Not to Miss: Intronic Variants, Treatment, and Review of the Phenotypic Spectrum in VPS13D-Related Disorder
Abstract
1. Introduction
2. Results
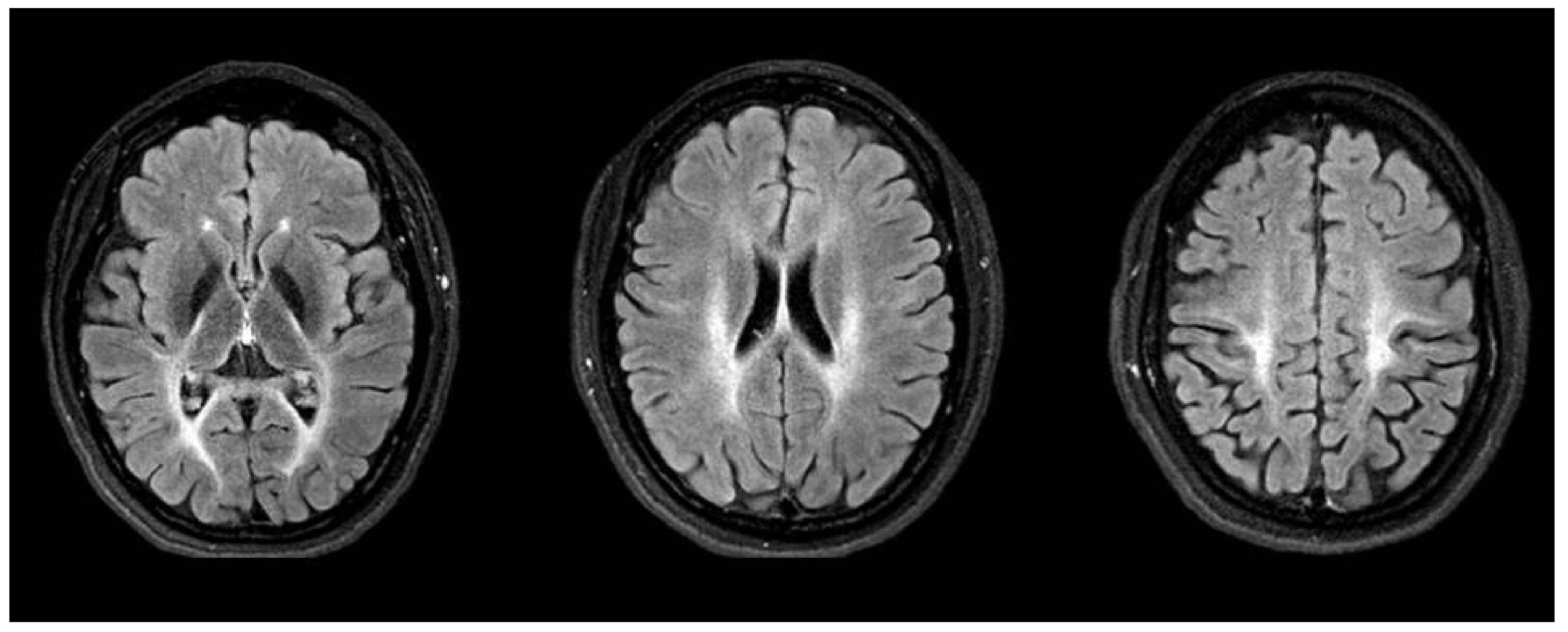
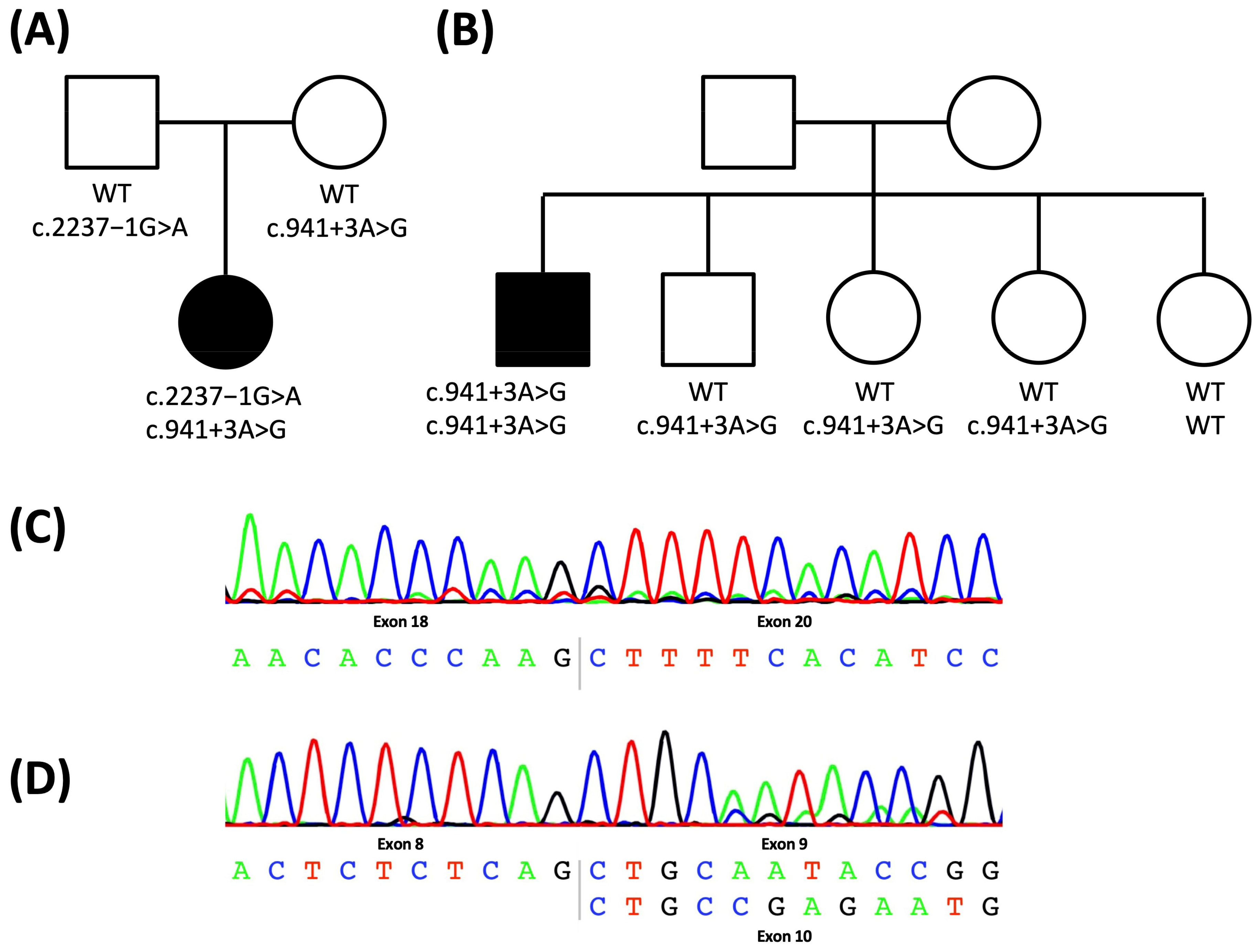
2.1. Case Report: Patient 1
2.2. Case Report: Patient 2
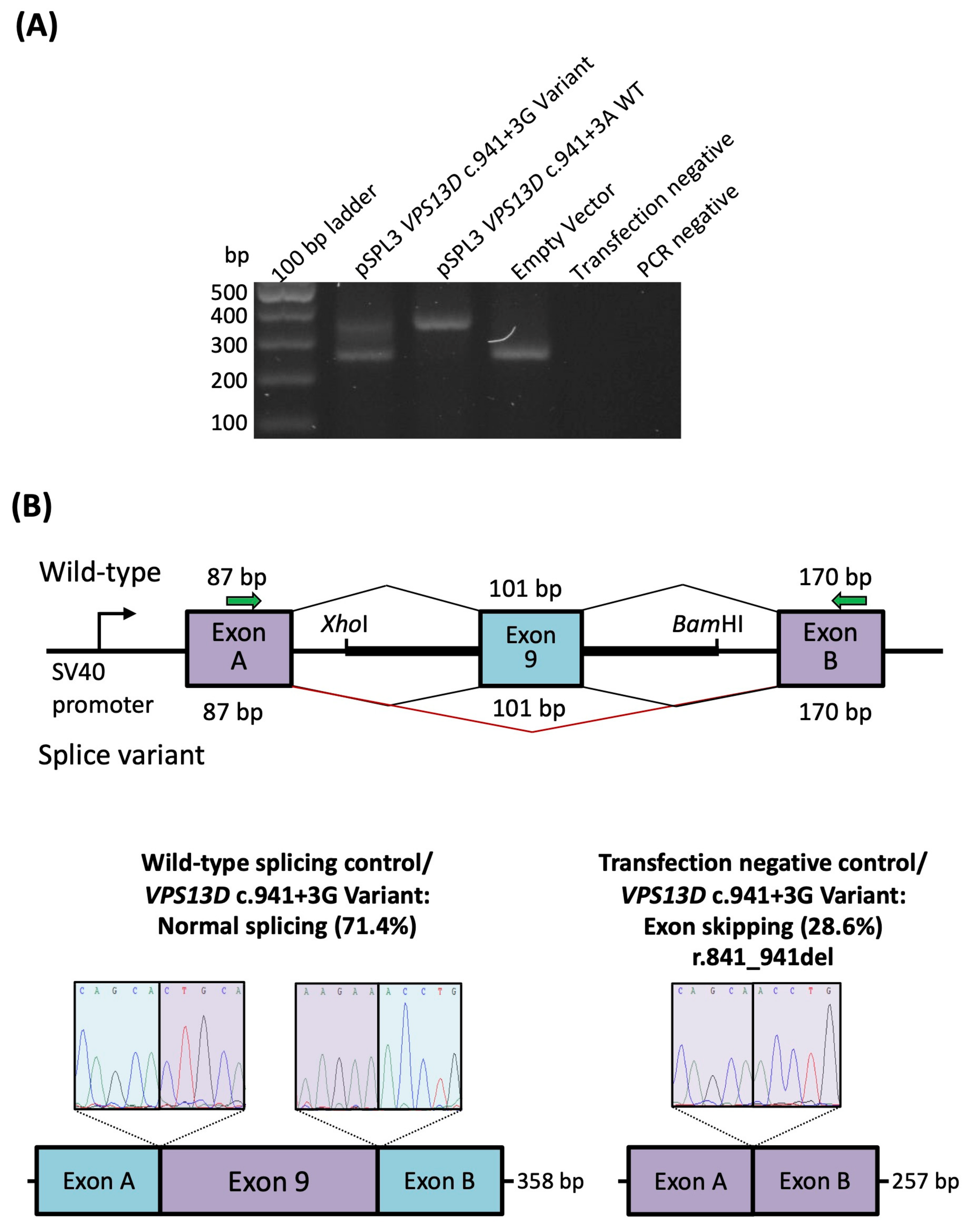
2.3. Characterization of the Intronic Variants for a Splicing Effect
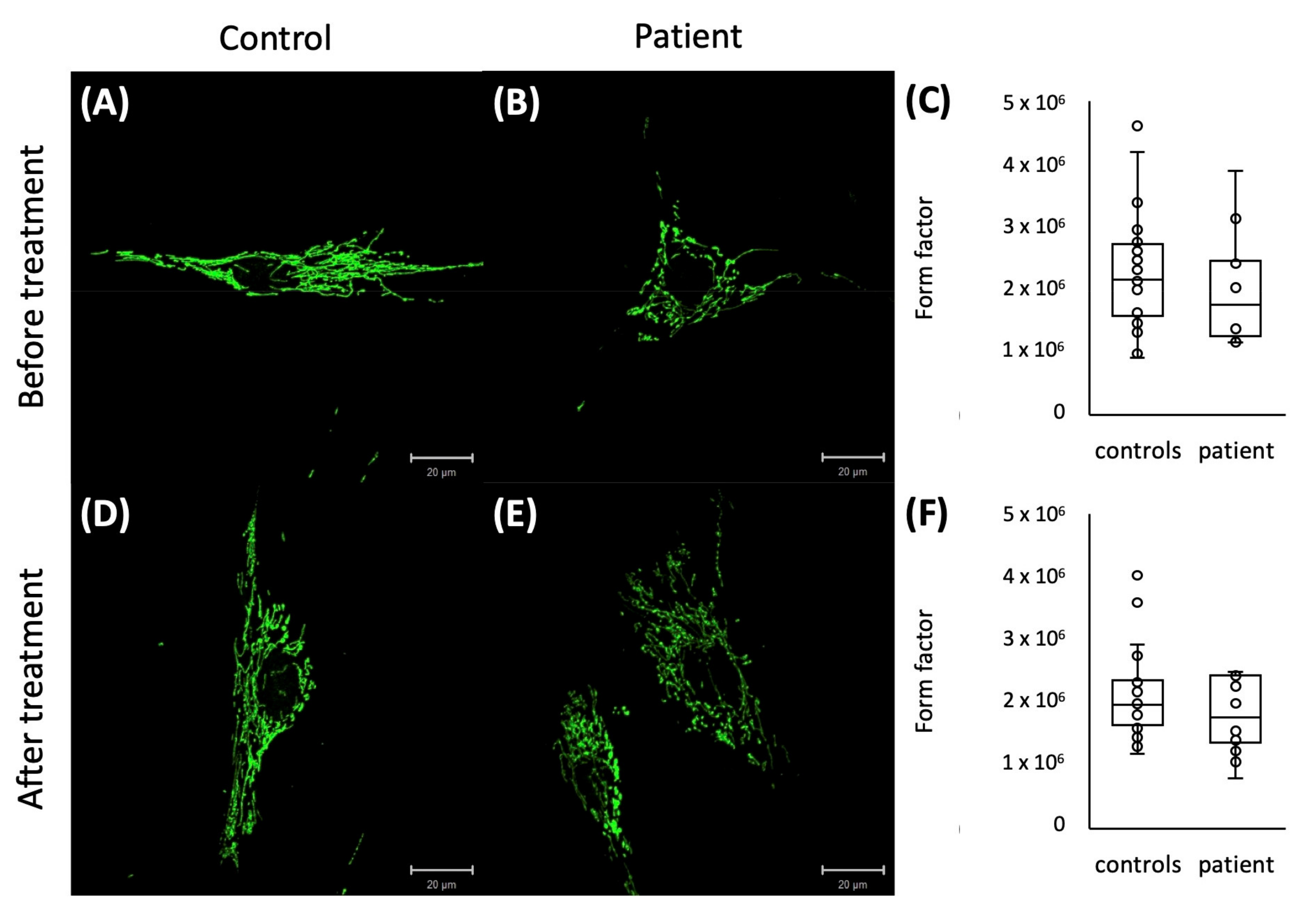
2.4. Mitochondrial Network Analysis
3. Discussion
4. Materials and Methods
4.1. Identification of Variants and Additional Patient
4.2. Sanger Sequencing and Sequencing of cDNA
4.3. Quantification of Exon Skipping
4.4. Form Factor of the Mitochondrial Network
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Dziurdzik, S.K.; Conibear, E. The Vps13 Family of Lipid Transporters and Its Role at Membrane Contact Sites. Int. J. Mol. Sci. 2021, 22, 2905. [Google Scholar] [CrossRef] [PubMed]
- Velayos-Baeza, A.; Vettori, A.; Copley, R.R.; Dobson-Stone, C.; Monaco, A.P. Analysis of the Human VPS13 Gene Family. Genomics 2004, 84, 536–549. [Google Scholar] [CrossRef] [PubMed]
- Peikert, K.; Danek, A.; Hermann, A. Current State of Knowledge in Chorea-Acanthocytosis as Core Neuroacanthocytosis Syndrome. Eur. J. Med. Genet. 2018, 61, 699–705. [Google Scholar] [CrossRef] [PubMed]
- Rodrigues, J.M.; Fernandes, H.D.; Caruthers, C.; Braddock, S.R.; Knutsen, A.P. Cohen Syndrome: Review of the Literature. Cureus 2018, 10, e3330. [Google Scholar] [CrossRef]
