Lee, E.W. Physio-ecological studies on stevia (Stevia rebaudiana Bertoni). Korean J. Crop. Sci. 1981, 26, 69–89. [Google Scholar]
- Tavarini, S.; Sgherri, C.; Ranieri, A.M.; Angelini, L.G. Effect of nitrogen fertilization and harvest time on steviol glycosides, flavonoid composition, and antioxidant properties in Stevia rebaudiana Bertoni. J. Agric. Food Chem. 2015, 63, 7041–7050. [Google Scholar] [CrossRef]
- Abdullateef, R.A.; Osman, M. Effects of visible light wavelengths on seed germinability in Stevia rebaudiana Bertoni. Int. J. Biol. 2011, 3, 83–91. [Google Scholar] [CrossRef]
- Kumar, R.; Sharma, S. Effect of light and temperature on seed germination of important medicinal and aromatic plants in north western Himalayas. Int. J. Med. Arom. Plants 2012, 2, 468–475. [Google Scholar]
- Simlat, M.; Ślęzak, P.; Moś, M.; Warchoł, M.; Skrzypek, E.; Ptak, A. The effect of light quality on seed germination, seedling growth and selected biochemical properties of Stevia rebaudiana Bertoni. Sci. Hortic. 2016, 211, 295–304. [Google Scholar] [CrossRef]
- Simlat, M.; Ptak, A.; Skrzypek, E.; Warchoł, M.; Morańska, E.; Piórkowska, E. Melatonin significantly influences seed germination and seedling growth of Stevia rebaudiana Bertoni. PeerJ 2018, 6, e5009. [Google Scholar] [CrossRef] [PubMed]
- Simlat, M.; Skrzypek, E.; Warchoł, M.; Maciaszek, I.; Ptak, A. Evaluation on Stevia rebaudiana Bertoni seed germination and seedling development under phytohormones treatment. Sci. Hortic. 2019, 257, 108717. [Google Scholar] [CrossRef]
- Simlat, M.; Szewczyk, A.; Ptak, A. Melatonin promotes seed germination under salinity and enhances the biosynthesis of steviol glycosides in Stevia rebaudiana Bertoni leaves. PLoS ONE 2020, 15, e0230755. [Google Scholar] [CrossRef]
- Puente, M.E.; Li, C.Y.; Bashan, Y. Endophytic bacteria in cacti seeds can improve the development of cactus seedlings. Environ. Exp. Bot. 2009, 66, 402–408. [Google Scholar] [CrossRef]
- Brader, G.; Compant, S.; Vescio, K.; Mitter, B.; Trognitz, F.; Ma, L.-J.; Sessitsch, A. Ecology and genomic insights into plant-pathogenic and plant-nonpathogenic endophytes. Annu. Rev. Phytopathol. 2017, 55, 61–83. [Google Scholar] [CrossRef]
- Smith, S.A.; Tank, D.C.; Boulanger, L.-A.; Bascom-Slack, C.A.; Eisenman, K.; Kingery, D.; Babbs, B.; Fenn, K.; Greene, J.S.; Hann, B.D.; et al. Bioactive endophytes warrant intensified exploration and conservation. PLoS ONE 2008, 3, e3052. [Google Scholar] [CrossRef] [PubMed]
- Coleman-Derr, D.; Tringe, S.G. Building the crops of tomorrow: Advantages of symbiont-based approaches to improving abiotic stress tolerance. Front. Microbiol. 2014, 5, 283. [Google Scholar] [CrossRef] [PubMed]
- Rosenblueth, M.; Martínez-Romero, E. Bacterial endophytes and their interactions with hosts. Mol. Plant Microbe. Interact. 2006, 19, 827–837. [Google Scholar] [CrossRef]
- Kobayashi, D.Y.; Palumbo, J.D. Bacterial endophytes and their effects on plants and uses in agriculture. In Microbial Endophytes; Bacon, C.W., White, J.F., Eds.; Marcel Dekker, Inc.: New York, NY, USA, 2000; pp. 199–233. [Google Scholar]
