A Paternal Methylation Error in the Congenital Hydrocephalic Texas (H-Tx) Rat Is Partially Rescued with Natural Folate Supplements
Abstract
1. Introduction
2. Results
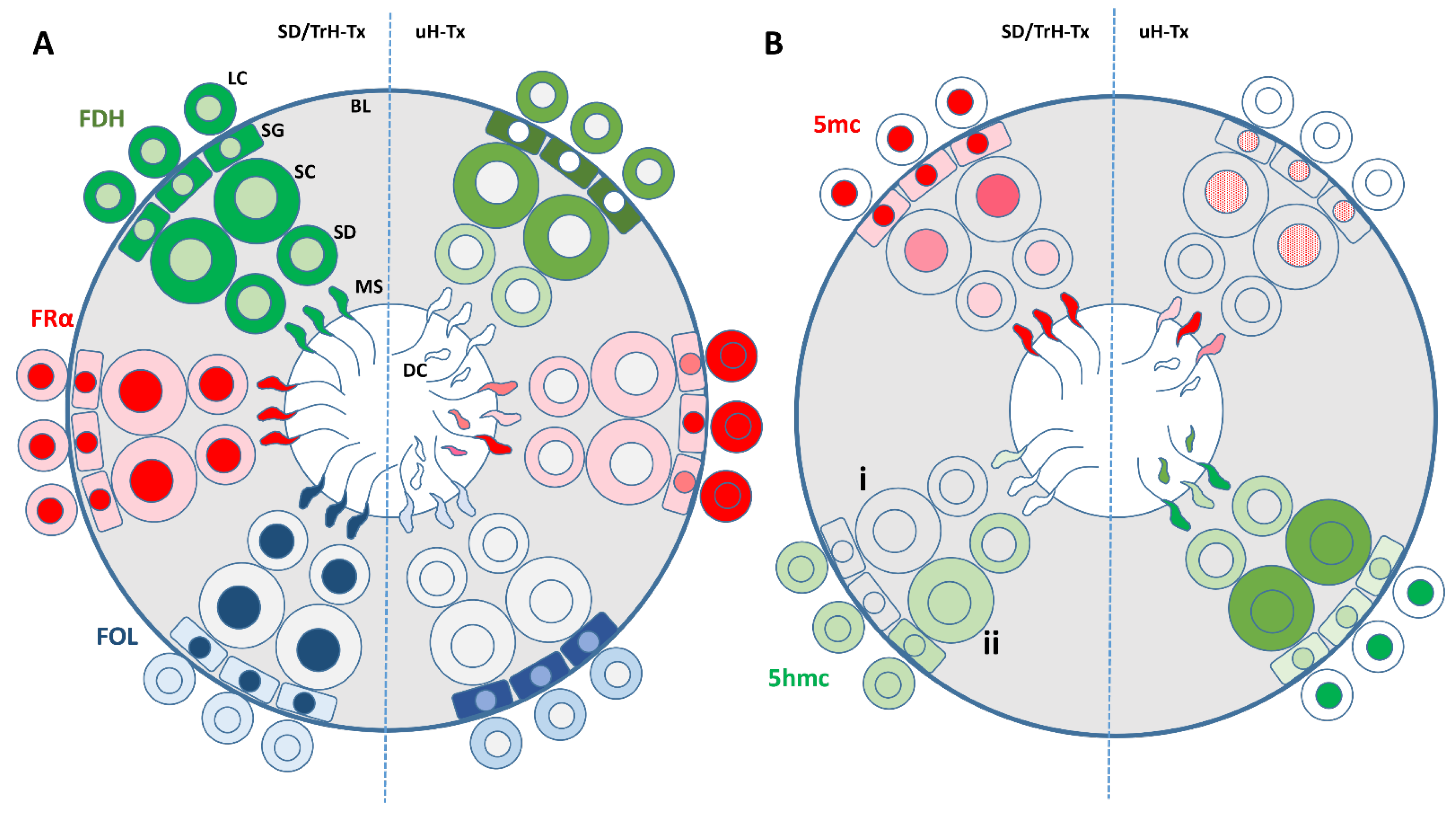
2.1. Histological and Morphological Analysis of Testes
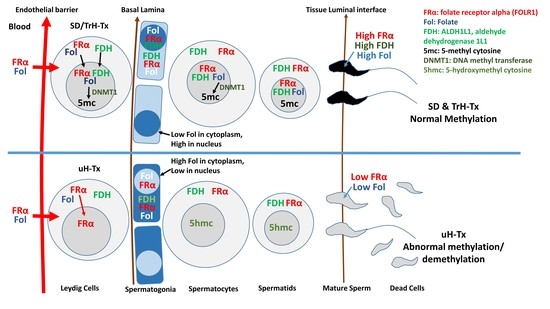
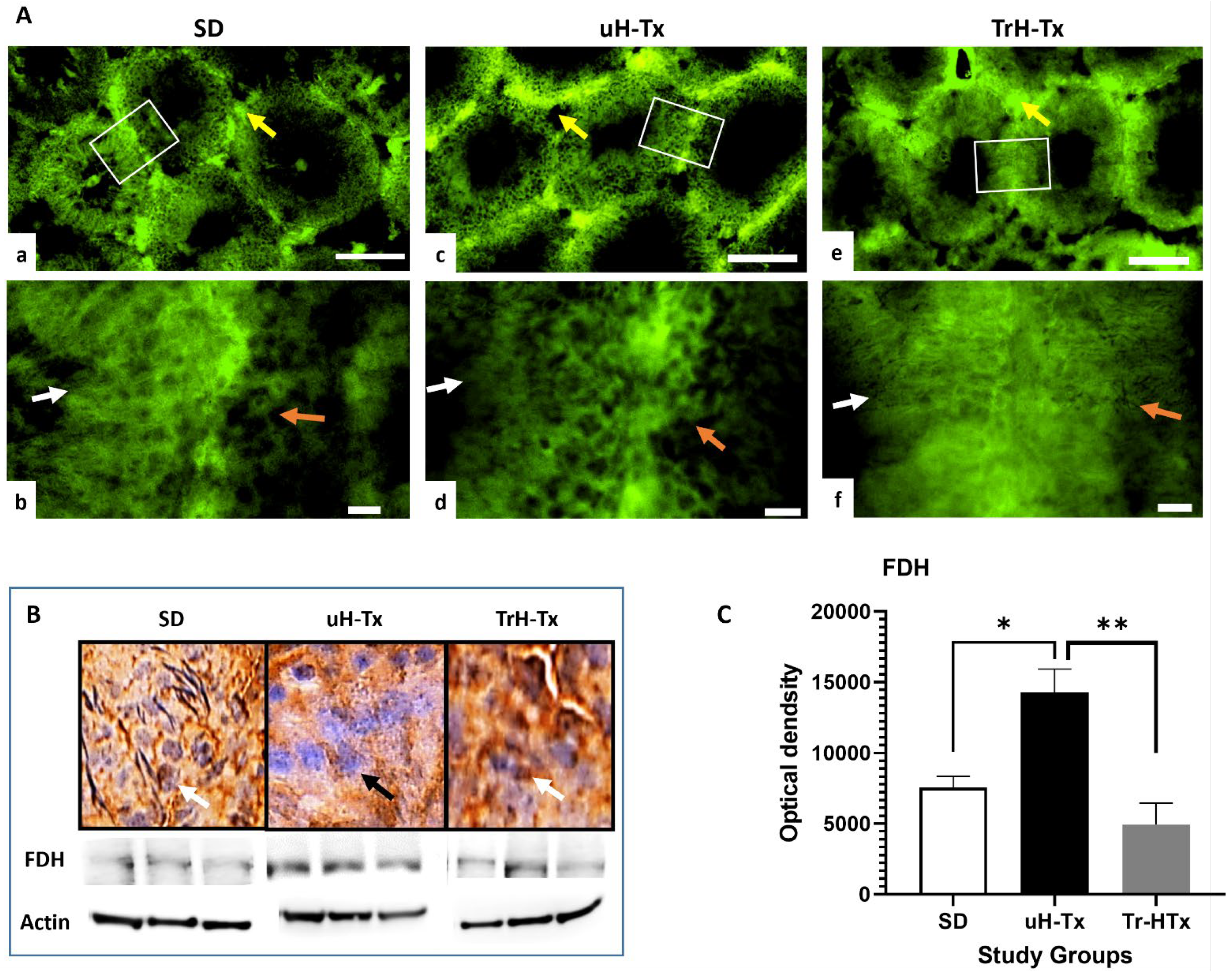
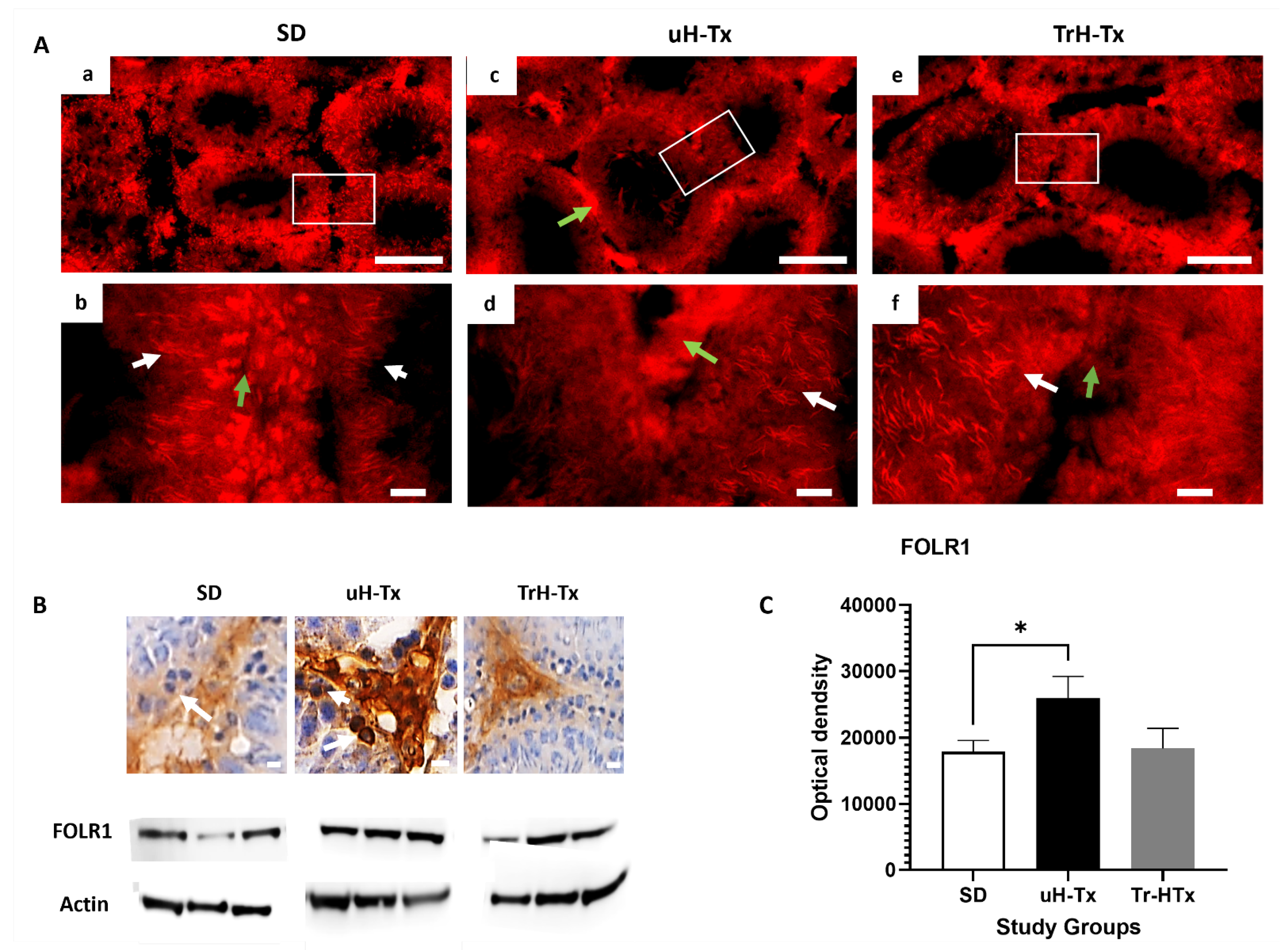
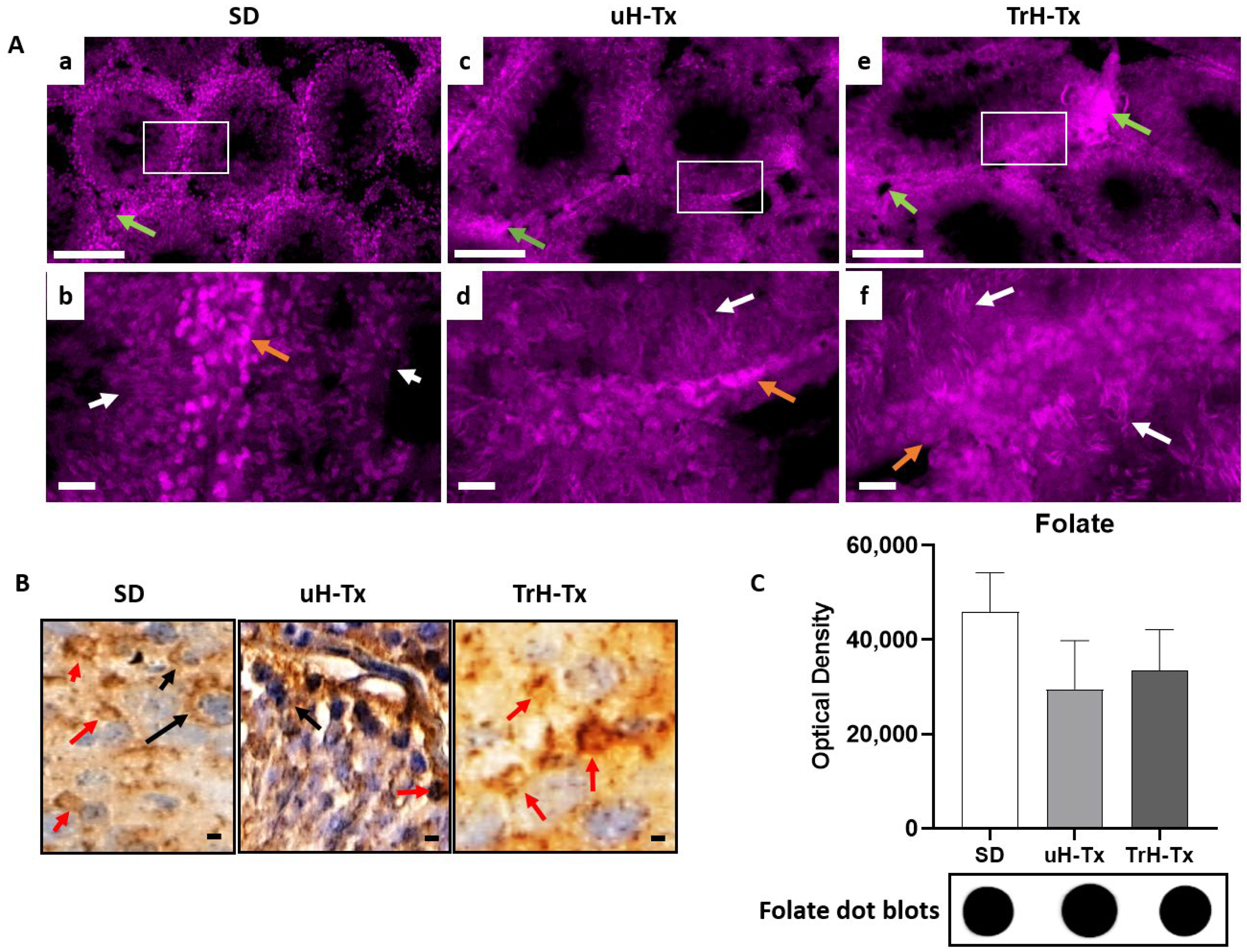
2.2. Changes in Folate Metabolism
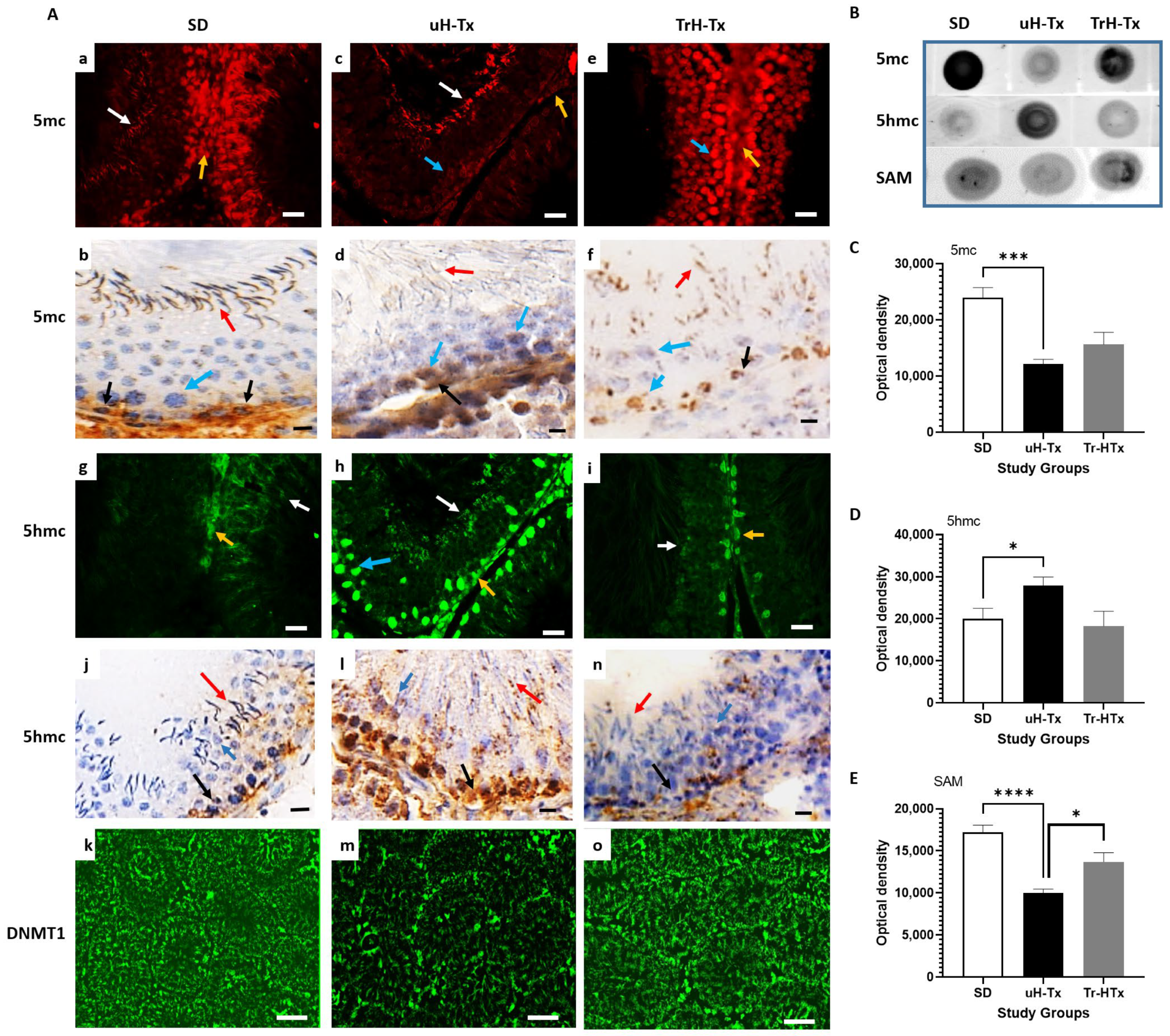
2.3. Changes in Methylation
3. Discussion
Limitations of the Current Study and Future Work
4. Materials and Methods
4.1. Animals
4.1.1. Animal Treatment
4.1.2. Animal Collection
4.2. Immunohistochemical Analysis
4.2.1. Antibodies Used in the Study
4.2.2. Tissue Preparation
4.3. Haematoxylin and Eosin Staining
4.4. Acridine Orange Staining
4.5. Peroxidase Staining
4.6. Immunofluorescence Staining
4.7. Total Protein Isolation
4.8. Protein Determination and Western Blots
4.9. Dot Blots
4.10. Statistical Analysis
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Eleftheriou, M.; Ruzov, A. Modified Forms of Cytosine in Eukaryotes: DNA (De)methylation and Beyond. Methods Mol. Biol. 2021, 2198, 3–13. [Google Scholar] [CrossRef] [PubMed]
- Avila-Barnard, S.; Volz, D.C. Rapid and Efficient Spatiotemporal Monitoring of Normal and Aberrant Cytosine Methylation within Intact Zebrafish Embryos. J. Vis. Exp. JoVE 2022, 186, e64190. [Google Scholar] [CrossRef] [PubMed]
- Petell, C.J.; Alabdi, L.; He, M.; San Miguel, P.; Rose, R.; Gowher, H. An epigenetic switch regulates de novo DNA methylation at a subset of pluripotency gene enhancers during embryonic stem cell differentiation. Nucleic Acids Res. 2016, 44, 7605–7617. [Google Scholar] [CrossRef] [PubMed]
- Grasso, L.; Suska, O.; Davidson, L.; Gonatopoulos-Pournatzis, T.; Williamson, R.; Wasmus, L.; Wiedlich, S.; Peggie, M.; Stavridis, M.P.; Cowling, V.H. mRNA Cap Methylation in Pluripotency and Differentiation. Cell Rep. 2016, 16, 1352–1365. [Google Scholar] [CrossRef] [PubMed]
- Lee, D.S.; Shin, J.Y.; Tonge, P.D.; Puri, M.C.; Lee, S.; Park, H.; Lee, W.C.; Hussein, S.M.; Bleazard, T.; Yun, J.Y.; et al. An epigenomic roadmap to induced pluripotency reveals DNA methylation as a reprogramming modulator. Nat. Commun. 2014, 5, 5619. [Google Scholar] [CrossRef]
- Farthing, C.R.; Ficz, G.; Ng, R.K.; Chan, C.F.; Andrews, S.; Dean, W.; Hemberger, M.; Reik, W. Global mapping of DNA methylation in mouse promoters reveals epigenetic reprogramming of pluripotency genes. PLoS Genet. 2008, 4, e1000116. [Google Scholar] [CrossRef]
- Alata Jimenez, N.; Strobl-Mazzulla, P.H. Folate Carrier Deficiency Drives Differential Methylation and Enhanced Cellular Potency in the Neural Plate Border. Front. Cell Dev. Biol. 2022, 10, 834625. [Google Scholar] [CrossRef]
- Miyashita, C.; Araki, A.; Miura, R.; Ait Bamai, Y.; Kobayashi, S.; Itoh, S.; Ito, K.; Tsai, M.S.; Kishi, R. Prevalence of childhood wheeze and modified DNA methylation at 7 years of age according to maternal folate levels during pregnancy in the Hokkaido Study. Pediatr. Allergy Immunol. 2021, 32, 514–523. [Google Scholar] [CrossRef]
