Identification of Compounds Preventing A. fumigatus Biofilm Formation by Inhibition of the Galactosaminogalactan Deacetylase Agd3
Abstract
1. Introduction
2. Results
2.1. Establishment of an Agd3 Enzymatic Activity Assay
2.2. Screening of Microbial Compounds Revealed Inhibition of Agd3 by Actinomycin D
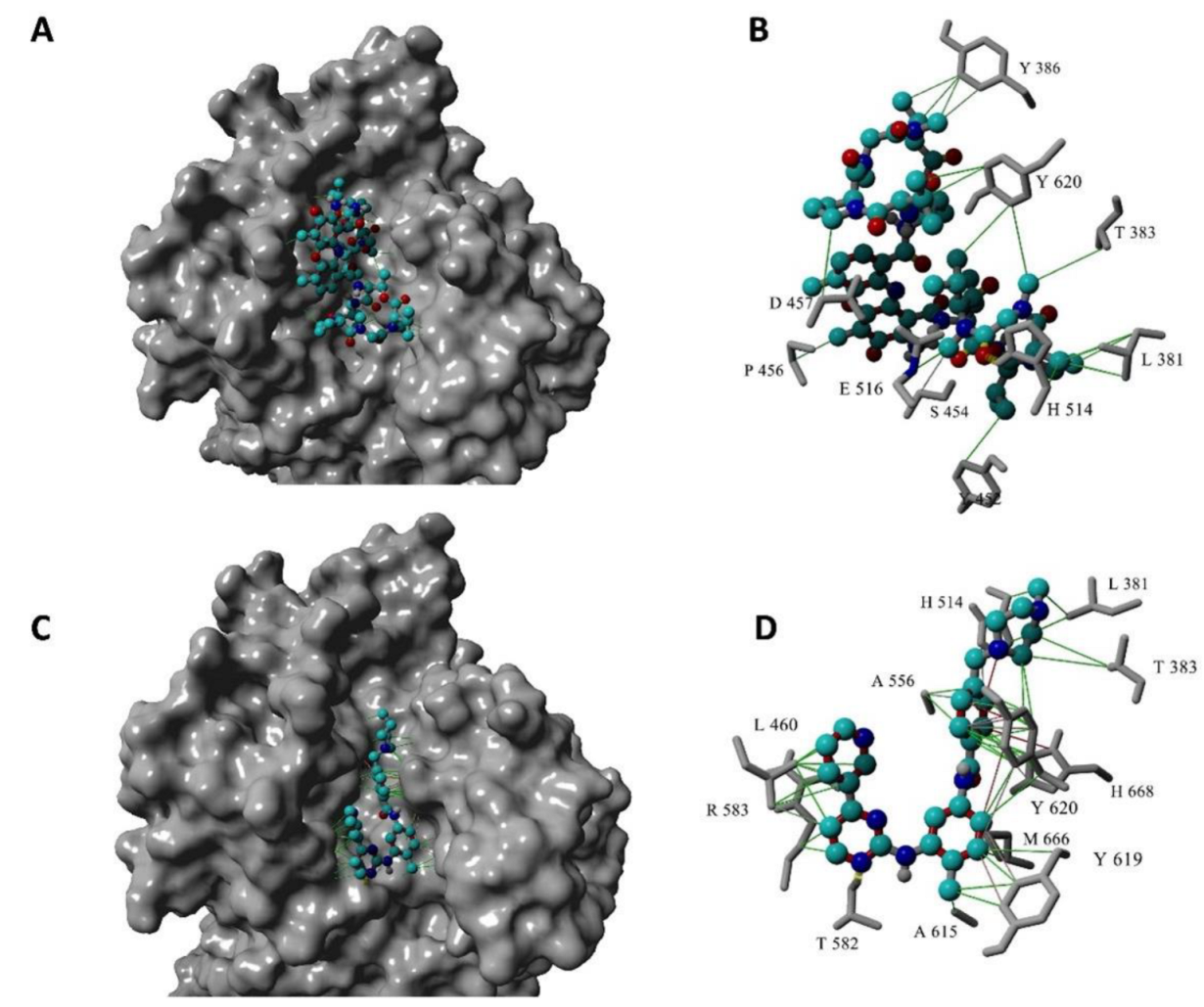
2.3. Structure-Based Virtual Screen Display Inhibition of Agd3 by Imatinib and Rifaximin
2.4. Imatinib Reduced A. fumigatus Virulence in a Galleria Mellonella Infection Model
3. Discussion
4. Materials and Methods
4.1. Strains and Growth Conditions of A. fumigatus
4.2. Expression and Purification of Agd3 from Pichia Pastoris X-33
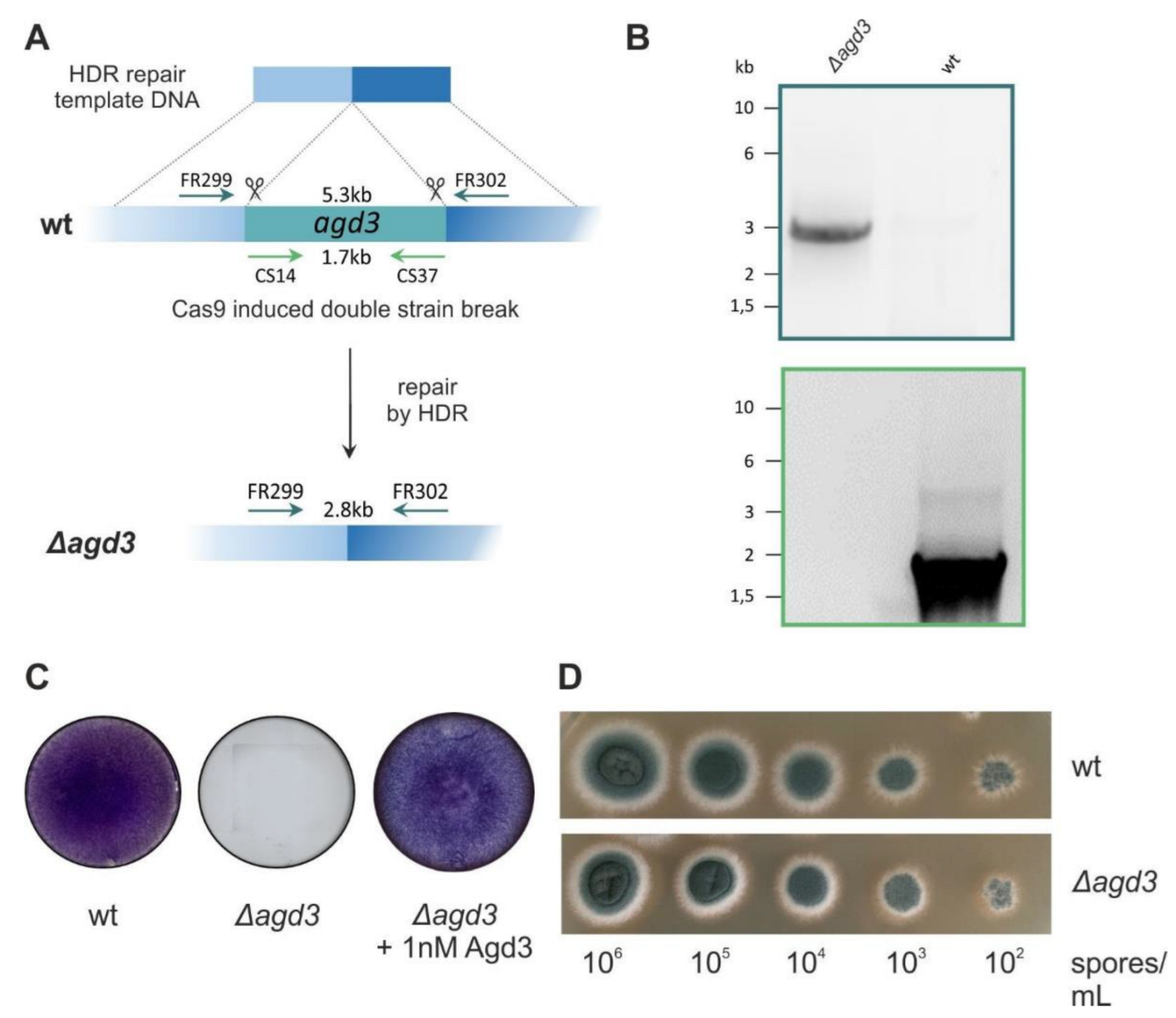
4.3. Generation of ∆agd3 Knockout
4.4. A. fumigatus Growth
4.5. Enzyme-Linked Lectin Activity Assay for Agd3
4.6. Molecular Docking of Agd3
4.7. Biofilm Staining of A. fumigatus Strains by Crystal Violet
4.8. Virulence Studies in Galleria Mellonella Larvae
4.9. Statistical Analysis
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Bongomin, F.; Gago, S.; Oladele, R.O.; Denning, D.W. Global and Multi-National Prevalence of Fungal Diseases-Estimate Precision. J. Fungi 2017, 3, 57. [Google Scholar] [CrossRef] [PubMed]
- Denning, D.W.; Pleuvry, A.; Cole, D.C. Global burden of allergic bronchopulmonary aspergillosis with asthma and its complication chronic pulmonary aspergillosis in adults. Med. Mycol. 2013, 51, 361–370. [Google Scholar] [CrossRef] [PubMed]
- Beswick, E.; Amich, J.; Gago, S. Factoring in the Complexity of the Cystic Fibrosis Lung to Understand Aspergillus fumigatus and Pseudomonas aeruginosa Interactions. Pathogens 2020, 9, 639. [Google Scholar] [CrossRef] [PubMed]
- Kousha, M.; Tadi, R.; Soubani, A.O. Pulmonary aspergillosis: A clinical review. Eur. Respir. Rev. 2011, 20, 156–174. [Google Scholar] [CrossRef] [PubMed]
