Specific Six-Transmembrane Epithelial Antigen of the Prostate 1 Capture with Gellan Gum Microspheres: Design, Optimization and Integration
Abstract
1. Introduction
2. Results
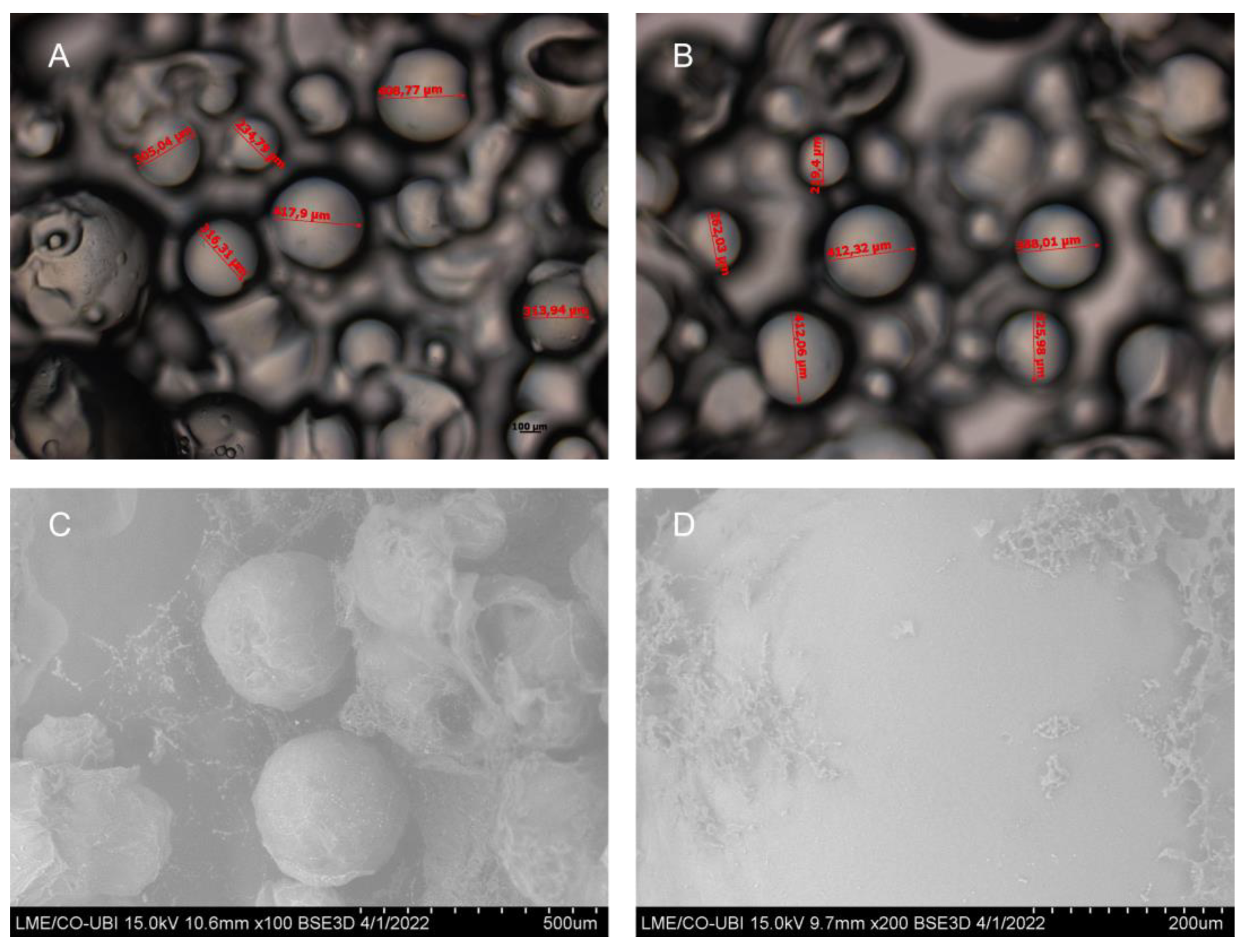
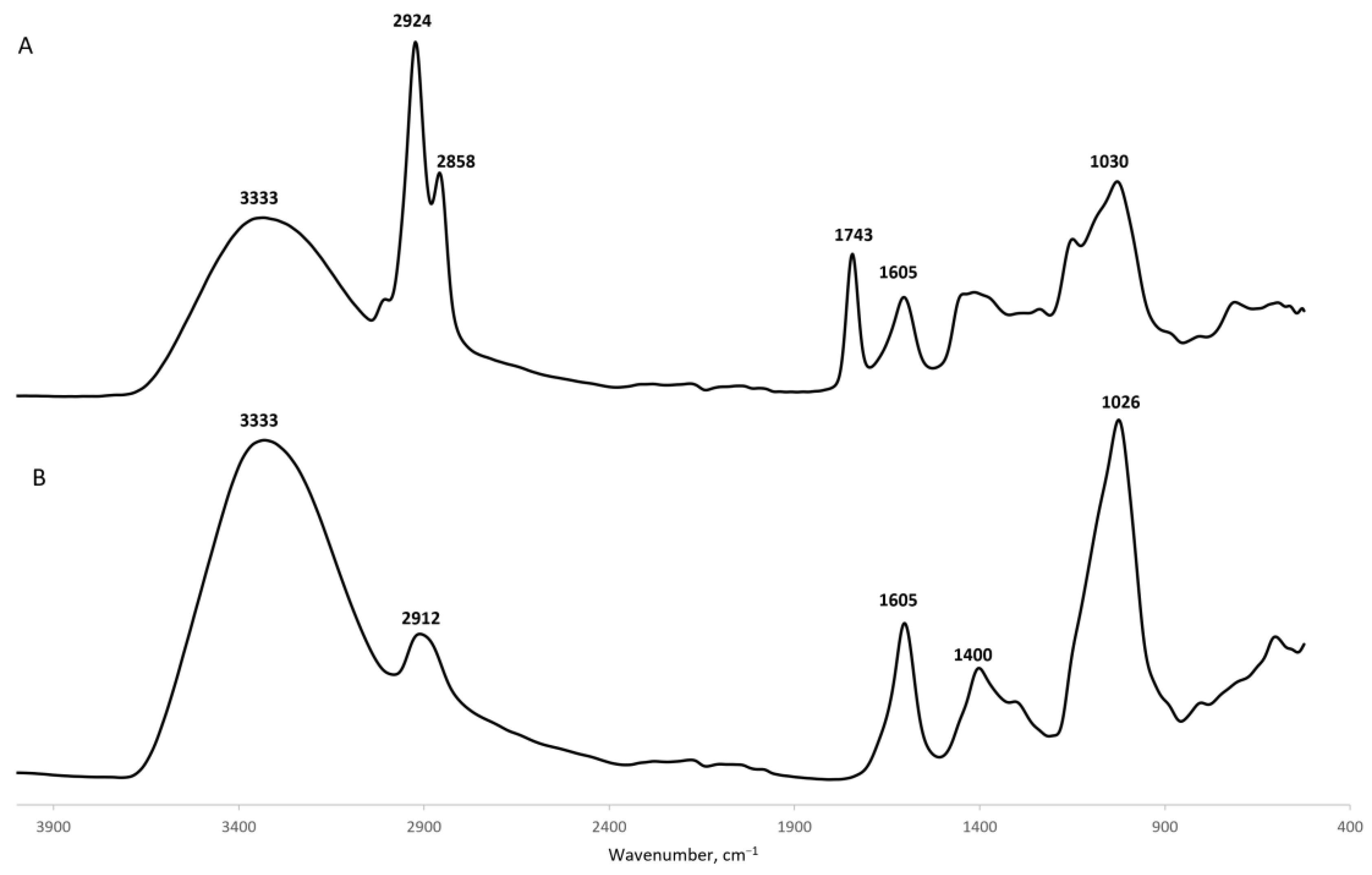
2.1. Characterization of Gellan Gum Microspheres
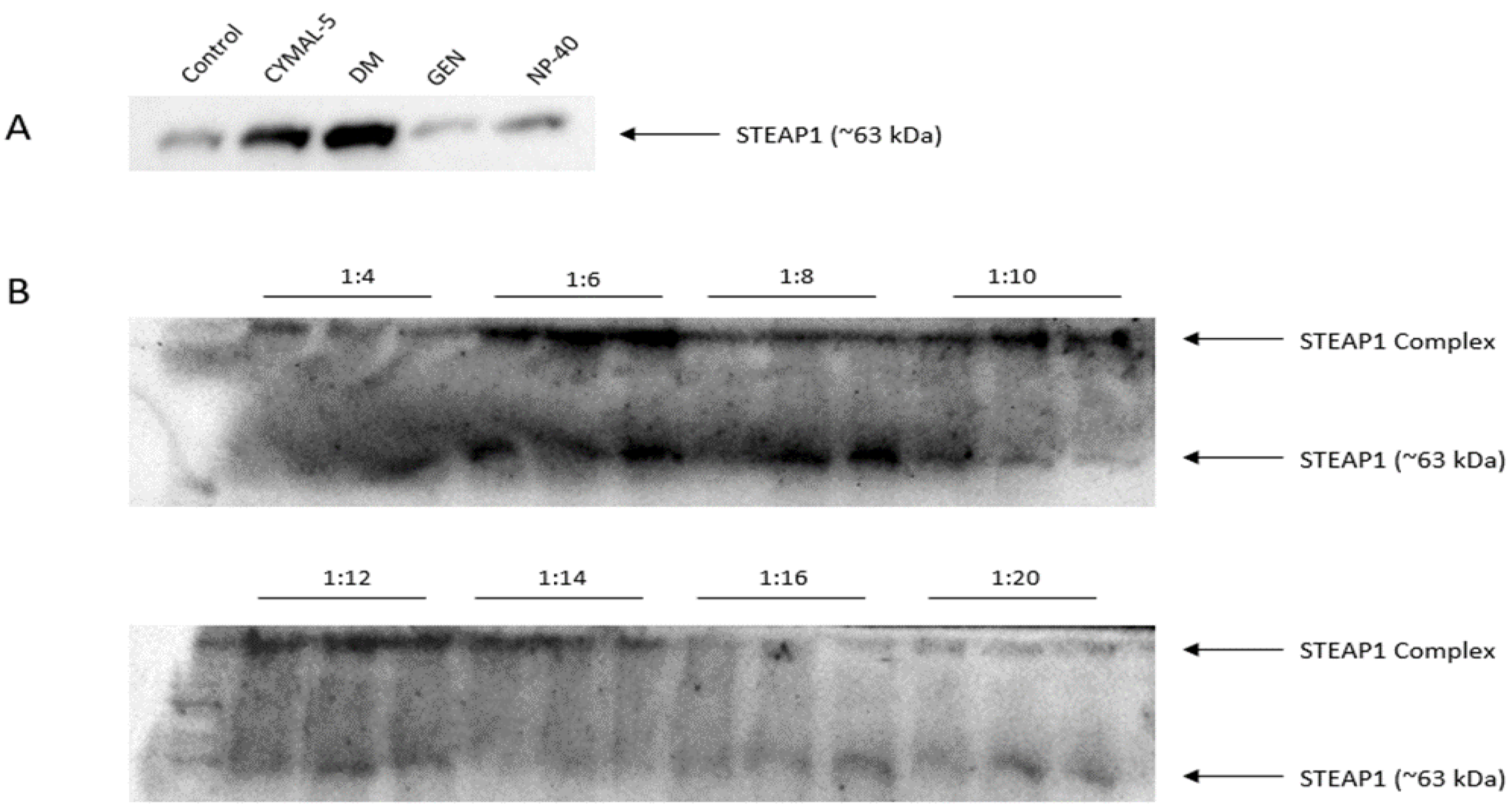
2.2. Optimization of the Batch Method for the Capture of STEAP1
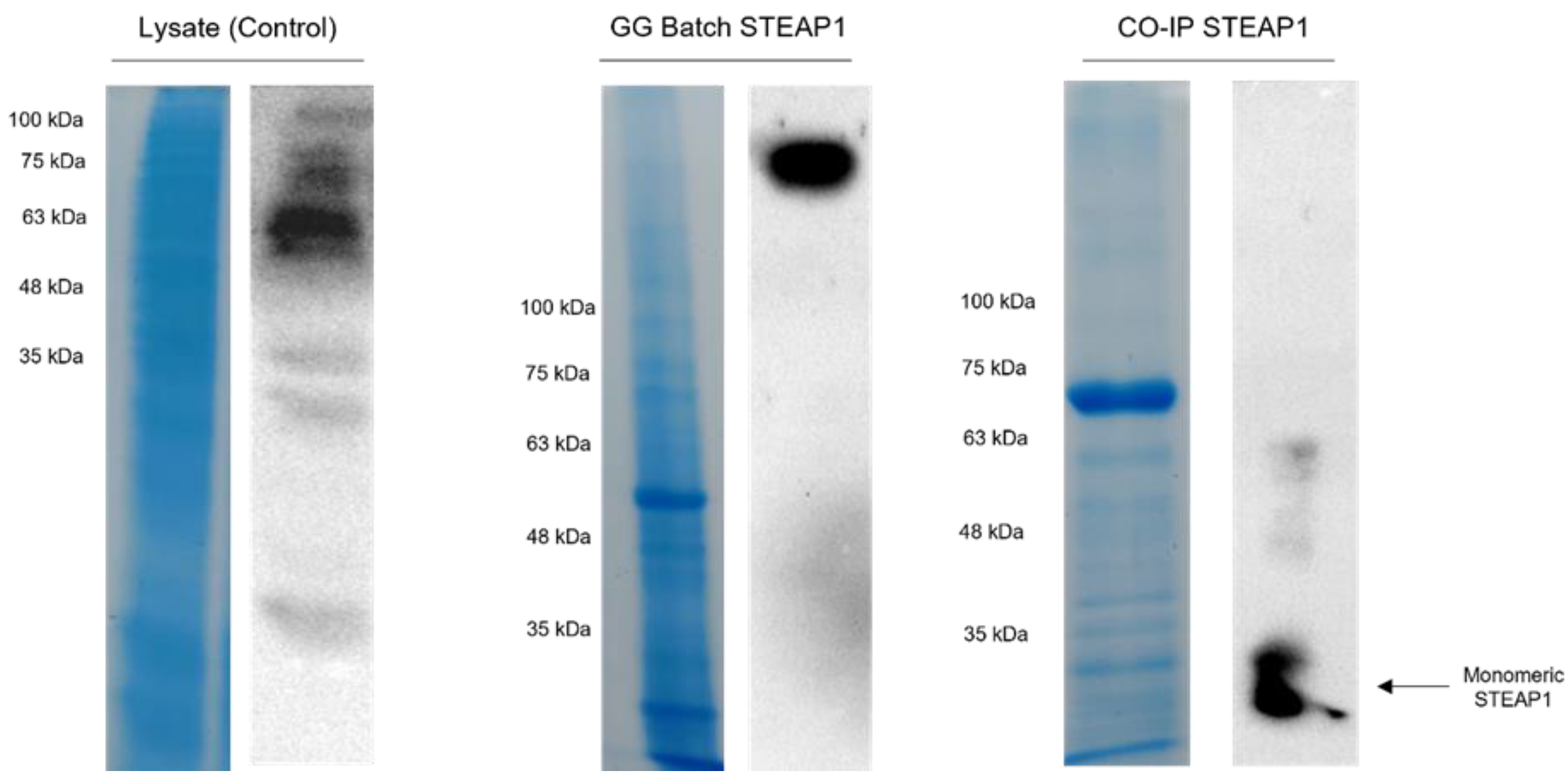
2.3. Batch Method for the Capture of STEAP1
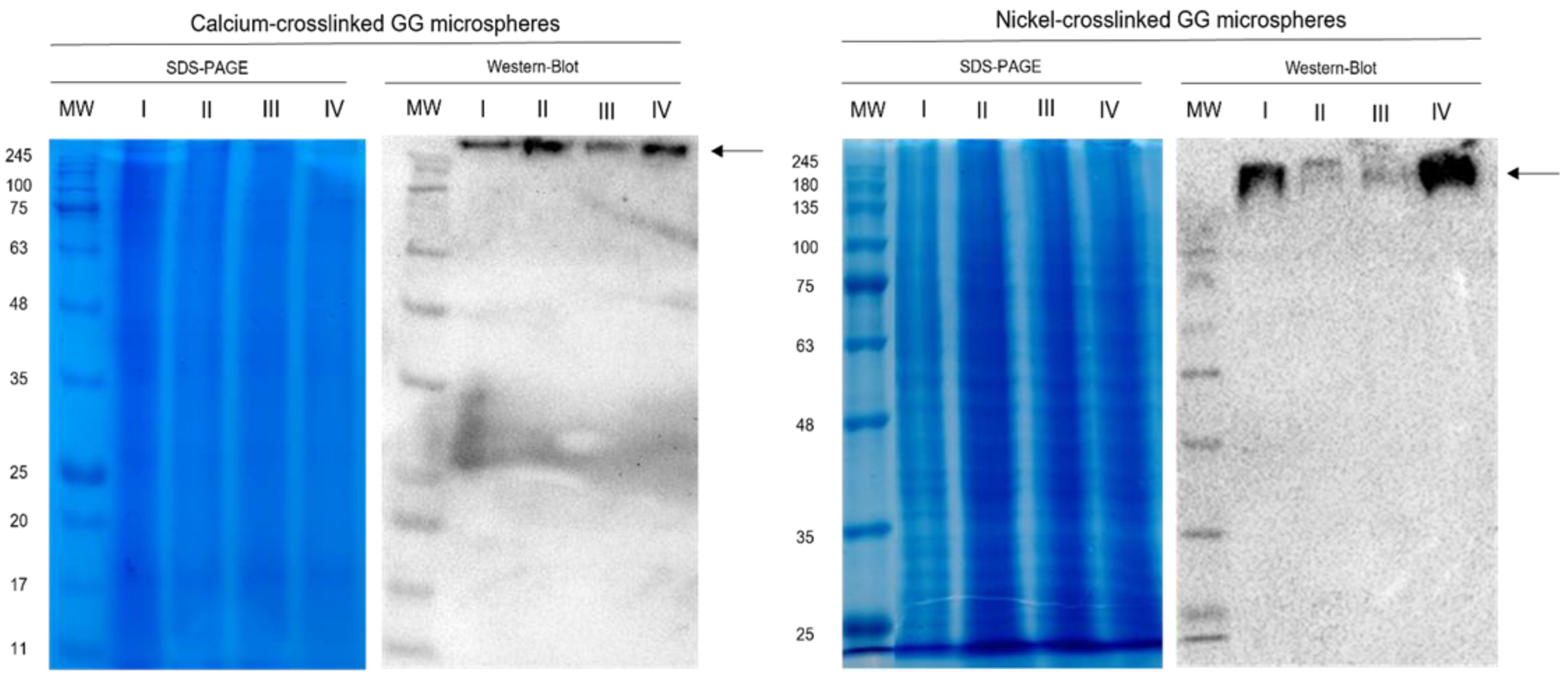
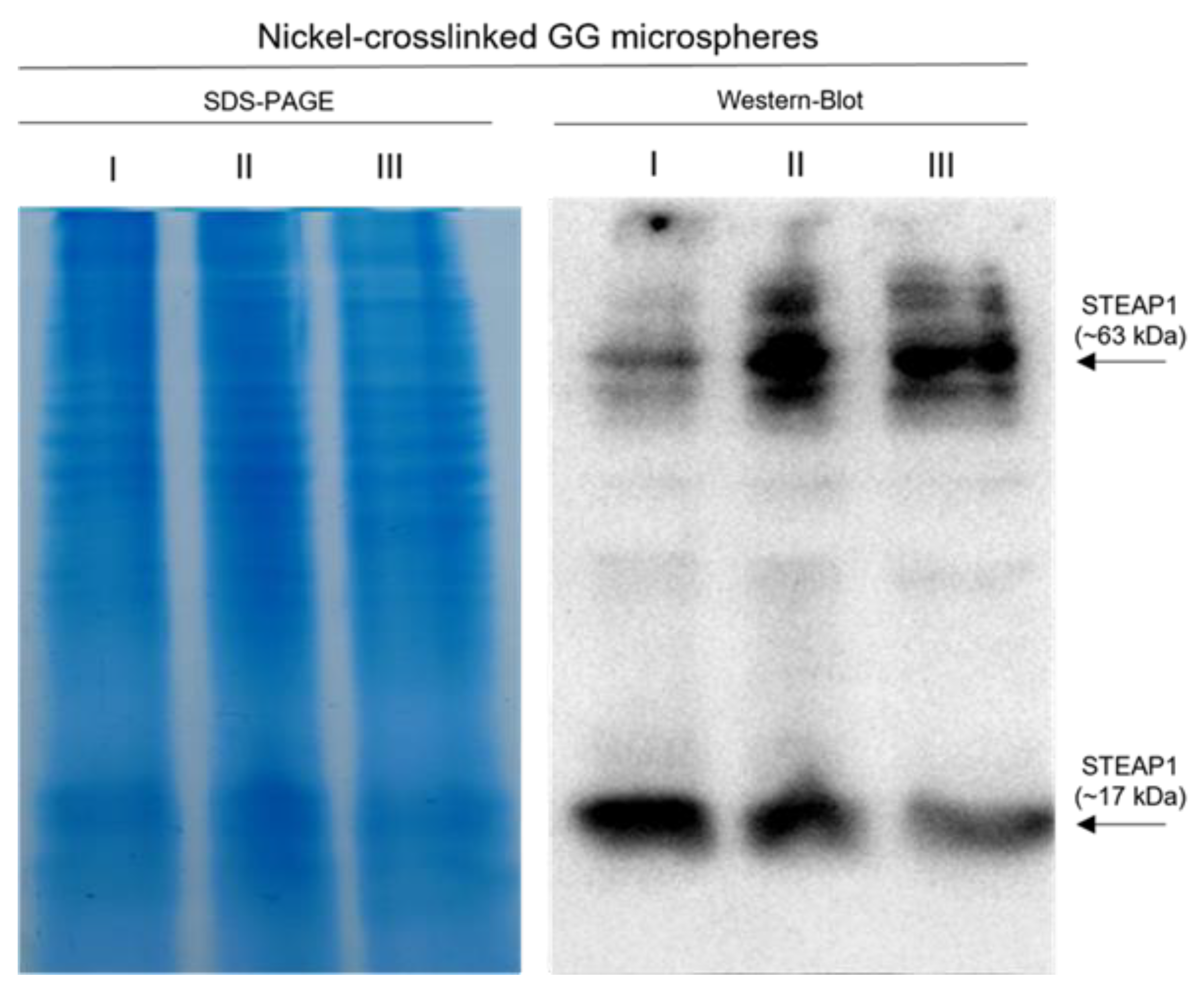
2.3.1. Affinity Strategy for STEAP1 Capture Using Nickel-Crosslinked GG Microspheres
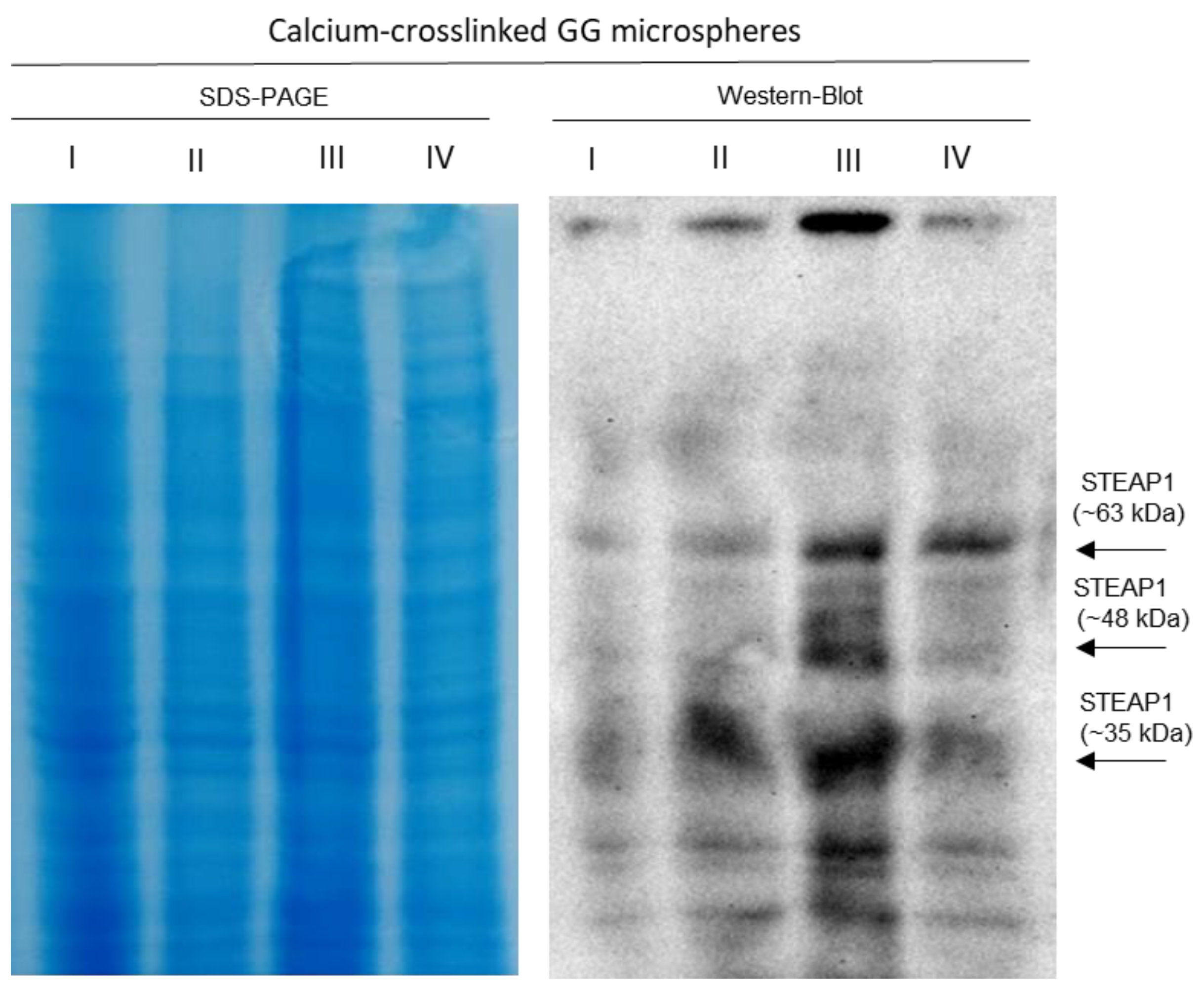
2.3.2. Ionic Strategy for STEAP1 Capture Using Calcium-Crosslinked GG Microspheres
3. Discussion
4. Materials and Methods
4.1. Materials
4.2. Gellan Microspheres Production
4.3. Gellan Microspheres Characterization
4.3.1. Semi-Optical Microscopy
4.3.2. Scanning Electron Microscopy (SEM)
4.3.3. Elemental Analysis and Chemical Composition
4.3.4. Fourier-Transformed Infrared Spectroscopy (FTIR)
4.4. Mini-Bioreactor Production and Recovery of STEAP1
4.5. Batch Method for the STEAP1 Capture
4.6. Co-Immunoprecipitation
4.7. SDS-PAGE and Western Blot
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Sung, H.; Ferlay, J.; Siegel, R.L.; Laversanne, M.; Soerjomataram, I.; Jemal, A.; Bray, F. Global Cancer Statistics 2020: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA Cancer J. Clin. 2021, 71, 209–249. [Google Scholar] [CrossRef] [PubMed]
- Hubert, R.S.; Vivanco, I.; Chen, E.; Rastegar, S.; Leong, K.; Mitchell, S.C.; Madraswala, R.; Zhou, Y.; Kuo, J.; Raitano, A.B.; et al. STEAP: A Prostate-Specific Cell-Surface Antigen Highly Expressed in Human Prostate Tumors. Proc. Natl. Acad. Sci. USA 1999, 96, 14523–14528. [Google Scholar] [CrossRef] [PubMed]
- Gomes, I.M.; Arinto, P.; Lopes, C.; Santos, C.R.; Maia, C.J. STEAP1 Is Overexpressed in Prostate Cancer and Prostatic Intraepithelial Neoplasia Lesions, and It Is Positively Associated with Gleason Score. Urol. Oncol. Semin. Orig. Investig. 2014, 32, 53.e23–53.e29. [Google Scholar] [CrossRef] [PubMed]
- Ihlaseh-Catalano, S.M.; Drigo, S.A.; de Jesus, C.M.N.; Domingues, M.A.C.; Trindade Filho, J.C.S.; de Camargo, J.L.V.; Rogatto, S.R. STEAP1 Protein Overexpression Is an Independent Marker for Biochemical Recurrence in Prostate Carcinoma. Histopathology 2013, 63, 678–685. [Google Scholar] [CrossRef]
- Chen, W.J.; Wu, H.T.; Li, C.L.; Lin, Y.K.; Fang, Z.X.; Lin, W.T.; Liu, J. Regulatory Roles of Six-Transmembrane Epithelial Antigen of the Prostate Family Members in the Occurrence and Development of Malignant Tumors. Front. Cell Dev. Biol. 2021, 9, 752426. [Google Scholar] [CrossRef]