- De Castro, T.B.; Rodrigues-Fleming, G.H.; Oliveira-Cucolo, J.G.; Silva, J.; Silva, F.P.; Raposo, L.S.; Maniglia, J.V.; Pavarino, E.C.; Batista Arantes, L.M.R.; Galbiatti-Dias, A.L.S.; et al. Gene Polymorphisms Involved in Folate Metabolism and DNA Methylation with the Risk of Head and Neck Cancer. Asian Pac. J. Cancer Prev. 2020, 21, 3751–3759. [Google Scholar] [CrossRef]
- Mahajan, A.; Sapehia, D.; Thakur, S.; Mohanraj, P.S.; Bagga, R.; Kaur, J. Effect of imbalance in folate and vitamin B12 in maternal/parental diet on global methylation and regulatory miRNAs. Sci. Rep. 2019, 9, 17602. [Google Scholar] [CrossRef]
- Coppede, F.; Stoccoro, A.; Tannorella, P.; Migliore, L. Plasma Homocysteine and Polymorphisms of Genes Involved in Folate Metabolism Correlate with DNMT1 Gene Methylation Levels. Metabolites 2019, 9, 298. [Google Scholar] [CrossRef]
- Luo, T.; Li, K.; Ling, Z.; Zhao, G.; Li, B.; Wang, Z.; Wang, X.; Han, Y.; Xia, L.; Zhang, Y.; et al. De novo mutations in folate-related genes associated with common developmental disorders. Comput. Struct. Biotechnol. J. 2021, 19, 1414–1422. [Google Scholar] [CrossRef]
- Desai, A.; Sequeira, J.M.; Quadros, E.V. The metabolic basis for developmental disorders due to defective folate transport. Biochimie 2016, 126, 31–42. [Google Scholar] [CrossRef]
- Reynolds, E. Folate and ageing. Lancet 2007, 369, 1601, author reply 1601–1602. [Google Scholar] [CrossRef]
- Requena-Jimenez, A.; Nabiuni, M.; Miyan, J.A. Profound changes in cerebrospinal fluid proteome and metabolic profile are associated with congenital hydrocephalus. J. Cereb. Blood Flow Metab. 2021, 41, 3400–3414. [Google Scholar] [CrossRef]
- Jimenez, A.R.; Naz, N.; Miyan, J.A. Altered folate binding protein expression and folate delivery are associated with congenital hydrocephalus in the hydrocephalic Texas rat. J. Cereb. Blood Flow Metab. 2019, 39, 2061–2073. [Google Scholar] [CrossRef]
- Naz, N.; Jimenez, A.R.; Sanjuan-Vilaplana, A.; Gurney, M.; Miyan, J. Neonatal hydrocephalus is a result of a block in folate handling and metabolism involving 10-formyltetrahydrofolate dehydrogenase. J. Neurochem. 2016, 138, 610–623. [Google Scholar] [CrossRef]
- Cains, S.; Shepherd, A.; Nabiuni, M.; Owen-Lynch, P.J.; Miyan, J. Addressing a folate imbalance in fetal cerebrospinal fluid can decrease the incidence of congenital hydrocephalus. J. Neuropathol. Exp. Neurol. 2009, 68, 404–416. [Google Scholar] [CrossRef]
- Sullivan, W.; Reeves, B.C.; Duy, P.Q.; Nelson-Williams, C.; Dong, W.; Jin, S.C.; Kahle, K.T. Exome Sequencing as a Potential Diagnostic Adjunct in Sporadic Congenital Hydrocephalus. JAMA Pediatr. 2021, 175, 310–313. [Google Scholar] [CrossRef]
- Kundishora, A.J.; Singh, A.K.; Allington, G.; Duy, P.Q.; Ryou, J.; Alper, S.L.; Jin, S.C.; Kahle, K.T. Genomics of human congenital hydrocephalus. Childs Nerv. Syst. 2021, 37, 3325–3340. [Google Scholar] [CrossRef]