- Singh, N.; Paterson, D.L. Aspergillus infections in transplant recipients. Clin. Microbiol. Rev. 2005, 18, 44–69. [Google Scholar] [CrossRef]
- Garcia-Vidal, C.; Barba, P.; Arnan, M.; Moreno, A.; Ruiz-Camps, I.; Gudiol, C.; Ayats, J.; Ortí, G.; Carratalà, J. Invasive aspergillosis complicating pandemic influenza A (H1N1) infection in severely immunocompromised patients. Clin. Infect. Dis. 2011, 53, e16–e19. [Google Scholar] [CrossRef]
- Baddley, J.W.; Andes, D.R.; Marr, K.A.; Kontoyiannis, D.P.; Alexander, B.D.; Kauffman, C.A.; Oster, R.A.; Anaissie, E.J.; Walsh, T.J.; Schuster, M.G.; et al. Factors associated with mortality in transplant patients with invasive aspergillosis. Clin. Infect. Dis. 2010, 50, 1559–1567. [Google Scholar] [CrossRef]
- Alastruey-Izquierdo, A.; Cadranel, J.; Flick, H.; Godet, C.; Hennequin, C.; Hoenigl, M.; Kosmidis, C.; Lange, C.; Munteanu, O.; Page, I.; et al. Treatment of Chronic Pulmonary Aspergillosis: Current Standards and Future Perspectives. Respiration 2018, 96, 159–170. [Google Scholar] [CrossRef] [PubMed]
- Latgé, J.P.; Chamilos, G. Aspergillus fumigatus and Aspergillosis in 2019. Clin. Microbiol. Rev. 2019, 33. [Google Scholar] [CrossRef]
- Sheppard, D.C.; Howell, P.L. Biofilm Exopolysaccharides of Pathogenic Fungi: Lessons from Bacteria. J. Biol. Chem. 2016, 291, 12529–12537. [Google Scholar] [CrossRef]
- Loussert, C.; Schmitt, C.; Prevost, M.C.; Balloy, V.; Fadel, E.; Philippe, B.; Kauffmann-Lacroix, C.; Latgé, J.P.; Beauvais, A. In vivo biofilm composition of Aspergillus fumigatus. Cell. Microbiol. 2010, 12, 405–410. [Google Scholar] [CrossRef]
- Schmalhorst, P.S.; Krappmann, S.; Vervecken, W.; Rohde, M.; Müller, M.; Braus, G.H.; Contreras, R.; Braun, A.; Bakker, H.; Routier, F.H. Contribution of galactofuranose to the virulence of the opportunistic pathogen Aspergillus fumigatus. Eukaryot. Cell 2008, 7, 1268–1277. [Google Scholar] [CrossRef] [PubMed]