- Gomes, I.M.; Maia, C.J.; Santos, C.R. STEAP Proteins: From Structure to Applications in Cancer Therapy. Mol. Cancer Res. 2012, 10, 573–587. [Google Scholar] [CrossRef] [PubMed]
- Yamamoto, T.; Tamura, Y.; Kobayashi, J.; Kamiguchi, K.; Hirohashi, Y.; Miyazaki, A.; Torigoe, T.; Asanuma, H.; Hiratsuka, H.; Sato, N. Six-Transmembrane Epithelial Antigen of the Prostate-1 Plays a Role for in Vivo Tumor Growth via Intercellular Communication. Exp. Cell Res. 2013, 319, 2617–2626. [Google Scholar] [CrossRef] [PubMed]
- Esmaeili, S.-A.; Nejatollahi, F.; Sahebkar, A. Inhibition of Intercellular Communication between Prostate Cancer Cells by A Specific Anti-STEAP-1 Single Chain Antibody. Anticancer Agents Med. Chem. 2018, 18, 1674–1679. [Google Scholar] [CrossRef] [PubMed]
- Kim, K.; Mitra, S.; Wu, G.; Berka, V.; Song, J.; Yu, Y.; Poget, S.; Wang, D.N.; Tsai, A.L.; Zhou, M. Six-Transmembrane Epithelial Antigen of Prostate 1 (STEAP1) Has a Single b Heme and Is Capable of Reducing Metal Ion Complexes and Oxygen. Biochemistry 2016, 55, 6673–6684. [Google Scholar] [CrossRef]
- Oosterheert, W.; Gros, P. Cryo-Electron Microscopy Structure and Potential Enzymatic Function of Human Six-Transmembrane Epithelial Antigen of the Prostate 1 (STEAP1). J. Biol. Chem. 2020, 295, 9502–9512. [Google Scholar] [CrossRef]
- Ohgami, R.S.; Campagna, D.R.; McDonald, A.; Fleming, M.D. The Steap Proteins Are Metalloreductases. Blood 2006, 108, 1388–1394. [Google Scholar] [CrossRef] [PubMed]
- Grunewald, T.G.P.; Diebold, I.; Esposito, I.; Plehm, S.; Hauer, K.; Thiel, U.; Da Silva-Buttkus, P.; Neff, F.; Unland, R.; Müller-Tidow, C.; et al. STEAP1 Is Associated with the Invasive and Oxidative Stress Phenotype of Ewing Tumors. Mol. Cancer Res. 2012, 10, 52–65. [Google Scholar] [CrossRef] [PubMed]
- Carrasquillo, J.A.; Fine, B.M.; Pandit-Taskar, N.; Larson, S.M.; Fleming, S.E.; Fox, J.J.; Cheal, S.M.; O’Donoghue, J.A.; Ruan, S.; Ragupathi, G.; et al. Imaging Patients with Metastatic Castration-Resistant Prostate Cancer Using 89Zr-DFO-MSTP2109A Anti-STEAP1 Antibody. J. Nucl. Med. 2019, 60, 1517–1523. [Google Scholar] [CrossRef] [PubMed]
- Doran, M.G.; Watson, P.A.; Cheal, S.M.; Spratt, D.E.; Wongvipat, J.; Steckler, J.M.; Carrasquillo, J.A.; Evans, M.J.; Lewis, J.S. Annotating STEAP1 Regulation in Prostate Cancer with 89Zr Immuno-PET. J. Nucl. Med. 2014, 55, 2045–2049. [Google Scholar] [CrossRef]
- O’Donoghue, J.A.; Danila, D.C.; Pandit-Taskar, N.; Beylergil, V.; Cheal, S.M.; Fleming, S.E.; Fox, J.J.; Ruan, S.; Zanzonico, P.B.; Ragupathi, G.; et al. Pharmacokinetics and Biodistribution of a [89Zr]Zr-DFO-MSTP2109A Anti-STEAP1 Antibody in Metastatic Castration-Resistant Prostate Cancer Patients. Mol. Pharm. 2019, 16, 3083–3090. [Google Scholar] [CrossRef]