- Allington, G.; Duy, P.Q.; Ryou, J.; Singh, A.; Kiziltug, E.; Robert, S.M.; Kundishora, A.J.; King, S.; Haider, S.; Kahle, K.T.; et al. Genomic approaches to improve the clinical diagnosis and management of patients with congenital hydrocephalus. J. Neurosurg. Pediatr. 2021, 29, 168–177. [Google Scholar] [CrossRef] [PubMed]
- Jin, S.C.; Dong, W.; Kundishora, A.J.; Panchagnula, S.; Moreno-De-Luca, A.; Furey, C.G.; Allocco, A.A.; Walker, R.L.; Nelson-Williams, C.; Smith, H.; et al. Exome sequencing implicates genetic disruption of prenatal neuro-gliogenesis in sporadic congenital hydrocephalus. Nat. Med. 2020, 26, 1754–1765. [Google Scholar] [CrossRef] [PubMed]
- Jin, S.C.; Furey, C.G.; Zeng, X.; Allocco, A.; Nelson-Williams, C.; Dong, W.; Karimy, J.K.; Wang, K.; Ma, S.; Delpire, E.; et al. SLC12A ion transporter mutations in sporadic and familial human congenital hydrocephalus. Mol. Genet. Genom. Med. 2019, 7, e892. [Google Scholar] [CrossRef] [PubMed]
- Allocco, A.A.; Jin, S.C.; Duy, P.Q.; Furey, C.G.; Zeng, X.; Dong, W.; Nelson-Williams, C.; Karimy, J.K.; DeSpenza, T.; Hao, L.T.; et al. Recessive Inheritance of Congenital Hydrocephalus with Other Structural Brain Abnormalities Caused by Compound Heterozygous Mutations in ATP1A3. Front. Cell. Neurosci. 2019, 13, 425. [Google Scholar] [CrossRef] [PubMed]
- Furey, C.G.; Choi, J.; Jin, S.C.; Zeng, X.; Timberlake, A.T.; Nelson-Williams, C.; Mansuri, M.S.; Lu, Q.; Duran, D.; Panchagnula, S.; et al. De Novo Mutation in Genes Regulating Neural Stem Cell Fate in Human Congenital Hydrocephalus. Neuron 2018, 99, 302–314.e4. [Google Scholar] [CrossRef]
- Miyan, J.; Cains, S.; Larcombe, S.; Naz, N.; Jimenez, A.R.; Bueno, D.; Gato, A. Subarachnoid cerebrospinal fluid is essential for normal development of the cerebral cortex. Semin. Cell Dev. Biol. 2020, 102, 28–39. [Google Scholar] [CrossRef]
- Jouet, M. The role of genetics in understanding hydrocephalus and spina bifida. An update. Eur. J. Pediatr. Surg. 1996, 6 (Suppl. S1), 35–36. [Google Scholar]
- Jouet, M.; Kenwrick, S. Gene analysis of L1 neural cell adhesion molecule in prenatal diagnosis of hydrocephalus. Lancet 1995, 345, 161–162. [Google Scholar] [CrossRef]
- Zhang, J.; Williams, M.A.; Rigamonti, D. Genetics of human hydrocephalus. J. Neurol. 2006, 253, 1255–1266. [Google Scholar] [CrossRef]
- Cai, X.; Pattisapu, J.V.; Tarnuzzer, R.W.; Fernandez-Valle, C.; Gibson, J.S. TGF-beta1 expression is reduced in hydrocephalic H-Tx rat brain. Eur. J. Pediatr. Surg. 1999, 9 (Suppl. S1), 35–38. [Google Scholar] [CrossRef]
- Cai, X.; McGraw, G.; Pattisapu, J.V.; von Kalm, L.; Willingham, S.; Socci, D.; Gibson, J.S. Hydrocephalus in the H-Tx rat: A monogenic disease? Exp. Neurol. 2000, 163, 131–135. [Google Scholar] [CrossRef]
- Miller, J.M.; Kumar, R.; McAllister, J.P., 2nd; Krause, G.S. Gene expression analysis of the development of congenital hydrocephalus in the H-Tx rat. Brain Res. 2006, 1075, 36–47. [Google Scholar] [CrossRef]