- Engel, J.; Schmalhorst, P.S.; Dörk-Bousset, T.; Ferrières, V.; Routier, F.H. A single UDP-galactofuranose transporter is required for galactofuranosylation in Aspergillus fumigatus. J. Biol. Chem. 2009, 284, 33859–33868. [Google Scholar] [CrossRef] [PubMed]
- Engel, J.; Schmalhorst, P.S.; Routier, F.H. Biosynthesis of the fungal cell wall polysaccharide galactomannan requires intraluminal GDP-mannose. J. Biol. Chem. 2012, 287, 44418–44424. [Google Scholar] [CrossRef] [PubMed]
- Lee, D.J.; O’Donnell, H.; Routier, F.H.; Tiralongo, J.; Haselhorst, T. Glycobiology of Human Fungal Pathogens: New Avenues for Drug Development. Cells 2019, 8, 1348. [Google Scholar] [CrossRef]
- Oka, T. Biosynthesis of galactomannans found in filamentous fungi belonging to Pezizomycotina. Biosci. Biotechnol. Biochem. 2018, 82, 183–191. [Google Scholar] [CrossRef]
- Fontaine, T.; Latgé, J.P. Galactomannan Produced by Aspergillus fumigatus: An Update on the Structure, Biosynthesis and Biological Functions of an Emblematic Fungal Biomarker. J. Fungi 2020, 6, 283. [Google Scholar] [CrossRef]
- Kadooka, C.; Hira, D.; Tanaka, Y.; Miyazawa, K.; Bise, M.; Takatsuka, S.; Oka, T. Identification of an α-(1→6)-Mannosyltransferase Contributing To Biosynthesis of the Fungal-Type Galactomannan α-Core-Mannan Structure in Aspergillus fumigatus. mSphere 2022, 7, e0048422. [Google Scholar] [CrossRef]
- Lee, M.J.; Geller, A.M.; Bamford, N.C.; Liu, H.; Gravelat, F.N.; Snarr, B.D.; Le Mauff, F.; Chabot, J.; Ralph, B.; Ostapska, H.; et al. Deacetylation of Fungal Exopolysaccharide Mediates Adhesion and Biofilm Formation. mBio 2016, 7, e00252-16. [Google Scholar] [CrossRef]