- Danila, D.C.; Szmulewitz, R.Z.; Vaishampayan, U.; Higano, C.S.; Baron, A.D.; Gilbert, H.N.; Brunstein, F.; Milojic-Blair, M.; Wang, B.; Kabbarah, O.; et al. Phase i Study of DSTP3086S, an Antibody-Drug Conjugate Targeting Six-Transmembrane Epithelial Antigen of Prostate 1, in Metastatic Castration-Resistant Prostate Cancer. J. Clin. Oncol. 2019, 37, 3518–3527. [Google Scholar] [CrossRef]
- Williams, S.P.; Ogasawara, A.; Tinianow, J.N.; Flores, J.E.; Kan, D.; Lau, J.; Go, M.A.; Vanderbilt, A.N.; Gill, H.S.; Miao, L.; et al. ImmunoPET Helps Predicting the Efficacy of Antibody-Drug Conjugates Targeting TENB2 and STEAP1. Oncotarget 2016, 7, 25103–25112. [Google Scholar] [CrossRef]
- Boswell, C.A.; Mundo, E.E.; Zhang, C.; Bumbaca, D.; Valle, N.R.; Kozak, K.R.; Fourie, A.; Chuh, J.; Koppada, N.; Saad, O.; et al. Impact of Drug Conjugation on Pharmacokinetics and Tissue Distribution of Anti-STEAP1 Antibody-Drug Conjugates in Rats. Bioconjug. Chem. 2011, 22, 1994–2004. [Google Scholar] [CrossRef]
- Krupa, M.; Canamero, M.; Gomez, C.E.; Najera, J.L.; Gil, J.; Esteban, M. Immunization with Recombinant DNA and Modified Vaccinia Virus Ankara (MVA) Vectors Delivering PSCA and STEAP1 Antigens Inhibits Prostate Cancer Progression. Vaccine 2011, 29, 1504–1513. [Google Scholar] [CrossRef]
- Schober, S.J.; Thiede, M.; Gassmann, H.; Prexler, C.; Xue, B.; Schirmer, D.; Wohlleber, D.; Stein, S.; Grünewald, T.G.P.; Busch, D.H.; et al. MHC Class I-Restricted TCR-Transgenic CD4+ T Cells Against STEAP1 Mediate Local Tumor Control of Ewing Sarcoma In Vivo. Cells 2020, 9, 1581. [Google Scholar] [CrossRef]
- Chen, X.; Wang, R.; Chen, A.; Wang, Y.; Wang, Y.; Zhou, J.; Cao, R. Inhibition of Mouse RM-1 Prostate Cancer and B16F10 Melanoma by the Fusion Protein of HSP65 & STEAP1 186-193. Biomed. Pharmacother. 2019, 111, 1124–1131. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Z.; Wang, Z.; Song, Z.; Wu, Y.; Jin, Q.; Zhao, Z. Predictive Potential of STEAP Family for Survival, Immune Microenvironment and Therapy Response in Glioma. Int. Immunopharmacol. 2021, 101, 108183. [Google Scholar] [CrossRef] [PubMed]
- Gomes, I.M.; Santos, C.R.; Maia, C.J. Expression of Steap1 and Steap1b in Prostate Cell Lines, and the Putative Regulation of Steap1 by Post-Transcriptional and Post-Translational Mechanisms. Genes and Cancer 2014, 5, 142–151. [Google Scholar] [CrossRef] [PubMed]
- Löw, C.; Moberg, P.; Quistgaard, E.M.; Hedrén, M.; Guettou, F.; Frauenfeld, J.; Haneskog, L.; Nordlund, P. High-Throughput Analytical Gel Filtration Screening of Integral Membrane Proteins for Structural Studies. Biochim. Biophys. Acta-Gen. Subj. 2013, 1830, 3497–3508. [Google Scholar] [CrossRef]
