BIRC5 Gene Polymorphisms Are Associated with a Higher Stage of Local and Regional Disease in Oral and Oropharyngeal Squamous Cell Carcinomas
Abstract
:1. Introduction
2. Results
2.1. Subject Data and Classification
2.2. BIRC5 Polymorphisms in Patients with HNSCC and Healthy Controls
2.3. Linkage Disequilibrium
2.4. Association of BIRC5 Polymorphisms with Clinicopathological Variables in HNSCC Patients
3. Discussion
4. Materials and Methods
4.1. Patients and Clinical Samples
4.2. SNP Selection and Genotyping
4.3. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Sung, H.; Ferlay, J.; Siegel, R.L.; Laversanne, M.; Soerjomataram, I.; Jemal, A.; Bray, F. Global Cancer Statistics 2020: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA Cancer J. Clin. 2021, 71, 209–249. [Google Scholar] [CrossRef] [PubMed]
- Bugshan, A.; Farooq, I. Oral Squamous Cell Carcinoma: Metastasis, Potentially Associated Malignant Disorders, Etiology and Recent Advancements in Diagnosis. F1000Research 2020, 9, 229. [Google Scholar] [CrossRef]
- Neville, B.W.; Damm, D.D.; Allen, C.M.; Bouquot, J.E. Oral and Maxillofacial Pathology, 3rd ed.; Saunders/Elsevier: St. Louis, MO, USA, 2009; ISBN 978-1-4377-2197-3. [Google Scholar]
- Chow, L.Q.M. Head and Neck Cancer. N. Engl. J. Med. 2020, 382, 60–72. [Google Scholar] [CrossRef] [PubMed]
- Mesia, R.; Iglesias, L.; Lambea, J.; Martínez-Trufero, J.; Soria, A.; Taberna, M.; Trigo, J.; Chaves, M.; García-Castaño, A.; Cruz, J. SEOM Clinical Guidelines for the Treatment of Head and Neck Cancer (2020). Clin. Transl. Oncol. 2021, 23, 913–921. [Google Scholar] [CrossRef] [PubMed]
- Panarese, I.; Aquino, G.; Ronchi, A.; Longo, F.; Montella, M.; Cozzolino, I.; Roccuzzo, G.; Colella, G.; Caraglia, M.; Franco, R. Oral and Oropharyngeal Squamous Cell Carcinoma: Prognostic and Predictive Parameters in the Etiopathogenetic Route. Expert Rev. Anticancer. Ther. 2019, 19, 105–119. [Google Scholar] [CrossRef] [PubMed]
- Fäldt Beding, A.; Larsson, P.; Helou, K.; Einbeigi, Z.; Parris, T.Z. Pan-Cancer Analysis Identifies BIRC5 as a Prognostic Biomarker. BMC Cancer 2022, 22, 322. [Google Scholar] [CrossRef]
- Wheatley, S.P.; McNeish, I.A. Survivin: A Protein with Dual Roles in Mitosis and Apoptosis. Int. Rev. Cytol. 2005, 247, 35–88. [Google Scholar] [CrossRef]
- Ambrosini, G.; Adida, C.; Altieri, D.C. A Novel Anti-Apoptosis Gene, Survivin, Expressed in Cancer and Lymphoma. Nat. Med. 1997, 3, 917–921. [Google Scholar] [CrossRef]