- Morgan, F.W.; Stewart, J.A.; Smith, A.N.; Tarnuzzer, R.W. Differential expression of stress response genes in the H-Tx rat model of congenital hydrocephalus. Brain Res. Mol. Brain Res. 2005, 138, 273–290. [Google Scholar] [CrossRef]
- Li, X.; Miyajima, M.; Arai, H. Analysis of TGF-beta2 and TGF-beta3 expression in the hydrocephalic H-Tx rat brain. Childs Nerv. Syst. 2005, 21, 32–38. [Google Scholar] [CrossRef]
- Jones, H.C.; Chen, G.F.; Yehia, B.R.; Carter, B.J.; Akins, E.J.; Wolpin, L.C. Single and multiple congenic strains for hydrocephalus in the H-Tx rat. Mamm. Genome 2005, 16, 251–261. [Google Scholar] [CrossRef]
- Jones, H.C.; Depelteau, J.S.; Carter, B.J.; Somera, K.C. The frequency of inherited hydrocephalus is influenced by intrauterine factors in H-Tx rats. Exp. Neurol. 2002, 176, 213–220. [Google Scholar] [CrossRef]
- Jones, H.C.; Depelteau, J.S.; Carter, B.J.; Lopman, B.A.; Morel, L. Genome-wide linkage analysis of inherited hydrocephalus in the H-Tx rat. Mamm. Genome 2001, 12, 22–26. [Google Scholar] [CrossRef]
- Jones, H.C.; Carter, B.J.; Depelteau, J.S.; Roman, M.; Morel, L. Chromosomal linkage associated with disease severity in the hydrocephalic H-Tx rat. Behav. Genet. 2001, 31, 101–111. [Google Scholar] [CrossRef]
- Depelteau, J.S.; Jones, H.C. Lactation during gestation affects the expression of inherited hydrocephalus in H-Tx rats. Eur. J. Pediatr. Surg. 2001, 11, S49–S51. [Google Scholar]
- Jones, H.C.; Lopman, B.A.; Jones, T.W.; Morel, L.M. Breeding characteristics and genetic analysis of the H-Tx rat strain. Eur. J. Pediatr. Surg. 1999, 9 (Suppl. S1), 42–43. [Google Scholar]
- Al-Ghnaniem Abbadi, R.; Emery, P.; Pufulete, M. Short-term folate supplementation in physiological doses has no effect on ESR1 and MLH1 methylation in colonic mucosa of individuals with adenoma. J. Nutr. Nutr. 2012, 5, 327–338. [Google Scholar] [CrossRef] [PubMed]
- Al-Ghnaniem, R.; Peters, J.; Foresti, R.; Heaton, N.; Pufulete, M. Methylation of estrogen receptor alpha and mutL homolog 1 in normal colonic mucosa: Association with folate and vitamin B-12 status in subjects with and without colorectal neoplasia. Am. J. Clin. Nutr. 2007, 86, 1064–1072. [Google Scholar] [CrossRef] [PubMed]
- Pufulete, M.; Al-Ghnaniem, R.; Khushal, A.; Appleby, P.; Harris, N.; Gout, S.; Emery, P.W.; Sanders, T.A. Effect of folic acid supplementation on genomic DNA methylation in patients with colorectal adenoma. Gut 2005, 54, 648–653. [Google Scholar] [CrossRef] [PubMed]
- Pufulete, M.; Al-Ghnaniem, R.; Rennie, J.A.; Appleby, P.; Harris, N.; Gout, S.; Emery, P.W.; Sanders, T.A. Influence of folate status on genomic DNA methylation in colonic mucosa of subjects without colorectal adenoma or cancer. Br. J. Cancer 2005, 92, 838–842. [Google Scholar] [CrossRef] [PubMed]
- Mai, C.T.; Evans, J.; Alverson, C.J.; Yue, X.; Flood, T.; Arnold, K.; Nestoridi, E.; Denson, L.; Adisa, O.; Moore, C.A.; et al. Changes in Spina Bifida Lesion Level after Folic Acid Fortification in the US. J. Pediatr. 2022, 249, 59–66.e1. [Google Scholar] [CrossRef] [PubMed]