- Nji, E.; Li, D.; Doyle, D.A.; Caffrey, M. Cloning, Expression, Purification, Crystallization and Preliminary X-Ray Diffraction of a Lysine-Specific Permease from Pseudomonas Aeruginosa. Acta Crystallogr. Sect. F Struct. Biol. Commun. 2014, 70, 1362–1367. [Google Scholar] [CrossRef]
- Merilaïnen, G.; Wierenga, R.K. Crystallization and Preliminary X-Ray Diffraction Studies of the C-Terminal Domain of Chlamydia Trachomatis CdsD. Acta Crystallogr. Sect. F Struct. Biol. Commun. 2014, 70, 1431–1433. [Google Scholar] [CrossRef]
- Thangavelu Muthukumar, J.E.S. and G.K. Biological Role of Gellan Gum in Improving Sca Ff Old Drug Delivery, Cell Adhesion Properties for Tissue. Molecules 2019, 24, 22. [Google Scholar]
- Zia, K.M.; Tabasum, S.; Khan, M.F.; Akram, N.; Akhter, N.; Noreen, A.; Zuber, M. Recent Trends on Gellan Gum Blends with Natural and Synthetic Polymers: A Review. Int. J. Biol. Macromol. 2018, 109, 1068–1087. [Google Scholar] [CrossRef]
- Danalache, F.; Carvalho, C.Y.; Alves, V.D.; Moldão-Martins, M.; Mata, P. Optimisation of Gellan Gum Edible Coating for Ready-to-Eat Mango (Mangifera indica L.) Bars. Int. J. Biol. Macromol. 2016, 84, 43–53. [Google Scholar] [CrossRef]
- Vieira, S.; da Silva Morais, A.; Garet, E.; Silva-Correia, J.; Reis, R.L.; González-Fernández, Á.; Miguel Oliveira, J. Self-Mineralizing Ca-Enriched Methacrylated Gellan Gum Beads for Bone Tissue Engineering. Acta Biomater. 2019, 93, 74–85. [Google Scholar] [CrossRef]
- Park, H.; Kim, H.; Kim, G.Y.; Lee, M.Y.; Kim, Y.; Kang, S. Enhanced Biodegradation of Hydrocarbons by Pseudomonas aeruginosa-Encapsulated Alginate/Gellan Gum Microbeads. J. Hazard. Mater. 2021, 406, 124752. [Google Scholar] [CrossRef] [PubMed]
- Lyu, C.J.; Liu, L.; Huang, J.; Zhao, W.R.; Hu, S.; Mei, L.H.; Yao, S.J. Biosynthesis of γ-Aminobutyrate by Engineered Lactobacillus Brevis Cells Immobilized in Gellan Gum Gel Beads. J. Biosci. Bioeng. 2019, 128, 123–128. [Google Scholar] [CrossRef] [PubMed]
- Villarreal-Otalvaro, C.; Coburn, J.M. Fabrication Methods and Form Factors of Gellan Gum-Based Materials for Drug Delivery and Anti-Cancer Applications. ACS Biomater. Sci. Eng. 2021. [Google Scholar] [CrossRef]
- Racovita, S.; Lungan, M.A.; Bunia, I.; Popa, M.; Vasiliu, S. Adsorption and Release Studies of Cefuroxime Sodium from Acrylic Ion Exchange Resin Microparticles Coated with Gellan. React. Funct. Polym. 2016, 105, 103–113. [Google Scholar] [CrossRef]
- Gomes, D.; Gonçalves, C.; Gonçalves, A.M.; Queiroz, J.A.; Sousa, A.; Passarinha, L.A. Applications of Gellan Natural Polymer Microspheres in Recombinant Catechol-O-Methyltransferase Direct Capture from a Komagataella Pastoris Lysate. Int. J. Biol. Macromol. 2021, 172, 186–196. [Google Scholar] [CrossRef] [PubMed]