- Liu, S.; Shi, L.; Yang, X.; Ye, D.; Wang, T.; Dong, C.; Guo, W.; Liao, Y.; Song, H.; Xu, D.; et al. Nuclear Survivin Promoted by Acetylation Is Associated with the Aggressive Phenotype of Oral Squamous Cell Carcinoma. Cell Cycle 2017, 16, 894–902. [Google Scholar] [CrossRef]
- Yan, X.; Su, H. YM155 Down-Regulates Survivin and Induces P53 Up-Regulated Modulator of Apoptosis (PUMA)-Dependent in Oral Squamous Cell Carcinoma Cells. Med. Sci. Monit. 2017, 23, 1963–1972. [Google Scholar] [CrossRef] [PubMed]
- Yang, X.; Xiong, G.; Chen, X.; Xu, X.; Wang, K.; Fu, Y.; Yang, K.; Bai, Y. Polymorphisms of Survivin Promoter Are Associated with Risk of Esophageal Squamous Cell Carcinoma. J. Cancer Res. Clin. Oncol. 2009, 135, 1341–1349. [Google Scholar] [CrossRef]
- Pu, F.; Shao, Z.; Yang, S.; Liu, J.; Lin, S.; Ma, X.; Yang, H. Association between Functional Variants in BIRC5/Survivin Gene 3′ Untranslated Region and mRNA Expression in Lymphoblastoid Cell Lines. Oncol. Lett. 2015, 10, 2319–2322. [Google Scholar] [CrossRef] [PubMed]
- Jang, J.S.; Kim, K.M.; Kang, K.H.; Choi, J.E.; Lee, W.K.; Kim, C.H.; Kang, Y.M.; Kam, S.; Kim, I.-S.; Jun, J.E.; et al. Polymorphisms in the Survivin Gene and the Risk of Lung Cancer. Lung Cancer 2008, 60, 31–39. [Google Scholar] [CrossRef]
- Weng, C.J.; Hsieh, Y.H.; Chen, M.K.; Tsai, C.M.; Lin, C.W.; Yang, S.F. Survivin SNP-Carcinogen Interactions in Oral Cancer. J. Dent. Res. 2012, 91, 358–363. [Google Scholar] [CrossRef] [PubMed]
- Wang, N.; Li, Y.; Zhou, R.-M.; Cui, S.-J.; Cao, S.-R.; Huang, X.; Huo, X.-R.; Shan, B.-E. The Effect of Polymorphisms in the Promoter of the BIRC5 Gene on the Risk of Oesophageal Squamous Cell Carcinoma and Patient’s Outcomes. Mutagenesis 2019, 34, 307–313. [Google Scholar] [CrossRef] [PubMed]
- Antonacopoulou, A.G.; Floratou, K.; Bravou, V.; Kottorou, A.; Dimitrakopoulos, F.-I.; Marousi, S.; Stavropoulos, M.; Koutras, A.K.; Scopa, C.D.; Kalofonos, H.P. The Survivin-31 Snp in Human Colorectal Cancer Correlates with Survivin Splice Variant Expression and Improved Overall Survival. Anal. Cell. Pathol. 2010, 33, 177–189. [Google Scholar] [CrossRef]
- Dai, J.; Jin, G.; Dong, J.; Chen, Y.; Xu, L.; Hu, Z.; Shen, H. Prognostic Significance of Survivin Polymorphisms on Non-Small Cell Lung Cancer Survival. J. Thorac. Oncol. 2010, 5, 1748–1754. [Google Scholar] [CrossRef]