- Rodrigues, V.B.; Silva, E.N.D.; Santos, M.L.P. Cost-effectiveness of mandatory folic acid fortification of flours in prevention of neural tube defects: A systematic review. PLoS ONE 2021, 16, e0258488. [Google Scholar] [CrossRef]
- Craenen, K.; Verslegers, M.; Callaerts-Vegh, Z.; Craeghs, L.; Buset, J.; Govaerts, K.; Neefs, M.; Gsell, W.; Baatout, S.; D’Hooge, R.; et al. Folic Acid Fortification Prevents Morphological and Behavioral Consequences of X-ray Exposure During Neurulation. Front. Behav. Neurosci. 2020, 14, 609660. [Google Scholar] [CrossRef]
- Yang, L.; Wang, W.; Mao, B.; Qiu, J.; Guo, H.; Yi, B.; He, X.; Lin, X.; Lv, L.; Xu, X.; et al. Maternal Folic Acid Supplementation, Dietary Folate Intake, and Low Birth Weight: A Birth Cohort Study. Front. Public Health 2022, 10, 844150. [Google Scholar] [CrossRef]
- Li, S.; Liu, D.; Zhang, R.; Lei, F.; Liu, X.; Cheng, Y.; Li, C.; Xiao, M.; Guo, L.; Li, M.; et al. The association of maternal dietary folate intake and folic acid supplementation with small-for-gestational-age births: A cross-sectional study in Northwest China. Br. J. Nutr. 2019, 122, 459–467. [Google Scholar] [CrossRef]
- Li, B.; Chang, S.; Liu, C.; Zhang, M.; Zhang, L.; Liang, L.; Li, R.; Wang, X.; Qin, C.; Zhang, T.; et al. Low Maternal Dietary Folate Alters Retrotranspose by Methylation Regulation in Intrauterine Growth Retardation (IUGR) Fetuses in a Mouse Model. Med. Sci. Monit. 2019, 25, 3354–3365. [Google Scholar] [CrossRef]
- Mao, B.; Qiu, J.; Zhao, N.; Shao, Y.; Dai, W.; He, X.; Cui, H.; Lin, X.; Lv, L.; Tang, Z.; et al. Maternal folic acid supplementation and dietary folate intake and congenital heart defects. PLoS ONE 2017, 12, e0187996. [Google Scholar] [CrossRef]
- Li, D.; Pickell, L.; Liu, Y.; Wu, Q.; Cohn, J.S.; Rozen, R. Maternal methylenetetrahydrofolate reductase deficiency and low dietary folate lead to adverse reproductive outcomes and congenital heart defects in mice. Am. J. Clin. Nutr. 2005, 82, 188–195. [Google Scholar] [CrossRef]
- Liu, J.; Li, Z.; Ye, R.; Ren, A.; Liu, J. Folic acid supplementation and risk for congenital hydrocephalus in China. Public Health Nutr. 2021, 24, 4238–4244. [Google Scholar] [CrossRef]
- Liu, J.; Jin, L.; Li, Z.; Zhang, Y.; Zhang, L.; Wang, L.; Ren, A. Prevalence and trend of isolated and complicated congenital hydrocephalus and preventive effect of folic acid in northern China, 2005–2015. Metab. Brain Dis. 2018, 33, 837–842. [Google Scholar] [CrossRef]
- D’Souza, S.W.; Copp, A.J.; Greene, N.D.E.; Glazier, J.D. Maternal Inositol Status and Neural Tube Defects: A Role for the Human Yolk Sac in Embryonic Inositol Delivery? Adv. Nutr. 2021, 12, 212–222. [Google Scholar] [CrossRef]
- Greene, N.D.; Leung, K.Y.; Copp, A.J. Inositol, neural tube closure and the prevention of neural tube defects. Birth Defects Res. 2017, 109, 68–80. [Google Scholar] [CrossRef]