- Gomes, D.; Costa, D.; Queiroz, J.A.; Passarinha, L.A.; Sousa, A. A New Insight in Gellan Microspheres Application to Capture a Plasmid DNA Vaccine from an Escherichia Coli Lysate. Sep. Purif. Technol. 2021, 274, 119013. [Google Scholar] [CrossRef]
- Lei, L.; Wang, X.; Zhu, Y.; Su, W.; Lv, Q.; Li, D. Antimicrobial Hydrogel Microspheres for Protein Capture and Wound Healing. Mater. Des. 2022, 215, 110478. [Google Scholar] [CrossRef]
- Koubková, J.; Müller, P.; Hlídková, H.; Plichta, Z.; Proks, V.; Vojtěšek, B.; Horák, D. Magnetic Poly(Glycidyl Methacrylate) Microspheres for Protein Capture. New Biotechnol. 2014, 31, 482–491. [Google Scholar] [CrossRef]
- Zhou, J.; Su, Z.; Wang, M.; Wang, Y.; Wang, J.; Zhang, B.; Zhang, Q. Thiolactone-Based Conjugation Assisted Magnetic Imprinted Microspheres for Specific Capturing Target Proteins. Chem. Eng. J. 2020, 399, 125767. [Google Scholar] [CrossRef]
- Lei, L.; Zhu, Y.; Qin, X.; Chai, S.; Liu, G.; Su, W.; Lv, Q.; Li, D. Magnetic Biohybrid Microspheres for Protein Purification and Chronic Wound Healing in Diabetic Mice. Chem. Eng. J. 2021, 425, 130671. [Google Scholar] [CrossRef]
- Mahajan, H.S.; Gattani, S.G. Gellan Gum Based Microparticles of Metoclopromide Hydrochloride for Intranasal Delivery: Development and Evaluation. Chem. Pharm. Bull. 2009, 57, 388–392. [Google Scholar] [CrossRef]
- Lin, S.H.; Guidotti, G. Chapter 35 Purification of Membrane Proteins, 1st ed.; Elsevier Inc.: Amsterdam, The Netherlands, 2009; Volume 463. [Google Scholar]
- Abbas, Z.; Marihal, S. Gellan Gum-Based Mucoadhesive Microspheres of Almotriptan for Nasal Administration: Formulation Optimization Using Factorial Design, Characterization, and in Vitro Evaluation. J. Pharm. Bioallied Sci. 2014, 6, 267. [Google Scholar] [CrossRef] [PubMed]
- Fan, Y.; Yi, J.; Hua, X.; Zhang, Y.; Yang, R. Preparation and Characterization of Gellan Gum Microspheres Containing a Cold-Adapted β-Galactosidase from Rahnella Sp. R3. Carbohydr. Polym. 2017, 162, 10–15. [Google Scholar] [CrossRef] [PubMed]
- Babu, R.; Sathigari, S.; Kumar, M.; Pandit, J. Formulation of Controlled Release Gellan Gum Macro Beads of Amoxicillin. Curr. Drug Deliv. 2010, 7, 36–43. [Google Scholar] [CrossRef] [PubMed]
- Agnihotri, S.A.; Aminabhavi, T.M. Development of Novel Interpenetrating Network Gellan Gum-Poly(Vinyl Alcohol) Hydrogel Microspheres for the Controlled Release of Carvedilol. Drug Dev. Ind. Pharm. 2005, 31, 491–503. [Google Scholar] [CrossRef]
- Dhanka, M.; Shetty, C.; Srivastava, R. Methotrexate Loaded Gellan Gum Microparticles for Drug Delivery. Int. J. Biol. Macromol. 2018, 110, 346–356. [Google Scholar] [CrossRef]
- Kozlowski, L.P. Proteome-PI: Proteome Isoelectric Point Database. Nucleic Acids Res. 2017, 45, D1112–D1116. [Google Scholar] [CrossRef]