- Lesage, S.; Drouet, V.; Majounie, E.; Deramecourt, V.; Jacoupy, M.; Nicolas, A.; Cormier-Dequaire, F.; Hassoun, S.M.; Pujol, C.; Ciura, S.; et al. Loss of VPS13C Function in Autosomal-Recessive Parkinsonism Causes Mitochondrial Dysfunction and Increases PINK1/Parkin-Dependent Mitophagy. Am. J. Hum. Genet. 2016, 98, 500–513. [Google Scholar] [CrossRef]
- Seong, E.; Insolera, R.; Dulovic, M.; Kamsteeg, E.; Trinh, J.; Brüggemann, N.; Sandford, E.; Li, S.; Ozel, A.B.; Li, J.Z.; et al. Mutations in VPS13D Lead to a New Recessive Ataxia with Spasticity and Mitochondrial Defects. Ann. Neurol. 2018, 83, 1075–1088. [Google Scholar] [CrossRef]
- Gauthier, J.; Meijer, I.A.; Lessel, D.; Mencacci, N.E.; Krainc, D.; Hempel, M.; Tsiakas, K.; Prokisch, H.; Rossignol, E.; Helm, M.H.; et al. Recessive Mutations in VPS13D Cause Childhood Onset Movement Disorders. Ann. Neurol. 2018, 83, 1089–1095. [Google Scholar] [CrossRef]
- Koh, K.; Ishiura, H.; Shimazaki, H.; Tsutsumiuchi, M.; Ichinose, Y.; Nan, H.; Hamada, S.; Ohtsuka, T.; Tsuji, S.; Takiyama, Y. VPS13D-related Disorders Presenting as a Pure and Complicated Form of Hereditary Spastic Paraplegia. Mol. Genet. Genom. Med. 2020, 8, e1108. [Google Scholar] [CrossRef]
- Petry-Schmelzer, J.N.; Keller, N.; Karakaya, M.; Wirth, B.; Fink, G.R.; Wunderlich, G. VPS13D: One Family, Same Mutations, Two Phenotypes. Mov. Disord. Clin. Pract. 2021, 8, 803–806. [Google Scholar] [CrossRef]
- Durand, C.M.; Angelini, C.; Michaud, V.; Delleci, C.; Coupry, I.; Goizet, C.; Trimouille, A. Whole-Exome Sequencing Confirms Implication of VPS13D as a Potential Cause of Progressive Spastic Ataxia. BMC Neurol. 2022, 22, 53. [Google Scholar] [CrossRef]