- Han, C.H.; Wei, Q.; Lu, K.K.; Liu, Z.; Mills, G.B.; Wang, L.-E. Polymorphisms in the Survivin Promoter Are Associated with Age of Onset of Ovarian Cancer. Int. J. Clin. Exp. Med. 2009, 2, 289–299. [Google Scholar]
- Sušac, I.; Ozretić, P.; Gregorić, M.; Levačić Cvok, M.; Sabol, M.; Levanat, S.; Trnski, D.; Eljuga, D.; Seiwerth, S.; Aralica, G.; et al. Polymorphisms in Survivin (BIRC5 Gene) Are Associated with Age of Onset in Breast Cancer Patients. J. Oncol. 2019, 2019, 3483192. [Google Scholar] [CrossRef]
- Trnski, D.; Gregorić, M.; Levanat, S.; Ozretić, P.; Rinčić, N.; Vidaković, T.M.; Kalafatić, D.; Maurac, I.; Orešković, S.; Sabol, M.; et al. Regulation of Survivin Isoform Expression by GLI Proteins in Ovarian Cancer. Cells 2019, 8, 128. [Google Scholar] [CrossRef]
- Jaiswal, P.K.; Goel, A.; Mandhani, A.; Mittal, R.D. Functional Polymorphisms in Promoter Survivin Gene and Its Association with Susceptibility to Bladder Cancer in North Indian Cohort. Mol. Biol. Rep. 2012, 39, 5615–5621. [Google Scholar] [CrossRef] [PubMed]
- Shi, H.; Bevier, M.; Johansson, R.; Enquist-Olsson, K.; Henriksson, R.; Hemminki, K.; Lenner, P.; Försti, A. Prognostic Impact of Polymorphisms in the MYBL2 Interacting Genes in Breast Cancer. Breast Cancer Res. Treat. 2012, 131, 1039–1047. [Google Scholar] [CrossRef] [PubMed]
- Lee, J.-S.; Kim, J.-H.; Park, B.-L.; Cheong, H.-S.; Kim, J.-Y.; Park, T.-J.; Chun, J.-Y.; Bae, J.-S.; Lee, H.-S.; Kim, Y.-J.; et al. Lack of Association of BIRC5 Polymorphisms with Clearance of HBV Infection and HCC Occurrence in a Korean Population. Genom. Inform. 2009, 7, 195–202. [Google Scholar] [CrossRef]
- Wagner, M.; Schmelz, K.; Dörken, B.; Tamm, I. Epigenetic and Genetic Analysis of the Survivin Promoter in Acute Myeloid Leukemia. Leuk. Res. 2008, 32, 1054–1060. [Google Scholar] [CrossRef]
- Boidot, R.; Végran, F.; Jacob, D.; Chevrier, S.; Cadouot, M.; Feron, O.; Solary, E.; Lizard-Nacol, S. The Transcription Factor GATA-1 Is Overexpressed in Breast Carcinomas and Contributes to Survivin Upregulation via a Promoter Polymorphism. Oncogene 2010, 29, 2577–2584. [Google Scholar] [CrossRef]
- Xu, Y.; Fang, F.; Ludewig, G.; Jones, G.; Jones, D. A Mutation Found in the Promoter Region of the Human Survivin Gene Is Correlated to Overexpression of Survivin in Cancer Cells. DNA Cell Biol. 2004, 23, 527–537. [Google Scholar] [CrossRef] [PubMed]