- Lee, M.J.; Gravelat, F.N.; Cerone, R.P.; Baptista, S.D.; Campoli, P.V.; Choe, S.I.; Kravtsov, I.; Vinogradov, E.; Creuzenet, C.; Liu, H.; et al. Overlapping and distinct roles of Aspergillus fumigatus UDP-glucose 4-epimerases in galactose metabolism and the synthesis of galactose-containing cell wall polysaccharides. J. Biol. Chem. 2014, 289, 1243–1256. [Google Scholar] [CrossRef]
- Bamford, N.C.; Snarr, B.D.; Gravelat, F.N.; Little, D.J.; Lee, M.J.; Zacharias, C.A.; Chabot, J.C.; Geller, A.M.; Baptista, S.D.; Baker, P.; et al. Sph3 Is a Glycoside Hydrolase Required for the Biosynthesis of Galactosaminogalactan in Aspergillus fumigatus. J. Biol. Chem. 2015, 290, 27438–27450. [Google Scholar] [CrossRef] [PubMed]
- Bamford, N.C.; Le Mauff, F.; Subramanian, A.S.; Yip, P.; Millán, C.; Zhang, Y.; Zacharias, C.; Forman, A.; Nitz, M.; Codée, J.D.C.; et al. Ega3 from the fungal pathogen Aspergillus fumigatus is an endo-α-1,4-galactosaminidase that disrupts microbial biofilms. J. Biol. Chem. 2019, 294, 13833–13849. [Google Scholar] [CrossRef]
- Lee, M.J.; Liu, H.; Barker, B.M.; Snarr, B.D.; Gravelat, F.N.; Al Abdallah, Q.; Gavino, C.; Baistrocchi, S.R.; Ostapska, H.; Xiao, T.; et al. The Fungal Exopolysaccharide Galactosaminogalactan Mediates Virulence by Enhancing Resistance to Neutrophil Extracellular Traps. PLoS Pathog. 2015, 11, e1005187. [Google Scholar] [CrossRef] [PubMed]
- Speth, C.; Rambach, G.; Lass-Flörl, C.; Howell, P.L.; Sheppard, D.C. Galactosaminogalactan (GAG) and its multiple roles in Aspergillus pathogenesis. Virulence 2019, 10, 976–983. [Google Scholar] [CrossRef]
- Gravelat, F.N.; Beauvais, A.; Liu, H.; Lee, M.J.; Snarr, B.D.; Chen, D.; Xu, W.; Kravtsov, I.; Hoareau, C.M.; Vanier, G.; et al. Aspergillus galactosaminogalactan mediates adherence to host constituents and conceals hyphal β-glucan from the immune system. PLoS Pathog. 2013, 9, e1003575. [Google Scholar] [CrossRef] [PubMed]