- Huang, X.; Fan, D.-S. Autosomal Recessive Spinocerebellar Ataxia Type 4 with a VPS13D Mutation: A Case Report. World J. Clin. Cases 2022, 10, 703–708. [Google Scholar] [CrossRef] [PubMed]
- Öztop-Çakmak, Ö.; Şimşir, G.; Tekgül, Ş.; Aygün, M.S.; Gökler, O.; Kahyaoğlu, B.; Kaya, Z.E.; Palvadeau, R.; Başak, A.N.; Ertan, S. VPS13D-Based Disease: Expansion of the Clinical Phenotype in Two Brothers and Mutation Diversity in the Turkish Population. Rev. Neurol. 2022, 178, 907–913. [Google Scholar] [CrossRef] [PubMed]
- Whiting, B.B.; Whiting, A.C.; Whiting, D.M. Thalamic Deep Brain Stimulation. In Progress in Neurological Surgery; Niranjan, A., Lunsford, L.D., Richardson, R.M., Eds.; S. Karger AG: Basel, Switzerland, 2018; Volume 33, pp. 198–206. ISBN 978-3-318-06201-4. [Google Scholar]
- Bargiotas, P.; Nguyen, T.A.K.; Bracht, T.; Mürset, M.; Nowacki, A.; Debove, I.; Muellner, J.; Michelis, J.P.; Pollo, C.; Schüpbach, W.M.M.; et al. Long-Term Outcome and Neuroimaging of Deep Brain Stimulation in Holmes Tremor: A Case Series. Neuromodul. Technol. Neural. Interface 2021, 24, 392–399. [Google Scholar] [CrossRef] [PubMed]
- Wang, K.-L.; Wong, J.K.; Eisinger, R.S.; Carbunaru, S.; Smith, C.; Hu, W.; Shukla, A.W.; Hess, C.W.; Okun, M.S.; Ramirez-Zamora, A. Therapeutic Advances in the Treatment of Holmes Tremor: Systematic Review. Neuromodul. Technol. Neural. Interface 2022, 25, 796–803. [Google Scholar] [CrossRef] [PubMed]