- Blanchard, V.; Gadkari, R.A.; George, A.V.E.; Roy, S.; Gerwig, G.J.; Leeflang, B.R.; Dighe, R.R.; Boelens, R.; Kamerling, J.P. High-Level Expression of Biologically Active Glycoprotein Hormones in Pichia Pastoris Strains-Selection of Strain GS115, and Not X-33, for the Production of Biologically Active N-Glycosylated 15N-Labeled PhCG. Glycoconj. J. 2008, 25, 245–257. [Google Scholar] [CrossRef]
- Tsuji, Y. Transmembrane Protein Western Blotting: Impact of Sample Preparation on Detection of SLC11A2 (DMT1) and SLC40A1 (Ferroportin). PLoS ONE 2020, 15, e0235563. [Google Scholar] [CrossRef]
- Seddon, A.M.; Curnow, P.; Booth, P.J. Membrane Proteins, Lipids and Detergents: Not Just a Soap Opera. Biochim. Biophys. Acta-Biomembr. 2004, 1666, 105–117. [Google Scholar] [CrossRef]
- Lantez, V.; Nikolaidis, I.; Rechenmann, M.; Vernet, T.; Noirclerc-Savoye, M. Rapid Automated Detergent Screening for the Solubilization and Purification of Membrane Proteins and Complexes. Eng. Life Sci. 2015, 15, 39–50. [Google Scholar] [CrossRef]
- Pedro, A.Q.; Gonçalves, A.M.; Queiroz, J.A.; Passarinha, L.A. Purification of Histidine-Tagged Membrane-Bound Catechol-O-Methyltransferase from Detergent-Solubilized Pichia Pastoris Membranes. Chromatographia 2018, 81, 425–434. [Google Scholar] [CrossRef]
- Reyes-Alcaraz, A.; Martínez-Archundia, M.; Ramon, E.; Garriga, P. Salt Effects on the Conformational Stability of the Visual G-Protein-Coupled Receptor Rhodopsin. Biophys. J. 2011, 101, 2798–2806. [Google Scholar] [CrossRef] [PubMed]
- Bacelar, A.H.; Silva-Correia, J.; Oliveira, J.M.; Reis, R.L. Recent Progress in Gellan Gum Hydrogels Provided by Functionalization Strategies. J. Mater. Chem. B 2016, 4, 6164–6174. [Google Scholar] [CrossRef] [PubMed]
- Huang, C.J.; Damasceno, L.M.; Anderson, K.A.; Zhang, S.; Old, L.J.; Batt, C.A. A Proteomic Analysis of the Pichia Pastoris Secretome in Methanol-Induced Cultures. Appl. Microbiol. Biotechnol. 2011, 90, 235–247. [Google Scholar] [CrossRef]
- Huo, S.F.; Shang, W.L.; Yu, M.; Ren, X.P.; Wen, H.X.; Chai, C.Y.; Sun, L.; Hui, K.; Liu, L.H.; Wei, S.H.; et al. STEAP1 Facilitates Metastasis and Epithelial-Mesenchymal Transition of Lung Adenocarcinoma via the JAK2/STAT3 Signaling Pathway. Biosci. Rep. 2020, 40, BSR20193169. [Google Scholar] [CrossRef]
- Guo, Q.; Ke, X.X.; Liu, Z.; Gao, W.L.; Fang, S.X.; Chen, C.; Song, Y.X.; Han, H.; Lu, H.L.; Xu, G. Evaluation of the Prognostic Value of STEAP1 in Lung Adenocarcinoma and Insights Into Its Potential Molecular Pathways via Bioinformatic Analysis. Front. Genet. 2020, 11, 242. [Google Scholar] [CrossRef]