- Jaiswal, P.K.; Goel, A.; Mittal, R.D. Survivin: A Molecular Biomarker in Cancer. Indian J. Med. Res. 2015, 141, 389–397. [Google Scholar] [CrossRef] [PubMed]
- Moazeni-Roodi, A.; Ghavami, S.; Hashemi, M. Survivin Rs9904341 Polymorphism Significantly Increased the Risk of Cancer: Evidence from an Updated Meta-Analysis of Case-Control Studies. Int. J. Clin. Oncol. 2019, 24, 335–349. [Google Scholar] [CrossRef]
- Mazoochi, T.; Karimian, M.; Ehteram, H.; Karimian, A. Survivin c.-31G>C (Rs9904341) Gene Transversion and Urinary System Cancers Risk: A Systematic Review and a Meta-Analysis. Per. Med. 2019, 16, 67–78. [Google Scholar] [CrossRef]
- Xu, M.; Hu, X.; Zhang, M.; Ge, Y. What Is the Impact of BIRC5 Gene Polymorphisms on Urinary Cancer Susceptibility? Evidence from 9348 Subjects. Gene 2020, 733, 144268. [Google Scholar] [CrossRef]
- Mehdi, R.F.; Sheikh, F.; Khan, R.; Fawad, B.; Haq, A.U. Survivin Promoter Polymorphism (-31 C/G): A Genetic Risk Factor for Oral Cancer. Asian Pac. J. Cancer Prev. 2019, 20, 1289–1293. [Google Scholar] [CrossRef] [PubMed]
- Upadhyay, R.; Khurana, R.; Kumar, S.; Ghoshal, U.C.; Mittal, B. Role of Survivin Gene Promoter Polymorphism (-31G>C) in Susceptibility and Survival of Esophageal Cancer in Northern India. Ann. Surg. Oncol. 2011, 18, 880–887. [Google Scholar] [CrossRef] [PubMed]
- Fathy, W.; Amar, M.; Montaser, B.; Ahmed, M. Association between Survivin Gene Polymorphism and Colorectal Cancer. Menoufia Med. J. 2019, 32, 296–300. [Google Scholar] [CrossRef]
- Gazouli, M.; Tzanakis, N.; Rallis, G.; Theodoropoulos, G.; Papaconstantinou, I.; Kostakis, A.; Anagnou, N.P.; Nikiteas, N. Survivin -31G/C Promoter Polymorphism and Sporadic Colorectal Cancer. Int. J. Color. Dis. 2009, 24, 145–150. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.-H.; Chiou, H.-Y.; Lin, C.-T.; Hsieh, H.-Y.; Wu, C.-C.; Hsu, C.-D.; Shen, C.-H. Association Between Survivin Gene Promoter −31 C/G Polymorphism and Urothelial Carcinoma Risk in Taiwanese Population. Urology 2009, 73, 670–674. [Google Scholar] [CrossRef]
- Kawata, N.; Tsuchiya, N.; Horikawa, Y.; Inoue, T.; Tsuruta, H.; Maita, S.; Satoh, S.; Mitobe, Y.; Narita, S.; Habuchi, T. Two Survivin Polymorphisms Are Cooperatively Associated with Bladder Cancer Susceptibility. Int. J. Cancer 2011, 129, 1872–1880. [Google Scholar] [CrossRef]