- Tai, C.-H.; Tseng, S.-H. Cerebellar Deep Brain Stimulation for Movement Disorders. Neurobiol. Dis. 2022, 175, 105899. [Google Scholar] [CrossRef]
- Swartz, B.E.; Burmeister, M.; Somers, J.T.; Rottach, K.G.; Bespalova, I.N.; Leigh, R.J. A Form of Inherited Cerebellar Ataxia with Saccadic Intrusions, Increased Saccadic Speed, Sensory Neuropathy, and Myoclonus. Ann. N. Y. Acad. Sci. 2002, 956, 441–444. [Google Scholar] [CrossRef]
- Cingolani, P.; Platts, A.; Wang, L.L.; Coon, M.; Nguyen, T.; Wang, L.; Land, S.J.; Lu, X.; Ruden, D.M. A Program for Annotating and Predicting the Effects of Single Nucleotide Polymorphisms, SnpEff: SNPs in the Genome of Drosophila Melanogaster Strain w1118; Iso-2; Iso-3. Fly 2012, 6, 80–92. [Google Scholar] [CrossRef]
- Lochmüller, H.; Badowska, D.M.; Thompson, R.; Knoers, N.V.; Aartsma-Rus, A.; Gut, I.; Wood, L.; Harmuth, T.; Durudas, A.; Graessner, H.; et al. RD-Connect, NeurOmics and EURenOmics: Collaborative European Initiative for Rare Diseases. Eur. J. Hum. Genet. 2018, 26, 778–785. [Google Scholar] [CrossRef]
- Zurek, B.; Ellwanger, K.; Vissers, L.E.L.M.; Schüle, R.; Synofzik, M.; Töpf, A.; de Voer, R.M.; Laurie, S.; Matalonga, L.; Gilissen, C.; et al. Solve-RD: Systematic Pan-European Data Sharing and Collaborative Analysis to Solve Rare Diseases. Eur. J. Hum. Genet. 2021, 29, 1325–1331. [Google Scholar] [CrossRef]
- Matalonga, L.; Hernández-Ferrer, C.; Piscia, D.; Solve-RD SNV-indel working group; Cohen, E.; Cuesta, I.; Danis, D.; Denommé-Pichon, A.-S.; Duffourd, Y.; Gilissen, C.; et al. Solving Patients with Rare Diseases through Programmatic Reanalysis of Genome-Phenome Data. Eur. J. Hum. Genet. 2021, 29, 1337–1347. [Google Scholar] [CrossRef]