- Elmagzob, A.A.H.; Ibrahim, M.M.; Zhang, G.F. Seasonal diversity of endophytic bacteria associated with Cinnamomum camphora (L.). Presl. Diversity 2019, 11, 112. [Google Scholar] [CrossRef]
- Anjum, N.; Chandra, R. Endophytic bacteria: Optimization of isolation procedure from various medicinal plants and their preliminary characterization. Asian J. Pharm. Clin. Res. 2015, 8, 233–238. [Google Scholar]
- Oviedo-Pereira, D.G.; López-Meyer, M.; Evangelista-Lozano, S.; Sarmiento-López, L.G.; Sepúlveda-Jiménez, G.; Rodríguez-Monroy, M. Enhanced specialized metabolite, trichome density, and biosynthetic gene expression in Stevia rebaudiana (Bertoni) Bertoni plants inoculated with endophytic bacteria Enterobacter hormaechei. PeerJ 2022, 10, e13675. [Google Scholar] [CrossRef] [PubMed]
- Yu, X.; Yang, J.; Wang, E.; Li, B.; Yuan, H. Effects of growth stage and fulvic acid on the diversity and dynamics of endophytic bacterial community in Stevia rebaudiana Bertoni leaves. Front. Microbiol. 2015, 6, 867. [Google Scholar] [CrossRef] [PubMed]
- Ferreira, A.; Quecine, M.C.; Lacava, P.T.; Oda, S.; Azevedo, J.L.; Araújo, W.L. Diversity of endophytic bacteria from Eucalyptus species seeds and colonization of seedlings by Pantoea agglomerans. FEMS Microbiol. Lett. 2008, 287, 8–14. [Google Scholar] [CrossRef] [PubMed]
- Johnston-Monje, D.; Raizada, M.N. Conservation and diversity of seed associated endophytes in Zea across boundaries of evolution, ethnography and ecology. PLoS ONE 2011, 6, e20396. [Google Scholar] [CrossRef]
- Rijavec, T.; Lapanje, A.; Dermastia, M.; Rupnik, M. Isolation of bacterial endophytes from germinated maize kernels. Can. J. Microbiol. 2007, 53, 802–808. [Google Scholar] [CrossRef]
- Walitang, D.I.; Kim, K.; Madhaiyan, M.K.; Kim, Y.K.; Kang, Y.; Sa, T. Characterizing endophytic competence and plant growth promotion of bacterial endophytes inhabiting the seed endosphere of Rice. BMC Microbiol. 2017, 17, 209. [Google Scholar] [CrossRef]
- Shahzad, R.; Khan, A.L.; Saqib, B.; Sajjad, A.; In-Jung, L. What is there in seeds? Vertically transmitted endophytic resources for sustainable improvement in plant growth. Front. Plant Sci. 2018, 9, 24. [Google Scholar] [CrossRef]
- Tyc, O.; Putra, R.; Gols, R.; Harvey, J.A.; Garbeva, P. The ecological role of bacterial seed endophytes associated with wild cabbage in the United Kingdom. MicrobiologyOpen 2020, 9, e00954. [Google Scholar] [CrossRef]
- Zhu, B.; Chen, M.; Lin, L.; Yang, L.; Li, Y.; An, Q. Genome sequence of Enterobacter sp. strain SP1, an endophytic nitrogen-fixing bacterium isolated from sugarcane. J. Bacteriol. 2012, 194, 6963–6964. [Google Scholar] [CrossRef] [PubMed]
- Lin, L.; Wei, C.; Chen, M.; Wang, H.; Li, Y.; Li, Y.; Yang, L.; An, Q. Complete genome sequence of endophytic nitrogen-fixing Klebsiella variicola strain DX120E. Stand. Genom. Sci. 2015, 10, 22. [Google Scholar] [CrossRef]
- Ji, S.H.; Gururanib, M.A.; Chun, S.C. Isolation and characterization of plant growth promoting endophytic diazotrophic bacteria from Korean rice cultivars. Microbiol. Res. 2014, 169, 83–98. [Google Scholar] [CrossRef] [PubMed]
- Qin, S.; Zhang, Y.J.; Yuan, B.; Xu, Y.P.; Xing, K.; Wang, J.; Jiang, J.-H. Isolation of ACC deaminase-producing habitat-adapted symbiotic bacteria associated with halophyte Limonium sinense (Girard) Kuntze and evaluating their plant growth-promoting activity under salt stress. Plant Soil. 2014, 374, 753–766. [Google Scholar] [CrossRef]