- Qin, C.; Cao, Q.; Li, P.; Ju, X.; Wang, M.; Chen, J.; Wu, Y.; Meng, X.; Zhu, J.; Zhang, Z.; et al. Functional Promoter-31G>C Variant in Survivin Gene Is Associated with Risk and Progression of Renal Cell Cancer in a Chinese Population. PLoS ONE 2012, 7, e28829. [Google Scholar] [CrossRef]
- Bogdanovic, L.; Lazic, M.; Bogdanovic, J.; Soldatovic, I.; Nikolic, N.; Radunovic, M.; Radojevic-Skodric, S.; Milasin, J.; Basta-Jovanovic, G. Polymorphisms of Survivin -31 G/C Gene Are Associated with Risk of Urothelial Carcinoma in Serbian Population. J. BUON 2017, 22, 270–277. [Google Scholar] [PubMed]
- Radojevic-Skodric, S.; Basta-Jovanovic, G.; Brasanac, D.; Nikolic, N.; Bogdanovic, L.; Milicic, B.; Milasin, J. Survivin Gene Promoter -31 G/C Polymorphism Is Associated with Wilms Tumor Susceptibility in Serbian Children. J. Pediatr. Hematol. Oncol. 2012, 34, e310–e314. [Google Scholar] [CrossRef]
- Szczyrek, M.; Mlak, R.; Szudy-Szczyrek, A.; Wojas-Krawczyk, K.; Kędziora, K.; Milanowski, J. Polymorphism of Baculoviral Inhibitor of Apoptosis Repeat-Containing 5 (BIRC5) Can Be Associated with Clinical Outcome of Non-Small Cell Lung Cancer. Cells 2022, 11, 956. [Google Scholar] [CrossRef] [PubMed]
- Karimian, M.; Aftabi, Y.; Mazoochi, T.; Babaei, F.; Khamechian, T.; Boojari, H.; Nikzad, H. Survivin Polymorphisms and Susceptibility to Prostate Cancer: A Genetic Association Study and an in Silico Analysis. EXCLI J. 2018, 17, 479–491. [Google Scholar] [CrossRef] [PubMed]
- Cacına, C.; Akgün, A.; Kayhan, K.B.; Yaylım, İ.; Çakmakoğlu, B. The Analysis of Survivin Promoter (-31G/C) Gene Variation in Oral Squamous Cell Carcinoma Risk and Prognosis. J. Stomatol. Oral Maxillofac. Surg. 2023, 124, 101494. [Google Scholar] [CrossRef] [PubMed]
- Mostaan, L.V.; Tabari, A.; Amiri, P.; Ashtiani, M.K.; Mahdkhah, A.; Yazdani, N.; Khaniki, M.; Tabari, A.; Tavakkoly-Bazzaz, J.; Amoli, M.M. Survivin Gene Polymorphism Association with Tongue Squamous Cell Carcinoma. Genet. Test. Mol. Biomark. 2013, 17, 74–77. [Google Scholar] [CrossRef] [PubMed]
- Theodoropoulos, G.E.; Michalopoulos, N.V.; Panoussopoulos, S.-G.; Taka, S.; Gazouli, M. Effects of Caspase-9 and Survivin Gene Polymorphisms in Pancreatic Cancer Risk and Tumor Characteristics. Pancreas 2010, 39, 976–980. [Google Scholar] [CrossRef] [PubMed]