- Chibba, A.; Poloczek, J.; Little, D.J.; Howell, P.L.; Nitz, M. Synthesis and evaluation of inhibitors of E. coli PgaB, a polysaccharide de-N-acetylase involved in biofilm formation. Org. Biomol. Chem. 2012, 10, 7103–7107. [Google Scholar] [CrossRef]
- Ariyakumaran, R.; Pokrovskaya, V.; Little, D.J.; Howell, P.L.; Nitz, M. Direct Staudinger-Phosphonite Reaction Provides Methylphosphonamidates as Inhibitors of CE4 De-N-acetylases. Chembiochem 2015, 16, 1350–1356. [Google Scholar] [CrossRef] [PubMed]
- Baumann, A.M.; Bakkers, M.J.; Buettner, F.F.; Hartmann, M.; Grove, M.; Langereis, M.A.; de Groot, R.J.; Mühlenhoff, M. 9-O-Acetylation of sialic acids is catalysed by CASD1 via a covalent acetyl-enzyme intermediate. Nat. Commun. 2015, 6, 7673. [Google Scholar] [CrossRef] [PubMed]
- Blair, D.E.; Schüttelkopf, A.W.; MacRae, J.I.; van Aalten, D.M. Structure and metal-dependent mechanism of peptidoglycan deacetylase, a streptococcal virulence factor. Proc. Natl. Acad. Sci. USA 2005, 102, 15429–15434. [Google Scholar] [CrossRef]
- Krappmann, S.; Sasse, C.; Braus, G.H. Gene targeting in Aspergillus fumigatus by homologous recombination is facilitated in a nonhomologous end- joining-deficient genetic background. Eukaryot. Cell 2006, 5, 212–215. [Google Scholar] [CrossRef]
- Bamford, N.C.; Le Mauff, F.; Van Loon, J.C.; Ostapska, H.; Snarr, B.D.; Zhang, Y.; Kitova, E.N.; Klassen, J.S.; Codée, J.D.C.; Sheppard, D.C.; et al. Structural and biochemical characterization of the exopolysaccharide deacetylase Agd3 required for Aspergillus fumigatus biofilm formation. Nat. Commun. 2020, 11, 2450. [Google Scholar] [CrossRef]
- Krieger, E.; Vriend, G. YASARA View-molecular graphics for all devices-from smartphones to workstations. Bioinformatics 2014, 30, 2981–2982. [Google Scholar] [CrossRef] [PubMed]
- Available online: https://pubchem.ncbi.nlm.nih.gov/compound/2019 (accessed on 19 December 2022).