- Khan, A.L.; Waqas, M.; Kang, S.M.; Al-Harrasi, A.; Hussain, J.; Al-Rawahi, A.; Al-Khiziri, S.; Ullah, I.; Ali, L.; Jung, H.Y.; et al. Bacterial endophyte Sphingomonas sp. LK11 produces gibberellins and IAA and promotes tomato plant growth. J. Microbiol. 2014, 52, 689–695. [Google Scholar] [CrossRef] [PubMed]
- Yan, X.; Wang, Z.; Mei, Y.; Wang, L.; Wang, X.; Xu, Q.; Peng, S.; Zhou, Y.; Wei, C. Isolation, diversity, and growth-promoting activities of endophytic bacteria from tea cultivars of Zijuan and Yunkang-10. Front. Microbiol. 2018, 9, 1848. [Google Scholar] [CrossRef]
- Chen, C.; Xin, K.; Liu, H.; Cheng, J.; Shen, X.; Wang, Y.; Zhang, L. Pantoea alhagi, a novel endophytic bacterium with ability to improve growth and drought tolerance in wheat. Sci. Rep. 2017, 7, 41564. [Google Scholar] [CrossRef]
- Wu, W.; Chen, W.; Liu, S.; Wu, J.; Zhu, Y.; Qin, L.; Zhu, B. Beneficial relationships between endophytic bacteria and medicinal plants. Front. Plant Sci. 2021, 12, 646146. [Google Scholar] [CrossRef]
- Lacava, P.T.; Azevedo, J.L. Endophytic bacteria: A biotechnological potential in agrobiology system. In Bacteria in Agrobiology: Crop Productivity; Maheshwari, D.K., Saraf, M., Aeron, A., Eds.; Springer-Verlag: Berlin/Heidelberg, Germany, 2013; pp. 1–44. [Google Scholar]
- Walterson, A.M.; Stavrinides, J. Pantoea: Insights into a highly versatile and diverse genus within the Enterobacteriaceae. FEMS Microbiol. Rev. 2015, 39, 968–984. [Google Scholar] [CrossRef]
- Pusey, P.L. Biological control agents for fire blight of apple compared under conditions limiting natural dispersal. Plant Dis. 2002, 86, 639–644. [Google Scholar] [CrossRef]
- Herrera, S.D.; Grossi, C.; Zawoznik, M.; Groppa, M.D. Wheat seeds harbour bacterial endophytes with potential as plant growth promoters and biocontrol agents of Fusarium graminearum. Microbiol. Res. 2016, 186–187, 37–43. [Google Scholar] [CrossRef] [PubMed]
- Lorenzi, A.S.; Bonatelli, M.L.; Chia, M.A.; Peressim, L.; Quecine, M.C. Opposite sides of Pantoea agglomerans and its associated commercial outlook. Microorganisms 2022, 10, 2072. [Google Scholar] [CrossRef] [PubMed]
- Lv, L.; Luo, J.; Ahmed, T.; Zaki, H.E.M.; Tian, Y.; Shahid, M.S.; Chen, J.; Li, B. Beneficial effect and potential risk of Pantoea on rice production. Plants 2022, 11, 2608. [Google Scholar] [CrossRef] [PubMed]
- Tambong, J.T. Taxogenomics and systematics of the genus Pantoea. Front. Microbiol. 2019, 10, 2463. [Google Scholar] [CrossRef] [PubMed]
- Palmer, M.; de Maayer, P.; Poulsen, M.; Steenkamp, E.T.; van Zyl, E.; Coutinho, T.A.; Venter, S.N. Draft genome sequences of Pantoea agglomerans and Pantoea vagans isolates associated with termites. Stand Genomic. Sci. 2016, 11, 23. [Google Scholar] [CrossRef]
- Sheibani-Tezerji, R.; Naveed, M.; Jehl, M.A.; Sessitsch, A.; Rattei, T.; Mitter, B. The genomes of closely related Pantoea ananatis maize seed endophytes having different effects on the host plant differ in secretion system genes and mobile genetic elements. Front. Microbiol. 2015, 6, 440. [Google Scholar] [CrossRef]