- Karczewski, K.J.; Francioli, L.C.; Tiao, G.; Cummings, B.B.; Alföldi, J.; Wang, Q.; Collins, R.L.; Laricchia, K.M.; Ganna, A.; Birnbaum, D.P.; et al. The Mutational Constraint Spectrum Quantified from Variation in 141,456 Humans. Nature 2020, 581, 434–443. [Google Scholar] [CrossRef] [PubMed]
- Kopanos, C.; Tsiolkas, V.; Kouris, A.; Chapple, C.E.; Albarca Aguilera, M.; Meyer, R.; Massouras, A. VarSome: The Human Genomic Variant Search Engine. Bioinformatics 2019, 35, 1978–1980. [Google Scholar] [CrossRef] [PubMed]
- Rad, A.; Schade-Mann, T.; Gamerdinger, P.; Yanus, G.A.; Schulte, B.; Müller, M.; Imyanitov, E.N.; Biskup, S.; Löwenheim, H.; Tropitzsch, A.; et al. Aberrant COL11A1 Splicing Causes Prelingual Autosomal Dominant Nonsyndromic Hearing Loss in the DFNA37 Locus. Hum. Mutat. 2021, 42, 25–30. [Google Scholar] [CrossRef]
- Tompson, S.W.; Young, T.L. Assaying the Effects of Splice Site Variants by Exon Trapping in a Mammalian Cell Line. Bio Protoc. 2017, 7, e2281. [Google Scholar] [CrossRef] [PubMed]
- Diaw, S.H.; Ganos, C.; Zittel, S.; Plötze-Martin, K.; Kulikovskaja, L.; Vos, M.; Westenberger, A.; Rakovic, A.; Lohmann, K.; Dulovic-Mahlow, M. Mutant WDR45 Leads to Altered Ferritinophagy and Ferroptosis in β-Propeller Protein-Associated Neurodegeneration. Int. J. Mol. Sci. 2022, 23, 9524. [Google Scholar] [CrossRef] [PubMed]