- Sander, T.; Freyss, J.; von Korff, M.; Rufener, C. DataWarrior: An open-source program for chemistry aware data visualization and analysis. J. Chem. Inf. Model. 2015, 55, 460–473. [Google Scholar] [CrossRef] [PubMed]
- Jemel, S.; Guillot, J.; Kallel, K.; Botterel, F.; Dannaoui, E. Galleria mellonella for the Evaluation of Antifungal Efficacy against Medically Important Fungi, a Narrative Review. Microorganisms 2020, 8, 390. [Google Scholar] [CrossRef] [PubMed]
- Day, C.J.; Bailly, B.; Guillon, P.; Dirr, L.; Jen, F.E.; Spillings, B.L.; Mak, J.; von Itzstein, M.; Haselhorst, T.; Jennings, M.P. Multidisciplinary Approaches Identify Compounds that Bind to Human ACE2 or SARS-CoV-2 Spike Protein as Candidates to Block SARS-CoV-2-ACE2 Receptor Interactions. mBio 2021, 12, e03681-20. [Google Scholar] [CrossRef] [PubMed]
- Koo, H.; Allan, R.N.; Howlin, R.P.; Stoodley, P.; Hall-Stoodley, L. Targeting microbial biofilms: Current and prospective therapeutic strategies. Nat. Rev. Microbiol. 2017, 15, 740–755. [Google Scholar] [CrossRef] [PubMed]
- Le Mauff, F.; Bamford, N.C.; Alnabelseya, N.; Zhang, Y.; Baker, P.; Robinson, H.; Codée, J.D.C.; Howell, P.L.; Sheppard, D.C. Molecular mechanism of Aspergillus fumigatus biofilm disruption by fungal and bacterial glycoside hydrolases. J. Biol. Chem. 2019, 294, 10760–10772. [Google Scholar] [CrossRef] [PubMed]
- Ostapska, H.; Raju, D.; Lehoux, M.; Lacdao, I.; Gilbert, S.; Sivarajah, P.; Bamford, N.C.; Baker, P.; Nguyen, T.T.M.; Zacharias, C.A.; et al. Preclinical Evaluation of Recombinant Microbial Glycoside Hydrolases in the Prevention of Experimental Invasive Aspergillosis. mBio 2021, 12, e0244621. [Google Scholar] [CrossRef]
- Nazik, H.; Kotta-Loizou, I.; Sass, G.; Coutts, R.H.A.; Stevens, D.A. Virus Infection of Aspergillus fumigatus Compromises the Fungus in Intermicrobial Competition. Viruses 2021, 13, 686. [Google Scholar] [CrossRef]
- Colvin, K.M.; Alnabelseya, N.; Baker, P.; Whitney, J.C.; Howell, P.L.; Parsek, M.R. PelA deacetylase activity is required for Pel polysaccharide synthesis in Pseudomonas aeruginosa. J. Bacteriol. 2013, 195, 2329–2339. [Google Scholar] [CrossRef]
- Jennings, L.K.; Storek, K.M.; Ledvina, H.E.; Coulon, C.; Marmont, L.S.; Sadovskaya, I.; Secor, P.R.; Tseng, B.S.; Scian, M.; Filloux, A.; et al. Pel is a cationic exopolysaccharide that cross-links extracellular DNA in the Pseudomonas aeruginosa biofilm matrix. Proc. Natl. Acad. Sci. USA 2015, 112, 11353–11358. [Google Scholar] [CrossRef] [PubMed]