- Greene, N.D.; Leung, K.Y.; Gay, V.; Burren, K.; Mills, K.; Chitty, L.S.; Copp, A.J. Inositol for the prevention of neural tube defects: A pilot randomised controlled trial. Br. J. Nutr. 2016, 115, 974–983. [Google Scholar] [CrossRef]
- Wilson, M.P.; Hugge, C.; Bielinska, M.; Nicholas, P.; Majerus, P.W.; Wilson, D.B. Neural tube defects in mice with reduced levels of inositol 1,3,4-trisphosphate 5/6-kinase. Proc. Natl. Acad. Sci. USA 2009, 106, 9831–9835. [Google Scholar] [CrossRef]
- Greene, N.D.; Copp, A.J. Inositol prevents folate-resistant neural tube defects in the mouse. Nat. Med. 1997, 3, 60–66. [Google Scholar] [CrossRef]
- Kancherla, V.; Botto, L.D.; Rowe, L.A.; Shlobin, N.A.; Caceres, A.; Arynchyna-Smith, A.; Zimmerman, K.; Blount, J.; Kibruyisfaw, Z.; Ghotme, K.A.; et al. Preventing birth defects, saving lives, and promoting health equity: An urgent call to action for universal mandatory food fortification with folic acid. Lancet Glob. Health 2022, 10, e1053–e1057. [Google Scholar] [CrossRef]
- Urdinguio, R.G.; Bayon, G.F.; Dmitrijeva, M.; Torano, E.G.; Bravo, C.; Fraga, M.F.; Bassas, L.; Larriba, S.; Fernandez, A.F. Aberrant DNA methylation patterns of spermatozoa in men with unexplained infertility. Hum. Reprod. 2015, 30, 1014–1028. [Google Scholar] [CrossRef] [PubMed]
- Kohn, D.F.; Chinookoswong, N.; Chou, S.M. Animal model of human disease. Congenital hydrocephalus. Am. J. Pathol. 1984, 114, 184–185. [Google Scholar] [PubMed]
- Kohn, D.F.; Chinookoswong, N.; Chou, S.M. A new model of congenital hydrocephalus in the rat. Acta Neuropathol. 1981, 54, 211–218. [Google Scholar] [CrossRef] [PubMed]
- Boillat, C.A.; Jones, H.C.; Kaiser, G.L. Inherited hydrocephalus in the H-Tx rat: The ventricular system in late-gestation and neonatal aqueduct stenosis. Eur. J. Pediatr. Surg. Z. Kinderchir. 2001, 11 (Suppl. S1), S43–S44. [Google Scholar]
- Jones, H.C.; Lopman, B.A.; Jones, T.W.; Carter, B.J.; Depelteau, J.S.; Morel, L. The expression of inherited hydrocephalus in H-Tx rats. Childs Nerv. Syst. 2000, 16, 578–584. [Google Scholar] [CrossRef]
- Harris, N.G.; Jones, H.C.; Williams, S.C. MR imaging for measurements of ventricles and cerebral cortex in postnatal rats (H-Tx strain) with progressive inherited hydrocephalus. Exp. Neurol. 1992, 118, 1–6. [Google Scholar] [CrossRef]
- Jones, H.C.; Bucknall, R.M. Inherited prenatal hydrocephalus in the H-Tx rat: A morphological study. Neuropathol. Appl. Aeurobiol. 1988, 14, 263–274. [Google Scholar] [CrossRef]
- Kelly, T.L.; Neaga, O.R.; Schwahn, B.C.; Rozen, R.; Trasler, J.M. Infertility in 5,10-methylenetetrahydrofolate reductase (MTHFR)-deficient male mice is partially alleviated by lifetime dietary betaine supplementation. Biol. Reprod. 2005, 72, 667–677. [Google Scholar] [CrossRef]
- Amir, R.E.; Zoghbi, H.Y. Rett syndrome: Methyl-CpG-binding protein 2 mutations and phenotype-genotype correlations. Am. J. Med. Genet. 2000, 97, 147–152. [Google Scholar] [CrossRef]