- Grünewald, A.; Voges, L.; Rakovic, A.; Kasten, M.; Vandebona, H.; Hemmelmann, C.; Lohmann, K.; Orolicki, S.; Ramirez, A.; Schapira, A.H.V.; et al. Mutant Parkin Impairs Mitochondrial Function and Morphology in Human Fibroblasts. PLoS ONE 2010, 5, e12962. [Google Scholar] [CrossRef]
| Category | P1 | P2 | Literature (n = 31 [5,6,7,8,9,10,11]) 1 |
|---|---|---|---|
| Sex | Female | Male | male 14/29 |
| Origin | Germany | UK | Europe: 18/31 |
| Asia: 10/31 | |||
| Other: 3/31 | |||
| Age of Onset | 6 years | 45 years | Mean: 20 years (range: 0–63 years) |
| Age at Examination | 31 years | 47 years | Mean: 35 years (range: 2–72 years) |
| First signs and symptom | Gait disturbance (unsteady gait) | Gait disturbance (unable to run) | Gait disturbance: 18/26 Developmental delay: 3/26 Tremor: 2/26 Other 2: 3/26 |
| Ataxia Upper; Lower Limb | yes; yes | yes; yes | 21/24 10/14; 13/15 |
| Spasticity Upper; Lower Limb Babinski Sign | yes; yes yes | yes; yes yes | 9/13 1/8; 9/13 22/27 |
| Hyperreflexia Upper; Lower Limb | yes; yes | yes; yes | 24/27 8/12; 24/27 |
| Paresis Upper; Lower Limb | no; yes | yes; yes | 8/12 4/7; 8/12 |
| Ambulation | Wheelchair user | Balance issues | Wheelchair: 6/13 Walker: 2/13 Independent: 5/13 |
| Tremor | yes | no | 2/4 |
| Oculomotoric Square Waves Jerks Nystagmus Suppresion of VOR | yes yes pathological | no no normal | 7/10 0/2 0/1 |
| Dysarthria | yes | no | 10/13 |
| MRI abnormalities | Leukencephalopathy | none | Normal: 6/20 Cerebellar atrophy: 11/20 Leukencephalopathie: 3/20 |