- Kim-Dura, S.; Lowman, S.; Zhang, S.; Mei, C. Growth promotion of switchgrass by bacterial endophyte Pantoea agglomerans strain PaKM isolated from seeds. J. Path. Microbiol. 2016, 1, 1007. [Google Scholar]
- Smits, T.H.; Rezzonico, F.; Kamber, T.; Goesmann, A.; Ishimaru, C.A.; Stockwell, V.O.; Frey, J.E.; Duffy, B. Genome sequence of the biocontrol agent Pantoea vagans strain C9-1. J. Bacteriol. 2010, 192, 6486–6487. [Google Scholar] [CrossRef]
- Feng, Y.; Shen, D.; Song, W. Rice endophyte Pantoea agglomerans YS19 promotes host plant growth and affects allocations of host photosynthates. J. Appl. Microbiol. 2006, 100, 938–945. [Google Scholar] [CrossRef]
- Omer, Z.S.; Björkman, P.-O.; Nicander, B.; Tillberg, E.; Gerhardson, B. 5′-Deoxyisopentenyladenosine and other cytokinins in culture filtrates of the bacterium Pantoea agglomerans. Physiol. Plant 2004, 121, 439–447. [Google Scholar] [CrossRef]
- Trifi, H.; Ben Salem, I.; Benzina, K.; Fourati, A.; Costa, M.C.; Achouak, W.; Saidi, M. Effectiveness of the plant growth-promoting rhizobacterium Pantoea sp. BRM17 in enhancing Brassica napus growth in phosphogypsum-amended soil. Pedosphere 2017, 30, 4. [Google Scholar] [CrossRef]
- Kim, S.-N.; Cho, W.-K.; Kim, W.-I.; Jee, H.-J.; Park, C.-S. Growth promotion of pepper plants by Pantoea ananatis B1-9 and its efficient endophytic colonization capacity in pant tissues. Plant Pathol. J. 2012, 28, 270–281. [Google Scholar] [CrossRef]
- Atzorn, R.; Crozier, A.; Wheeler, C.T.; Sandberg, G. Production of gibberellins and indole-3-acetic acid by Rhizobium phaseoli in relation to nodulation of Phaseolus vulgaris roots. Planta 1988, 175, 532–538. [Google Scholar] [CrossRef] [PubMed]
- Bottini, R.; Fulchieri, M.; Pearce, D.; Pharis, R.P. Identification of gibberellins A1, A3, and Iso-A3 in cultures of Azospirillum lipoferum. Plant Physiol. 1989, 90, 45–47. [Google Scholar] [CrossRef]
- Bastian, F.; Cohen, A.; Piccoli, P.; Luna, V.; Baraldi, R.; Bottini, R. Production of indole-3-acetic acid and gibberellins A1 and A3 by Acetobacter diazotrophicus and Herbaspirillum seropedicae in chemically-defined culture media. Plant Growth Regul. 1998, 24, 7–11. [Google Scholar] [CrossRef]
- Kang, S.M.; Joo, G.J.; Hamayun, M.; Na, C.I.; Shin, D.H.; Kim, H.Y.; Lee, I.J. Gibberellin production and phosphate solubilization by newly isolated strain of Acinetobacter calcoaceticus and its effect on plant growth. Biotechnol. Lett. 2009, 31, 277–281. [Google Scholar] [CrossRef]
- Kang, S.M.; Khan, A.L.; You, Y.H.; Kim, J.G.; Kamran, M.; Lee, I.J. Gibberellin production by newly isolated strain Leifsonia soli SE134 and its potential to promote plant growth. J. Microbiol. Biotechnol. 2014, 24, 106–112. [Google Scholar] [CrossRef]
- Joo, G.J.; Kim, Y.M.; Kim, J.T.; Rhee, I.K.; Kim, J.H.; Lee, I.J. Gibberellins-producing rhizobacteria increase endogenous gibberellins content and promote growth of red peppers. J. Microbiol. 2005, 43, 510–515. [Google Scholar]
- Joo, G.J.; Kang, S.M.; Hamayun, M.; Kim, S.K.; Na, C.I.; Shin, D.H.; Lee, I.J. Burkholderia sp. KCTC 11096BP as a newly isolated gibberellin producing bacterium. J. Microbiol. 2009, 47, 167–171. [Google Scholar] [CrossRef]