- Liu, T.-C.; Hsieh, M.-J.; Wu, W.-J.; Chou, Y.-E.; Chiang, W.-L.; Yang, S.-F.; Su, S.-C.; Tsao, T.C.-Y. Association between Survivin Genetic Polymorphisms and Epidermal Growth Factor Receptor Mutation in Non-Small-Cell Lung Cancer. Int. J. Med. Sci. 2016, 13, 929–935. [Google Scholar] [CrossRef] [PubMed]
- Ma, F.; Zhang, H.; Zhai, Y.; Huang, W.; Zhao, C.; Ou, S.; Zhou, H.; Yuan, W.; Wang, Z.; Wang, H.; et al. Functional Polymorphism -31C/G in the Promoter of BIRC5 Gene and Risk of Nasopharyngeal Carcinoma among Chinese. PLoS ONE 2011, 6, e16748. [Google Scholar] [CrossRef]
- Aynaci, E.; Coskunpinar, E.; Eren, A.; Kum, O.; Oltulu, Y.M.; Akkaya, N.; Turna, A.; Yaylim, I.; Yildiz, P. Association between Survivin Gene Promoter -31G/C and -644C/T Polymorphisms and Non-Small Cell Lung Cancer. Genet. Mol. Res. 2013, 12, 3975–3982. [Google Scholar] [CrossRef]
- Kostić, M.; Nikolić, N.; Ilić, B.; Carkić, J.; Milenković, S.; Vukadinović, M. Analysis of Polymorphism in the Survivin Gene Promoter as a Potential Risk Factor for Head and Neck Cancers Development. Srp. Arh. Celok. Lek. 2013, 141, 304–307. [Google Scholar] [CrossRef]
- Hmeljak, J.; Erčulj, N.; Dolžan, V.; Kern, I.; Cör, A. BIRC5 Promoter SNPs Do Not Affect Nuclear Survivin Expression and Survival of Malignant Pleural Mesothelioma Patients. J. Cancer Res. Clin. Oncol. 2011, 137, 1641–1651. [Google Scholar] [CrossRef]
- Wang, X.; Huang, L.; Xu, Y.; Shi, Z.; Wang, Y.; Zhang, J.; Wang, X.; Cao, L.; Luo, H.; Chen, J.; et al. Association between Survivin -31G>C Promoter Polymorphism and Cancer Risk: A Meta-Analysis. Eur. J. Hum. Genet. 2012, 20, 790–795. [Google Scholar] [CrossRef]
- Kabisch, M.; Lorenzo Bermejo, J.; Dünnebier, T.; Ying, S.; Michailidou, K.; Bolla, M.K.; Wang, Q.; Dennis, J.; Shah, M.; Perkins, B.J.; et al. Inherited Variants in the Inner Centromere Protein (INCENP) Gene of the Chromosomal Passenger Complex Contribute to the Susceptibility of ER-Negative Breast Cancer. Carcinogenesis 2015, 36, 256–271. [Google Scholar] [CrossRef] [PubMed]
- Zu, Y.; Ban, J.; Xia, Z.; Wang, J.; Cai, Y.; Ping, W.; Sun, W. Genetic Variation in a miR-335 Binding Site in BIRC5 Alters Susceptibility to Lung Cancer in Chinese Han Populations. Biochem. Biophys. Res. Commun. 2013, 430, 529–534. [Google Scholar] [CrossRef] [PubMed]
- Leovic, D.; Sabol, M.; Ozretic, P.; Musani, V.; Car, D.; Marjanovic, K.; Zubcic, V.; Sabol, I.; Sikora, M.; Grce, M.; et al. Hh-Gli Signaling Pathway Activity in Oral and Oropharyngeal Squamous Cell Carcinoma. Head Neck 2012, 34, 104–112. [Google Scholar] [CrossRef] [PubMed]