| Response to treatment | Levodopa (mild), VIM- DBS (good) | Poor to Baclofen | n. a. |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Pauly, M.G.; Brüggemann, N.; Efthymiou, S.; Grözinger, A.; Diaw, S.H.; Chelban, V.; Turchetti, V.; Vona, B.; Tadic, V.; Houlden, H.; et al. Not to Miss: Intronic Variants, Treatment, and Review of the Phenotypic Spectrum in VPS13D-Related Disorder. Int. J. Mol. Sci. 2023, 24, 1874. https://doi.org/10.3390/ijms24031874
Pauly MG, Brüggemann N, Efthymiou S, Grözinger A, Diaw SH, Chelban V, Turchetti V, Vona B, Tadic V, Houlden H, et al. Not to Miss: Intronic Variants, Treatment, and Review of the Phenotypic Spectrum in VPS13D-Related Disorder. International Journal of Molecular Sciences. 2023; 24(3):1874. https://doi.org/10.3390/ijms24031874
Chicago/Turabian StylePauly, Martje G., Norbert Brüggemann, Stephanie Efthymiou, Anne Grözinger, Sokhna Haissatou Diaw, Viorica Chelban, Valentina Turchetti, Barbara Vona, Vera Tadic, Henry Houlden, and et al. 2023. "Not to Miss: Intronic Variants, Treatment, and Review of the Phenotypic Spectrum in VPS13D-Related Disorder" International Journal of Molecular Sciences 24, no. 3: 1874. https://doi.org/10.3390/ijms24031874
APA StylePauly, M. G., Brüggemann, N., Efthymiou, S., Grözinger, A., Diaw, S. H., Chelban, V., Turchetti, V., Vona, B., Tadic, V., Houlden, H., Münchau, A., & Lohmann, K. (2023). Not to Miss: Intronic Variants, Treatment, and Review of the Phenotypic Spectrum in VPS13D-Related Disorder. International Journal of Molecular Sciences, 24(3), 1874. https://doi.org/10.3390/ijms24031874