- Amir, R.E.; Van den Veyver, I.B.; Wan, M.; Tran, C.Q.; Francke, U.; Zoghbi, H.Y. Rett syndrome is caused by mutations in X-linked MECP2, encoding methyl-CpG-binding protein 2. Nat. Genet. 1999, 23, 185–188. [Google Scholar] [CrossRef]
- Klein, C.J.; Botuyan, M.V.; Wu, Y.; Ward, C.J.; Nicholson, G.A.; Hammans, S.; Hojo, K.; Yamanishi, H.; Karpf, A.R.; Wallace, D.C.; et al. Mutations in DNMT1 cause hereditary sensory neuropathy with dementia and hearing loss. Nat. Genet. 2011, 43, 595–600. [Google Scholar] [CrossRef]
- Urdinguio, R.G.; Sanchez-Mut, J.V.; Esteller, M. Epigenetic mechanisms in neurological diseases: Genes, syndromes, and therapies. Lancet. Neurol. 2009, 8, 1056–1072. [Google Scholar] [CrossRef]
- Urdinguio, R.G.; Torro, M.I.; Bayon, G.F.; Alvarez-Pitti, J.; Fernandez, A.F.; Redon, P.; Fraga, M.F.; Lurbe, E. Longitudinal study of DNA methylation during the first 5 years of life. J. Transl. Med. 2016, 14, 160. [Google Scholar] [CrossRef]
- Waligora, A.; Waligora, S.; Kozarska, M.; Damasiewicz-Bodzek, A.; Gorczyca, P.; Tyrpien-Golder, K. Autism spectrum disorder (ASD)—Biomarkers of oxidative stress and methylation and transsulfuration cycle. Psychiatr. Pol. 2019, 53, 771–788. [Google Scholar] [CrossRef]
- Sofer, Y.; Harel, L.; Sharkia, M.; Amir, J.; Schoenfeld, T.; Straussberg, R. Neurological manifestations of folate transport defect: Case report and review of the literature. J. Child. Neurol. 2007, 22, 783–786. [Google Scholar] [CrossRef]
- Uysal, F.; Ozturk, S. The loss of global DNA methylation due to decreased DNMT expression in the postnatal mouse ovaries may associate with infertility emerging during ovarian aging. Histochem. Cell Biol. 2020, 154, 301–314. [Google Scholar] [CrossRef]
- Li, E.; Bestor, T.H.; Jaenisch, R. Targeted mutation of the DNA methyltransferase gene results in embryonic lethality. Cell 1992, 69, 915–926. [Google Scholar] [CrossRef]
- Rotondo, J.C.; Lanzillotti, C.; Mazziotta, C.; Tognon, M.; Martini, F. Epigenetics of Male Infertility: The Role of DNA Methylation. Front. Cell Dev. Biol. 2021, 9, 689624. [Google Scholar] [CrossRef]
- Sujit, K.M.; Singh, V.; Trivedi, S.; Singh, K.; Gupta, G.; Rajender, S. Increased DNA methylation in the spermatogenesis-associated (SPATA) genes correlates with infertility. Andrology 2020, 8, 602–609. [Google Scholar] [CrossRef]
- Tang, Q.; Pan, F.; Yang, J.; Fu, Z.; Lu, Y.; Wu, X.; Han, X.; Chen, M.; Lu, C.; Xia, Y.; et al. Idiopathic male infertility is strongly associated with aberrant DNA methylation of imprinted loci in sperm: A case-control study. Clin. Epigenet. 2018, 10, 134. [Google Scholar] [CrossRef]
- Sujit, K.M.; Sarkar, S.; Singh, V.; Pandey, R.; Agrawal, N.K.; Trivedi, S.; Singh, K.; Gupta, G.; Rajender, S. Genome-wide differential methylation analyses identifies methylation signatures of male infertility. Hum. Reprod. 2018, 33, 2256–2267. [Google Scholar] [CrossRef] [PubMed]
- Nasri, F.; Gharesi-Fard, B.; Namavar Jahromi, B.; Farazi-Fard, M.A.; Banaei, M.; Davari, M.; Ebrahimi, S.; Anvar, Z. Sperm DNA methylation of H19 imprinted gene and male infertility. Andrologia 2017, 49, e12766. [Google Scholar] [CrossRef] [PubMed]