- Nakamura, H.; Takada, K.; Arihara, Y.; Hayasaka, N.; Murase, K.; Iyama, S.; Kobune, M.; Miyanishi, K.; Kato, J. Six-Transmembrane Epithelial Antigen of the Prostate 1 Protects against Increased Oxidative Stress via a Nuclear Erythroid 2-Related Factor Pathway in Colorectal Cancer. Cancer Gene Ther. 2019, 26, 313–322. [Google Scholar] [CrossRef]
- Zhang, Z.; Hou, W.; Zhang, C.; Tan, Y.; Zhang, D.; An, W.; Pan, S.; Wu, W.; Chen, Q.; Xu, H. A Research of STEAP1 Regulated Gastric Cancer Cell Proliferation, Migration and Invasion in Vitro and in Vivos. J. Cell. Mol. Med. 2020, 24, 14217–14230. [Google Scholar] [CrossRef]
- Duarte, D.R.; Barroca-Ferreira, J.; Gonçalves, A.M.; Santos, F.M.; Rocha, S.M.; Pedro, A.Q.; Maia, C.J.; Passarinha, L.A. Impact of Glycerol Feeding Profiles on STEAP1 Biosynthesis by Komagataella Pastoris Using a Methanol-Inducible Promoter. Appl. Microbiol. Biotechnol. 2021, 105, 4635–4648. [Google Scholar] [CrossRef]
- Narkar, M.; Sher, P.; Pawar, A. Stomach-Specific Controlled Release Gellan Beads of Acid-Soluble Drug Prepared by Ionotropic Gelation Method. AAPS PharmSciTech 2010, 11, 267–277. [Google Scholar] [CrossRef] [PubMed]
- Kawaguchi, H. Functional Polymer Microspheres. Prog. Polym. Sci. 2000, 25, 1171–1210. [Google Scholar] [CrossRef]
- Cassanelli, M.; Prosapio, V.; Norton, I.; Mills, T. Acidified/Basified Gellan Gum Gels: The Role of the Structure in Drying/Rehydration Mechanisms. Food Hydrocoll. 2018, 82, 346–354. [Google Scholar] [CrossRef]
- Kermani, A.A. A Guide to Membrane Protein X-ray Crystallography. FEBS J. 2021, 288, 5788–5804. [Google Scholar] [CrossRef] [PubMed]
- Stetsenko, A.; Guskov, A. An Overview of the Top Ten Detergents Used for Membrane Protein Crystallization. Crystals 2017, 7, 197. [Google Scholar] [CrossRef]
- Barroca-Ferreira, J.; Pais, J.P.; Santos, M.M.; Goncalves, A.M.; Gomes, I.M.; Sousa, I.; Rocha, S.M.; Passarinha, L.A.; Maia, C.J. Targeting STEAP1 Protein in Human Cancer: Current Trends and Future Challenges. Curr. Cancer Drug Targets 2018, 18, 222–230. [Google Scholar] [CrossRef] [PubMed]
- Muñoz, A.; Costa, M. Elucidating the Mechanisms of Nickel Compound Uptake: A Review of Particulate and Nano-Nickel Endocytosis and Toxicity. Toxicol. Appl. Pharmacol. 2012, 260, 1–16. [Google Scholar] [CrossRef]
- Barroca-Ferreira, J.; Cruz-Vicente, P.; Santos, M.F.A.; Rocha, S.M.; Santos-Silva, T.; Maia, C.J.; Passarinha, L.A. Enhanced Stability of Detergent-Free Human Native STEAP1 Protein from Neoplastic Prostate Cancer Cells upon an Innovative Isolation Procedure. Int. J. Mol. Sci. 2021, 22, 10012. [Google Scholar] [CrossRef]
- Barroca-Ferreira, J.; Gonçalves, A.M.; Santos, M.F.A.; Santos-Silva, T.; Maia, C.J.; Passarinha, L.A. A Chromatographic Network for the Purification of Detergent-Solubilized Six-Transmembrane Epithelial Antigen of the Prostate 1 from Komagataella Pastoris Mini-Bioreactor Lysates. J. Chromatogr. A 2022, 1685, 463576. [Google Scholar] [CrossRef]