- Salazar-Cerezo, S.; Martínez-Montiel, N.; García-Sánchez, J.; Pérez-y-Terrón, R.; Martínez-Contreras, R.D. Gibberellin biosynthesis and metabolism: A convergent route for plants, fungi and bacteria. Microbiol. Res. 2018, 208, 85–98. [Google Scholar] [CrossRef]
- Nett, R.S.; Montanares, M.; Marcassa, A.; Lu, X.; Nagel, R.; Charles, T.C.; Hedden, P.; Rojas, M.C.; Peters, R.J. Elucidation of gibberellin biosynthesis in bacteria reveals convergent evolution. Nat. Chem. Biol. 2017, 13, 69–74. [Google Scholar] [CrossRef] [PubMed]
- Olsson, K.; Carlsen, S.; Semmler, A.; Simón, E.; Mikkelsen, M.D.; Møller, B.L. Microbial production of next-generation stevia sweeteners. Microb. Cell Fact. 2016, 15, 207. [Google Scholar] [CrossRef]
- Wang, J.; Li, S.; Xiong, Z.; Wang, Y. Pathway mining-based integration of critical enzyme parts for de novo biosynthesis of steviol glycosides sweetener in Escherichia coli. Cell Res. 2016, 26, 258–261. [Google Scholar] [CrossRef] [PubMed]
- Moon, J.H.; Lee, K.; Lee, J.H.; Lee, P.C. Redesign and reconstruction of a steviol-biosynthetic pathway for enhanced production of steviol in Escherichia coli. Microb. Cell Fact. 2020, 19, 20. [Google Scholar] [CrossRef]
- Guo, B.; Hou, X.; Zhang, Y.; Deng, Z.; Ping, Q.; Fu, K.; Yuan, Z.; Rao, Y. Highly efficient production of rebaudioside D enabled by structure-guided engineering of bacterial glycosyltransferase YojK. Front. Bioeng. Biotechnol. 2022, 10, 985826. [Google Scholar] [CrossRef] [PubMed]
- Eljounaidi, K.; Lee, S.K.; Bae, H. Bacterial endophytes as potential biocontrol agents of vascular wilt diseases—Review and future prospects. Biol. Control. 2016, 103, 62–68. [Google Scholar] [CrossRef]
- Ptak, A.; Morańska, E.; Warchoł, M.; Gurgul, A.; Skrzypek, E.; Dziurka, M.; Laurain-Mattar, D.; Spina, R.; Jaglarz, A.; Simlat, M. Endophytic bacteria from in vitro culture of Leucojum aestivum L. a new source of galanthamine and elicitor of alkaloid biosynthesis. Sci. Rep. 2022, 12, 13700. [Google Scholar] [CrossRef]
- Ali, S.; Charles, T.C.; Glick, B.R. Amelioration of high salinity stress damage by growth-promoting bacterial endophytes that contain ACC deaminase. Plant Physiol. Biochem. 2014, 80, 160–167. [Google Scholar] [CrossRef]
- Fikri, A.S.I.; Rahman, I.A.; Nor, N.S.M.; Hamzah, A. Isolation and identification of local bacteria endophyte and screening of its antimicrobial property against pathogenic bacteria and fungi. AIP Conf. Proc. 2018, 1940, 020072. [Google Scholar] [CrossRef]
- Ek-Ramos, M.J.; Gomez-Flores, R.; Orozco-Flores, A.A.; Rodríguez-Padilla, C.; González-Ochoa, G.; Tamez-Guerra, P. Bioactive products from plant-endophytic gram-positive bacteria. Front. Microbiol. 2019, 10, 463. [Google Scholar] [CrossRef]
- Munakata, Y.; Gavira, C.; Genestier, J.; Bourgaud, F.; Hehn, A.; Slezack-Deschaumes, S. Composition and functional comparison of vetiver root endophytic microbiota originating from different geographic locations that show antagonistic activity towards Fusarium graminearum. Microbiol. Res. 2021, 243, 126650. [Google Scholar] [CrossRef] [PubMed]