- Dong, H.; Wang, Y.; Zou, Z.; Chen, L.; Shen, C.; Xu, S.; Zhang, J.; Zhao, F.; Ge, S.; Gao, Q.; et al. Abnormal Methylation of Imprinted Genes and Cigarette Smoking: Assessment of Their Association with the Risk of Male Infertility. Reprod. Sci. 2017, 24, 114–123. [Google Scholar] [CrossRef] [PubMed]
- Castillo-Fernandez, J.E.; Loke, Y.J.; Bass-Stringer, S.; Gao, F.; Xia, Y.; Wu, H.; Lu, H.; Liu, Y.; Wang, J.; Spector, T.D.; et al. DNA methylation changes at infertility genes in newborn twins conceived by in vitro fertilisation. Genome Med. 2017, 9, 28. [Google Scholar] [CrossRef]
- Cui, X.; Jing, X.; Wu, X.; Yan, M.; Li, Q.; Shen, Y.; Wang, Z. DNA methylation in spermatogenesis and male infertility. Exp. Ther. Med. 2016, 12, 1973–1979. [Google Scholar] [CrossRef]
- Crider, K.S.; Yang, T.P.; Berry, R.J.; Bailey, L.B. Folate and DNA methylation: A review of molecular mechanisms and the evidence for folate’s role. Adv. Nutr. 2012, 3, 21–38. [Google Scholar] [CrossRef]
- Crider, K.S.; Zhu, J.H.; Hao, L.; Yang, Q.H.; Yang, T.P.; Gindler, J.; Maneval, D.R.; Quinlivan, E.P.; Li, Z.; Bailey, L.B.; et al. MTHFR 677C->T genotype is associated with folate and homocysteine concentrations in a large, population-based, double-blind trial of folic acid supplementation. Am. J. Clin. Nutr. 2011, 93, 1365–1372. [Google Scholar] [CrossRef]
- Crider, K.S.; Quinlivan, E.P.; Berry, R.J.; Hao, L.; Li, Z.; Maneval, D.; Yang, T.P.; Rasmussen, S.A.; Yang, Q.; Zhu, J.H.; et al. Genomic DNA methylation changes in response to folic acid supplementation in a population-based intervention study among women of reproductive age. PLoS ONE 2011, 6, e28144. [Google Scholar] [CrossRef]

Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Naz, N.; Moshkdanian, G.; Miyan, S.; Eljabri, S.; James, C.; Miyan, J. A Paternal Methylation Error in the Congenital Hydrocephalic Texas (H-Tx) Rat Is Partially Rescued with Natural Folate Supplements. Int. J. Mol. Sci. 2023, 24, 1638. https://doi.org/10.3390/ijms24021638
Naz N, Moshkdanian G, Miyan S, Eljabri S, James C, Miyan J. A Paternal Methylation Error in the Congenital Hydrocephalic Texas (H-Tx) Rat Is Partially Rescued with Natural Folate Supplements. International Journal of Molecular Sciences. 2023; 24(2):1638. https://doi.org/10.3390/ijms24021638
Chicago/Turabian StyleNaz, Naila, Ghazaleh Moshkdanian, Salma Miyan, Sereen Eljabri, Charlotte James, and Jaleel Miyan. 2023. "A Paternal Methylation Error in the Congenital Hydrocephalic Texas (H-Tx) Rat Is Partially Rescued with Natural Folate Supplements" International Journal of Molecular Sciences 24, no. 2: 1638. https://doi.org/10.3390/ijms24021638
APA StyleNaz, N., Moshkdanian, G., Miyan, S., Eljabri, S., James, C., & Miyan, J. (2023). A Paternal Methylation Error in the Congenital Hydrocephalic Texas (H-Tx) Rat Is Partially Rescued with Natural Folate Supplements. International Journal of Molecular Sciences, 24(2), 1638. https://doi.org/10.3390/ijms24021638