- Fontaine, T.; Delangle, A.; Simenel, C.; Coddeville, B.; van Vliet, S.J.; van Kooyk, Y.; Bozza, S.; Moretti, S.; Schwarz, F.; Trichot, C.; et al. Galactosaminogalactan, a new immunosuppressive polysaccharide of Aspergillus fumigatus. PLoS Pathog. 2011, 7, e1002372. [Google Scholar] [CrossRef] [PubMed]
- Mu, Y.Q.; Xie, T.T.; Zeng, H.; Chen, W.; Wan, C.X.; Zhang, L.L. Streptomyces-derived actinomycin D inhibits biofilm formation via downregulating ica locus and decreasing production of PIA in Staphylococcus epidermidis. J. Appl. Microbiol. 2020, 128, 1201–1207. [Google Scholar] [CrossRef] [PubMed]
- Hochhaus, A.; Larson, R.A.; Guilhot, F.; Radich, J.P.; Branford, S.; Hughes, T.P.; Baccarani, M.; Deininger, M.W.; Cervantes, F.; Fujihara, S.; et al. Long-Term Outcomes of Imatinib Treatment for Chronic Myeloid Leukemia. N. Engl. J. Med. 2017, 376, 917–927. [Google Scholar] [CrossRef]
- Peng, B.; Lloyd, P.; Schran, H. Clinical pharmacokinetics of imatinib. Clin. Pharmacokinet. 2005, 44, 879–894. [Google Scholar] [CrossRef]
- Reece, E.; Segurado, R.; Jackson, A.; McClean, S.; Renwick, J.; Greally, P. Co-colonisation with Aspergillus fumigatus and Pseudomonas aeruginosa is associated with poorer health in cystic fibrosis patients: An Irish registry analysis. BMC Pulm. Med. 2017, 17, 70. [Google Scholar] [CrossRef] [PubMed]
- Marmont, L.S.; Whitfield, G.B.; Rich, J.D.; Yip, P.; Giesbrecht, L.B.; Stremick, C.A.; Whitney, J.C.; Parsek, M.R.; Harrison, J.J.; Howell, P.L. PelA and PelB proteins form a modification and secretion complex essential for Pel polysaccharide-dependent biofilm formation in Pseudomonas aeruginosa. J. Biol. Chem. 2017, 292, 19411–19422. [Google Scholar] [CrossRef]
- Seegers, C.I.I.; Ramón Roth, I.; Zarnovican, P.; Buettner, F.F.R.; Routier, F.H. Characterization of a gene cluster involved in Aspergillus fumigatus zwitterionic glycosphingolipid synthesis. Glycobiology 2022, 32, 814–824. [Google Scholar] [CrossRef] [PubMed]
- Labun, K.; Montague, T.G.; Krause, M.; Torres Cleuren, Y.N.; Tjeldnes, H.; Valen, E. CHOPCHOP v3: Expanding the CRISPR web toolbox beyond genome editing. Nucleic Acids Res. 2019, 47, W171–W174. [Google Scholar] [CrossRef] [PubMed]
- Song, L.; Ouedraogo, J.P.; Kolbusz, M.; Nguyen, T.T.M.; Tsang, A. Efficient genome editing using tRNA promoter-driven CRISPR/Cas9 gRNA in Aspergillus niger. PLoS ONE 2018, 13, e0202868. [Google Scholar] [CrossRef]
- van Leeuwe, T.M.; Arentshorst, M.; Ernst, T.; Alazi, E.; Punt, P.J.; Ram, A.F.J. Efficient marker free CRISPR/Cas9 genome editing for functional analysis of gene families in filamentous fungi. Fungal Biol. Biotechnol. 2019, 6, 13. [Google Scholar] [CrossRef] [PubMed]