- Joseph, B.; Mini Priya, R. Bioactive compounds from endophytes and their potential in pharmaceutical effect: A Review. Am. J. Biochem. Mol. Biol. 2011, 1, 291–309. [Google Scholar] [CrossRef]
- Singh, M.; Kumar, A.; Singh, R.; Pandey, K.D. Endophytic bacteria: A new source of bioactive compounds. 3 Biotech 2017, 7, 315. [Google Scholar] [CrossRef]
- Basit, A.; Shah, S.T.; Ullah, I.; Ullah, I.; Mohamed, H.I. Microbial bioactive compounds produced by endophytes (bacteria and fungi) and their uses in plant health. In Plant Growth-Promoting Microbes for Sustainable Biotic and Abiotic Stress Management; Mohamed, H.I., El-Beltagi, H.E.-D.S., Abd-Elsalam, K.A., Eds.; Springer: Cham, Switzerland, 2021; pp. 285–318. [Google Scholar] [CrossRef]
- Rochín-Hernández, L.S.; Rochín-Hernández, L.J.; Flores-Cotera, L.B. Endophytes, a potential source of bioactive compounds to curtail the formation–accumulation of advanced glycation end products: A Review. Molecules 2022, 27, 4469. [Google Scholar] [CrossRef]
- Periche, A.; Koutsidis, G.; Escriche, I. Composition of antioxidants and amino acids in stevia leaf infusions. Plant Foods Hum. Nutr. 2014, 69, 1–7. [Google Scholar] [CrossRef] [PubMed]
- Chandra, S.; Askari, K.; Kumari, M. Optimization of indole acetic acid production by isolated bacteria from Stevia rebaudiana rhizosphere and its effects on plant growth. J. Genet. Eng. Biotechnol. 2018, 16, 581–586. [Google Scholar] [CrossRef] [PubMed]
- Kim, Y.C.; Leveau, J.; Gardener, B.B.M.; Pierson, E.A.; Pierson, L.S.; Ryu, C.-M. The multifactorial basis for plant health promotion by plant-associated bacteria. Appl. Environ. Microb. 2011, 77, 1548–1555. [Google Scholar] [CrossRef]
- Strader, L.C.; Culler, A.H.; Cohen, J.D.; Bartel, B. Conversion of endogenous indole-3-butyric acid to indole-3-acetic acid drives cell expansion in Arabidopsis seedlings. Plant Physiol. 2010, 153, 1577–1586. [Google Scholar] [CrossRef]
- Avanci, N.C.; Luche, D.D.; Goldman, G.H.; Goldman, M.H. Jasmonates are phytohormones with multiple functions, including plant defense and reproduction. Genet. Mol. Res. 2010, 9, 484–505. [Google Scholar] [CrossRef]
- Eng, F.; Marin, J.E.; Zienkiewicz, K.; Gutiérrez-Rojas, M.; Favela-Torres, E.; Feussner, I. Jasmonic acid biosynthesis by fungi: Derivatives, first evidence on biochemical pathways and culture conditions for production. PeerJ 2021, 9, e10873. [Google Scholar] [CrossRef]
- Quecine, M.; Araújo, W.L.; Rossetto, P.B.; Ferreira, A.; Tsui, S.; Lacava, P.; Mondin, M.; Azevedo, J.L.; Pizzirani-Kleiner, A.A. Sugarcane growth promotion by the endophytic bacterium Pantoea agglomerans 33. 1. Appl. Environ. Microbiol. 2012, 78, 7511–7518. [Google Scholar] [CrossRef] [PubMed]
- Du Jardin, P. Plant biostimulants: Definition, concept, main categories and regulation. Sci. Hortic. 2015, 196, 3–14. [Google Scholar] [CrossRef]
- Angelini, L.G.; Martini, A.; Passera, B.; Tavarini, S. Cultivation of Stevia rebaudiana Bertoni and Associated Challenges. In Sweeteners; Mérillon, J.M., Ramawat, K., Eds.; Reference Series in Phytochemistry; Springer: Cham, Switzerland, 2018; pp. 35–85. [Google Scholar]
- Bharti, N.; Pandey, S.; Barnawal, D.; Patel, V.K.; Kalra, A. Plant growth promoting rhizobacteria Dietzia natronolimnaea modulates the expression of stress responsive genes providing protection of wheat from salinity stress. Sci. Rep. 2016, 6, 34768. [Google Scholar] [CrossRef] [PubMed]