- Cvok, M.L.; Cretnik, M.; Musani, V.; Ozretic, P.; Levanat, S. New Sequence Variants in BRCA1 and BRCA2 Genes Detected by High-Resolution Melting Analysis in an Elderly Healthy Female Population in Croatia. Clin. Chem. Lab. Med. 2008, 46, 1376–1383. [Google Scholar] [CrossRef] [PubMed]
- Shen, J.; Li, Z.; Chen, J.; Song, Z.; Zhou, Z.; Shi, Y. SHEsisPlus, a Toolset for Genetic Studies on Polyploid Species. Sci. Rep. 2016, 6, 24095. [Google Scholar] [CrossRef]
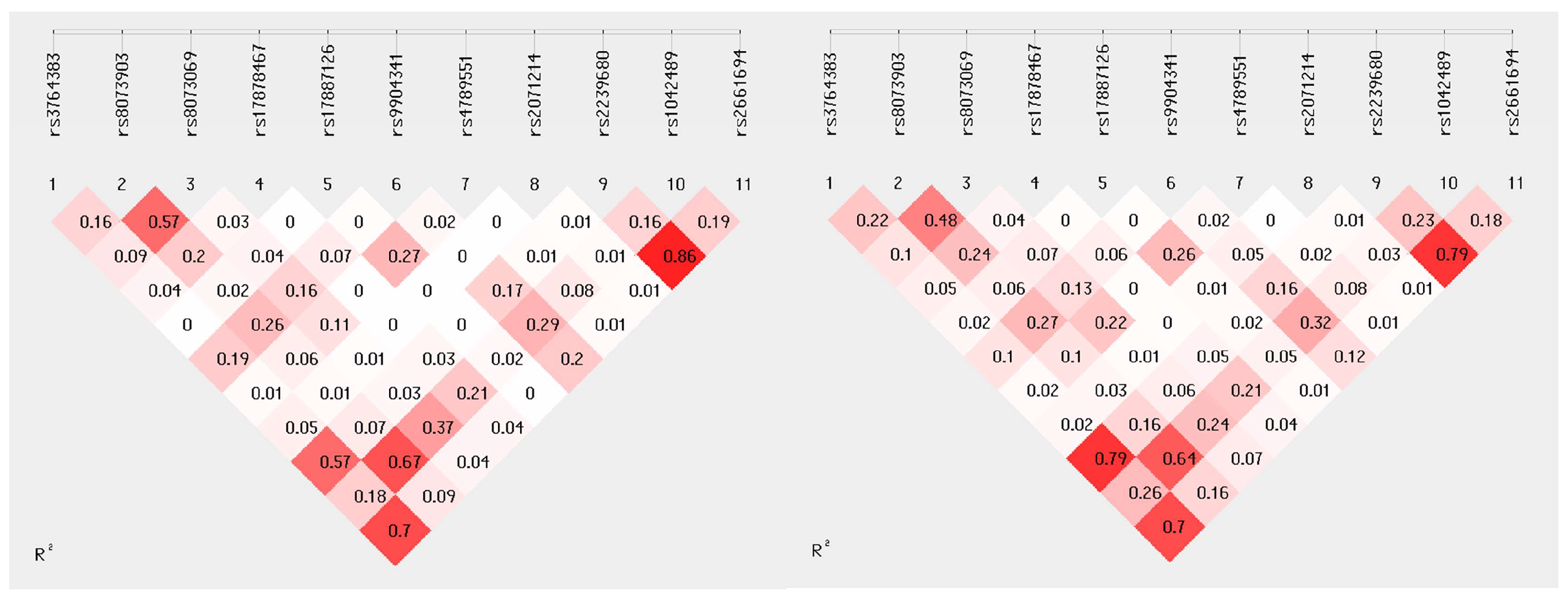
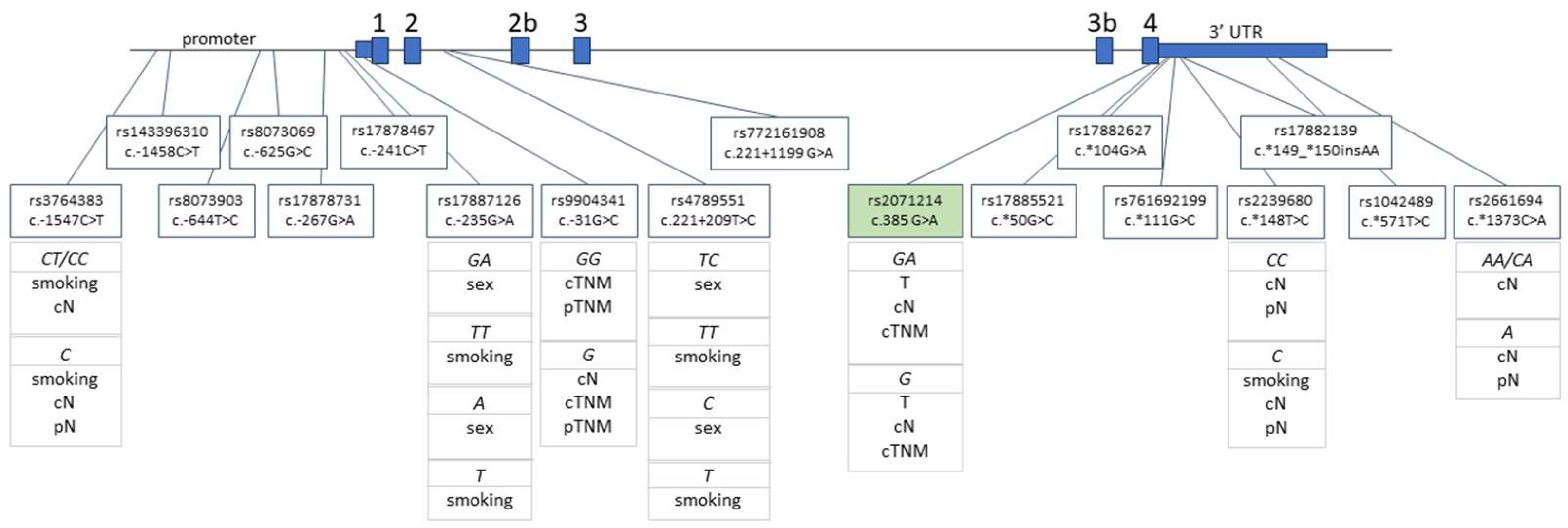
| Characteristic | No. of Patients (%) |
|---|---|
| Age, years | |
| median (range) | 59 (39–78) |
| Sex | |
| male | 41 (85.4%) |
| female | 7 (14.6%) |
| Tobacco consumption | |
| no | 6 (12.5%) |
| yes | 42 (87.5%) |
| Type | |
| oral | 31 (64.6%) |
| oropharyngeal | 17 (35.4%) |
| T | |
| 1 | 3 (6.2%) |
| 2 | 19 (39.6%) |
| 3 | 11 (22.9%) |
| 4 | 15 (31.2%) |
| cN | |
| 0 | 20 (41.7%) |
| 1 | 11 (22.9%) |
| 2 | 15 (31.2%) |
| 3 | 2 (4.2%) |
| cTNM | |
| I | 2 (4.2%) |
| II | 10 (20.8%) |
| III | 11 (22.9%) |
| IV | 25 (52.1%) |
| pN | |
| 0 | 16 (33.3%) |
| 1 | 5 (10.4%) |
| 2 | 25 (52.1%) |
| 3 | 2 (4.2%) |
| pTNM | |
| I | 3 (6.2%) |
| II | 7 (14.6%) |
| III | 7 (14.6%) |
| IV | 31 (64.6%) |
| Broders | |
| 1 | 19 (39.6%) |
| 2 | 22 (45.8%) |
| 3 | 5 (10.4%) |
| 4 | 2 (4.2%) |
| Survival | |
| alive | 8 (16.7%) |
| deceased | 35 (72.9%) |
| N.A. | 5 (10.4%) |
| Gene Region | SNP ID Number | Nucleotide Change | Minor Allele Frequency Controls (n/N, %) | Minor Allele Frequency OSCC Cases (n/N, %) | p-Value (for Genotype Frequencies) | p-Value (for Allele Frequencies) |
|---|---|---|---|---|---|---|
| promoter | rs3764383 | c.-1547C>T * | 37/148 (25.0) | 26/92 (28.3) | 0.327 | 0.651 |
| promoter | rs143396310 | c.-1458C>T | 2/148 (1.3) | 0/92 (0.0) | 0.523 | 0.525 |
| promoter | rs8073903 | c.-644T>C | 49/148 (33.1) | 35/96 (36.5) | 0.548 | 0.679 |
| promoter | rs8073069 | c.-625G>C | 33/148 (22.3) | 21/96 (21.9) | 0.977 | 1.000 |
| promoter | rs17878731 | c.-267G>A | 1/148 (0.7) | 0/96 (0.0) | 1.000 | 1.000 |
| promoter | rs17878467 | c.-241C>T | 16/148 (10.8) | 12/96 (12.5) | 0.666 | 0.686 |
| promoter | rs17887126 | c.-235G>A | 2/148 (1.4) | 6/96 (6.3) | 0.056 | 0.060 |