- Nødvig, C.S.; Nielsen, J.B.; Kogle, M.E.; Mortensen, U.H. A CRISPR-Cas9 System for Genetic Engineering of Filamentous Fungi. PLoS ONE 2015, 10, e0133085. [Google Scholar] [CrossRef] [PubMed]
- Punt, P.J.; van den Hondel, C.A. Transformation of filamentous fungi based on hygromycin B and phleomycin resistance markers. Methods Enzymol. 1992, 216, 447–457. [Google Scholar] [CrossRef] [PubMed]


| Name | Primer Sequence * | Description |
|---|---|---|
| CS27 | GTACGGTCTCGAATTCTCGATTGGCCTGTCCTCC | Forward agd3109–801 |
| CS29 | GCTATCTAGATGCAAAGCAATAGGAGTGGAAAGCG | Reverse agd3109–801 |
| FR294 | GACAGGTCTCAGATCTACTCCGCCGAACGTACTG | Forward CRISPR/CAS9 |
| FR297 | GACAGGTCTCCAAACTGGTGAAATACGGCCACATGGACGAGCTTACTCGTTTCGTCCTC | Reverse CRISPR/CAS9 |
| FR298 | GACAGGTCTCCAAACTCGTTACATCTGTGCCTCCAGACGAGCTTACTCGTTTCGTCCTC | Reverse CRISPR/CAS9 |
| FR299 | CATCGGATCCTTGGTTTGCCTT | Forward 5′UTR agd3 repair |
| FR300 | TGCAGTGTCAGGGAACAGAG | Reverse 5′UTR agd3 repair |
| FR301 | CTTGTCACCCTCTCTGTTCCCTGACACTGCACCGCCCTTATGCGGGTCG | Forward 3′UTR agd3 repair |
| FR302 | GCTAAAGCTTAGCGTGGATC | Reverse 3′UTR agd3 repair |
| CS14 | AGTTCATATGAACACCGGCGTGGAGCAG | Forward agd3 |
| CS37 | TCTAGATCACAAAGCAATAGGAGTGGAAAGCG | Reverse agd3 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Seegers, C.I.I.; Lee, D.J.; Zarnovican, P.; Kirsch, S.H.; Müller, R.; Haselhorst, T.; Routier, F.H. Identification of Compounds Preventing A. fumigatus Biofilm Formation by Inhibition of the Galactosaminogalactan Deacetylase Agd3. Int. J. Mol. Sci. 2023, 24, 1851. https://doi.org/10.3390/ijms24031851
Seegers CII, Lee DJ, Zarnovican P, Kirsch SH, Müller R, Haselhorst T, Routier FH. Identification of Compounds Preventing A. fumigatus Biofilm Formation by Inhibition of the Galactosaminogalactan Deacetylase Agd3. International Journal of Molecular Sciences. 2023; 24(3):1851. https://doi.org/10.3390/ijms24031851
Chicago/Turabian StyleSeegers, Carla I. I., Danielle J. Lee, Patricia Zarnovican, Susanne H. Kirsch, Rolf Müller, Thomas Haselhorst, and Françoise H. Routier. 2023. "Identification of Compounds Preventing A. fumigatus Biofilm Formation by Inhibition of the Galactosaminogalactan Deacetylase Agd3" International Journal of Molecular Sciences 24, no. 3: 1851. https://doi.org/10.3390/ijms24031851
APA StyleSeegers, C. I. I., Lee, D. J., Zarnovican, P., Kirsch, S. H., Müller, R., Haselhorst, T., & Routier, F. H. (2023). Identification of Compounds Preventing A. fumigatus Biofilm Formation by Inhibition of the Galactosaminogalactan Deacetylase Agd3. International Journal of Molecular Sciences, 24(3), 1851. https://doi.org/10.3390/ijms24031851