- Noyes, A.; Godavarti, R.; Titchener-Hooker, N.; Coffman, J.; Mukhopadhyay, T. Quantitative High Throughput Analytics to Support Polysaccharide Production Process Development. Vaccine 2014, 32, 2819–2828. [Google Scholar] [CrossRef]
- Coelho, J.; Eusébio, D.; Gomes, D.; Frias, F.; Passarinha, L.A.; Sousa, Â. Biosynthesis and Isolation of Gellan Polysaccharide to Formulate Microspheres for Protein Capture. Carbohydr. Polym. 2019, 220, 236–246. [Google Scholar] [CrossRef] [PubMed]
- Laemmli, U.K. Cleavage of Structural Proteins during the Assembly of the Head of Bacteriophage T4. Nature 1970, 227, 680–685. [Google Scholar] [CrossRef] [PubMed]
- Tokmakov, A.A.; Kurotani, A.; Sato, K.I. Protein PI and Intracellular Localization. Front. Mol. Biosci. 2021, 8, 775736. [Google Scholar] [CrossRef] [PubMed]

| Element | Calcium-Crosslinked GG Microspheres | Nickel-Crosslinked GG Microspheres | ||
|---|---|---|---|---|
| C Norm. [wt%] | C Atom. [at%] | C Norm. [wt%] | C Atom. [at%] | |
| Carbon | 31.87 | 38.72 | 39.19 | 47.13 |
| Oxygen | 66.54 | 60.70 | 57.73 | 52.12 |
| Calcium | 1.59 | 0.58 | - | - |
| Nickel | - | - | 3.08 | 0.76 |
| Total | 100.0 | 100.0 | 100.0 | 100.0 |
| Ref. | - | [35] | ||
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Batista-Silva, J.; Gomes, D.; Barroca-Ferreira, J.; Gallardo, E.; Sousa, Â.; Passarinha, L.A. Specific Six-Transmembrane Epithelial Antigen of the Prostate 1 Capture with Gellan Gum Microspheres: Design, Optimization and Integration. Int. J. Mol. Sci. 2023, 24, 1949. https://doi.org/10.3390/ijms24031949
Batista-Silva J, Gomes D, Barroca-Ferreira J, Gallardo E, Sousa Â, Passarinha LA. Specific Six-Transmembrane Epithelial Antigen of the Prostate 1 Capture with Gellan Gum Microspheres: Design, Optimization and Integration. International Journal of Molecular Sciences. 2023; 24(3):1949. https://doi.org/10.3390/ijms24031949
Chicago/Turabian StyleBatista-Silva, João, Diana Gomes, Jorge Barroca-Ferreira, Eugénia Gallardo, Ângela Sousa, and Luís A. Passarinha. 2023. "Specific Six-Transmembrane Epithelial Antigen of the Prostate 1 Capture with Gellan Gum Microspheres: Design, Optimization and Integration" International Journal of Molecular Sciences 24, no. 3: 1949. https://doi.org/10.3390/ijms24031949
APA StyleBatista-Silva, J., Gomes, D., Barroca-Ferreira, J., Gallardo, E., Sousa, Â., & Passarinha, L. A. (2023). Specific Six-Transmembrane Epithelial Antigen of the Prostate 1 Capture with Gellan Gum Microspheres: Design, Optimization and Integration. International Journal of Molecular Sciences, 24(3), 1949. https://doi.org/10.3390/ijms24031949