| 5′UTR | rs9904341 | c.-31G>C | 55/148 (37.2) | 30/92 (32.6) | 0.581 | 0.491 |
| intron 2 | rs4789551 | c.221+209T>C | 7/148 (4.7) | 5/92 (5.4) | 1.000 | 0.772 |
| intron 2 | rs772161908 | c.221+1199G>A | 0/148 (0.0) | 1/96 (1.0) | 0.393 | 0.393 |
| exon 4 | rs2071214 | c.385G>A ** (c.9194G>A) | 5/148 (3.4) | 5/96 (5.2) | 0.513 | 0.521 |
| 3′UTR | rs17885521 | c.*50G>C (c.9288G>C) | 3/148 (2.0) | 1/96 (1.0) | 1.000 | 1.000 |
| 3′UTR | rs17882627 | c.*104G>A (c.9342G>A) | 2/148 (1.3) | 0/96 (0.0) | 0.519 | 0.522 |
| 3′UTR | rs761692199 | c.*111G>C, (c.9349G>C) | 0/148 (0.0) | 1/96 (1.0) | 0.393 | 0.393 |
| 3′UTR | rs2239680 | c.*148T>C (c.9386T>C) | 34/148 (23.0) | 25/96 (26.0) | 0.824 | 0.647 |
| 3′UTR | rs17882139 | c.*149_*150insAA, (c.9387_9388insAA) | 3/148 (3.8) | 1/96 (1.0) | 1.000 | 1.000 |
| 3′UTR | rs1042489 | c.*571T>C (c.9809T>C) | 53/148 (35.8) | 38/96 (39.6) | 0.642 | 0.589 |
| 3′UTR | rs2661694 | c.*1373C>A (c.10611C>A) | 38/148 (25.7) | 21/96 (21.9) | 0.642 | 0.543 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Mumlek, I.; Ozretić, P.; Sabol, M.; Leović, M.; Glavaš-Obrovac, L.; Leović, D.; Musani, V. BIRC5 Gene Polymorphisms Are Associated with a Higher Stage of Local and Regional Disease in Oral and Oropharyngeal Squamous Cell Carcinomas. Int. J. Mol. Sci. 2023, 24, 17490. https://doi.org/10.3390/ijms242417490
Mumlek I, Ozretić P, Sabol M, Leović M, Glavaš-Obrovac L, Leović D, Musani V. BIRC5 Gene Polymorphisms Are Associated with a Higher Stage of Local and Regional Disease in Oral and Oropharyngeal Squamous Cell Carcinomas. International Journal of Molecular Sciences. 2023; 24(24):17490. https://doi.org/10.3390/ijms242417490
Chicago/Turabian StyleMumlek, Ivan, Petar Ozretić, Maja Sabol, Matko Leović, Ljubica Glavaš-Obrovac, Dinko Leović, and Vesna Musani. 2023. "BIRC5 Gene Polymorphisms Are Associated with a Higher Stage of Local and Regional Disease in Oral and Oropharyngeal Squamous Cell Carcinomas" International Journal of Molecular Sciences 24, no. 24: 17490. https://doi.org/10.3390/ijms242417490
APA StyleMumlek, I., Ozretić, P., Sabol, M., Leović, M., Glavaš-Obrovac, L., Leović, D., & Musani, V. (2023). BIRC5 Gene Polymorphisms Are Associated with a Higher Stage of Local and Regional Disease in Oral and Oropharyngeal Squamous Cell Carcinomas. International Journal of Molecular Sciences, 24(24), 17490. https://doi.org/10.3390/ijms242417490