- Marchesi, J.R.; Sato, T.; Weightman, A.J.; Martin, T.A.; Fry, J.C.; Hiom, S.J.; Dymock, D.; Wade, W.G. Design and evaluation of useful bacterium-specific PCR primers that amplify genes coding for bacterial 16S rRNA. Appl. Environ. Microbiol. 1998, 64, 795–799. [Google Scholar] [CrossRef] [PubMed]
- Bolger, A.M.; Lohse, M.; Usadel, B. Trimmomatic: A flexible trimmer for Illumina sequence data. Bioinformatics 2014, 30, 2114–2210. [Google Scholar] [CrossRef] [PubMed]
- Wick, R.R.; Judd, L.M.; Gorrie, C.L.; Holt, K.E. Unicycler: Resolving bacterial genome assemblies from short and long sequencing reads. PLoS Comput. Biol. 2017, 13, e1005595. [Google Scholar] [CrossRef]
- Bankevich, A.; Nurk, S.; Antipov, D.; Gurevich, A.A.; Dvorkin, M.; Kulikov, A.S.; Lesin, V.M.; Nikolenko, S.I.; Pham, S.; Prjibelski, A.D.; et al. SPAdes: A new genome assembly algorithm and its applications to single-cell sequencing. J. Comput. Biol. 2012, 19, 455–477. [Google Scholar] [CrossRef]
- Walker, B.; Abeel, T.; Shea, T.; Priest, M.; Abouelliel, A.; Sakthikumar, S.; Cuomo, C.A.; Zeng, Q.; Wortman, J.; Young, S.K.; et al. Pilon: An integrated tool for comprehensive microbial variant detection and genome assembly improvement. PLoS ONE 2014, 9, e112963. [Google Scholar] [CrossRef]
- Gurevich, A.; Saveliev, V.; Vyahhi, N.; Tesler, G. QUAST: Quality assessment tool for genome assemblies. Bioinformatics 2013, 29, 1072–1075. [Google Scholar] [CrossRef]
- Aziz, R.K.; Bartels, D.; Best, A.A.; DeJongh, M.; Disz, T.; Edwards, R.A.; Formsma, K.; Gerdes, S.; Glass, E.M.; Kubal, M.; et al. The RAST Server: Rapid annotations using subsystems technology. BMC Genomics. 2008, 8, 75. [Google Scholar] [CrossRef]
- Taboada, B.; Estrada, K.; Ciria, R.; Merino, E. Operon-mapper: A web server for precise operon identification in bacterial and archaeal genomes. Bioinformatics 2018, 34, 4118–4120. [Google Scholar] [CrossRef]
- Darling, A.C.; Mau, B.; Blattner, F.R.; Perna, N.T. Mauve: Multiple alignment of conserved genomic sequence with rearrangements. Genome Res. 2004, 14, 1394–1403. [Google Scholar] [CrossRef] [PubMed]
- Bosi, E.; Donati, B.; Galardini, M.; Brunetti, S.; Sagot, M.F.; Lió, P.; Crescenzi, P.; Fani, R.; Fondi, M. MeDuSa: A multi-draft based scaffolder. Bioinformatics 2015, 31, 2443–2451. [Google Scholar] [CrossRef]
- Chaudhari, N.; Gupta, V.; Dutta, C. BPGA- an ultra-fast pan-genome analysis pipeline. Sci. Rep. 2016, 6, 24373. [Google Scholar] [CrossRef]
- Hura, T.; Dziurka, M.; Hura, K.; Ostrowska, A.; Dziurka, K.; Gadzinowska, J. Wheat and rye genome confer specific phytohormone profile features and interplay under water stress in two phenotypes of triticale. Plant Physiol. Biochem. 2017, 118, 494–509. [Google Scholar] [CrossRef] [PubMed]
- Dziurka, K.; Dziurka, M.; Muszyńska, E.; Czyczyło-Mysza, I.; Warchoł, M.; Juzoń, K.; Laskoś, K.; Skrzypek, E. Anatomical and hormonal factors determining the development of haploid and zygotic embryos of oat (Avena sativa L.). Sci. Rep. 2022, 12, 1–13. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).