Computational Exploration of Minimum Energy Reaction Pathway of N2O Formation from Intermediate I of P450nor Using an Active Center Model
Abstract
:1. Introduction
2. Results and Discussion
2.1. Assessment of the Validity of Computational Model for the N2O Gereration from the Intermediate I of P450nor
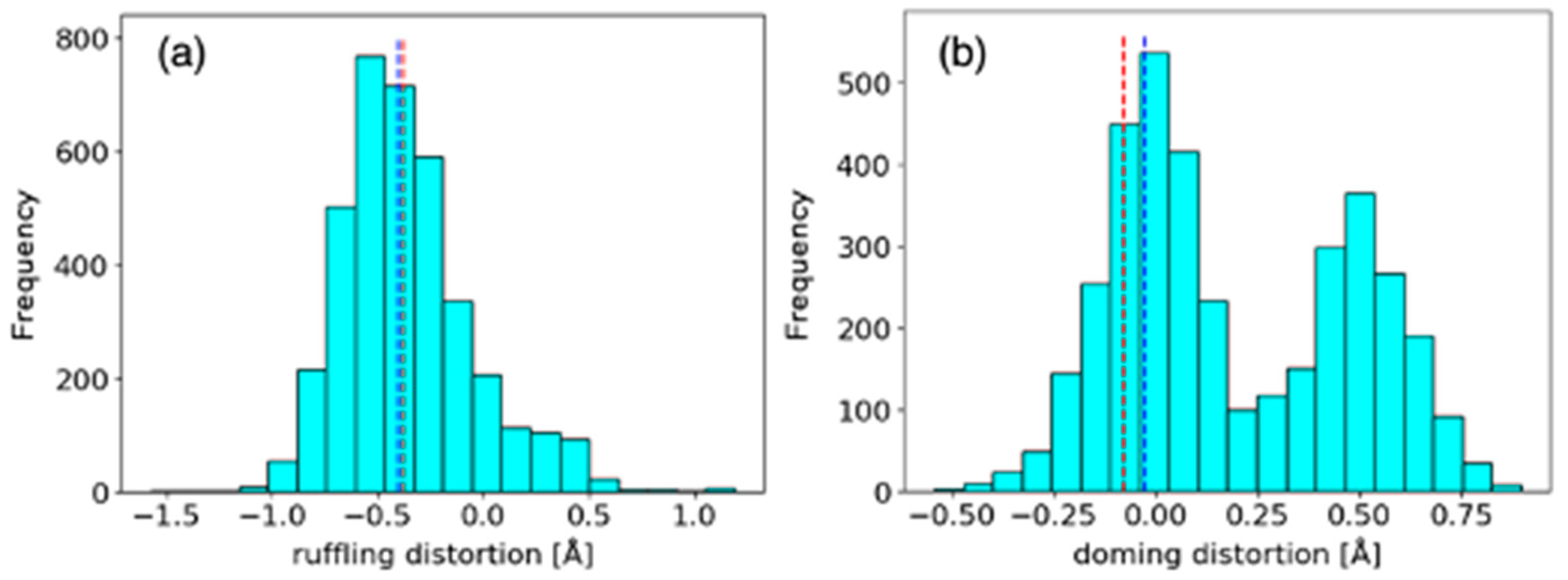
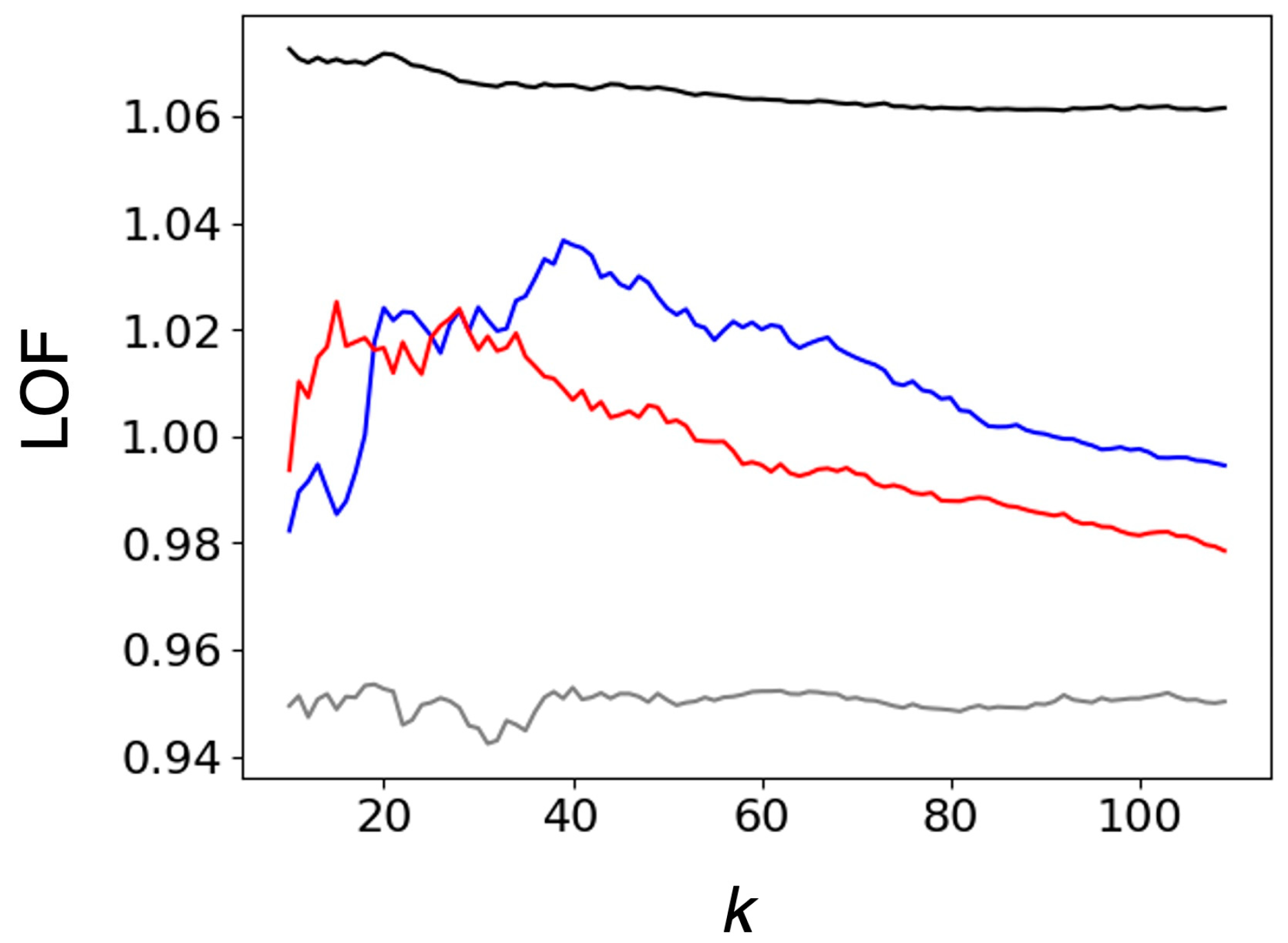
2.1.1. Comparison of Heme Distortions Using the Heme Database, PyDISH
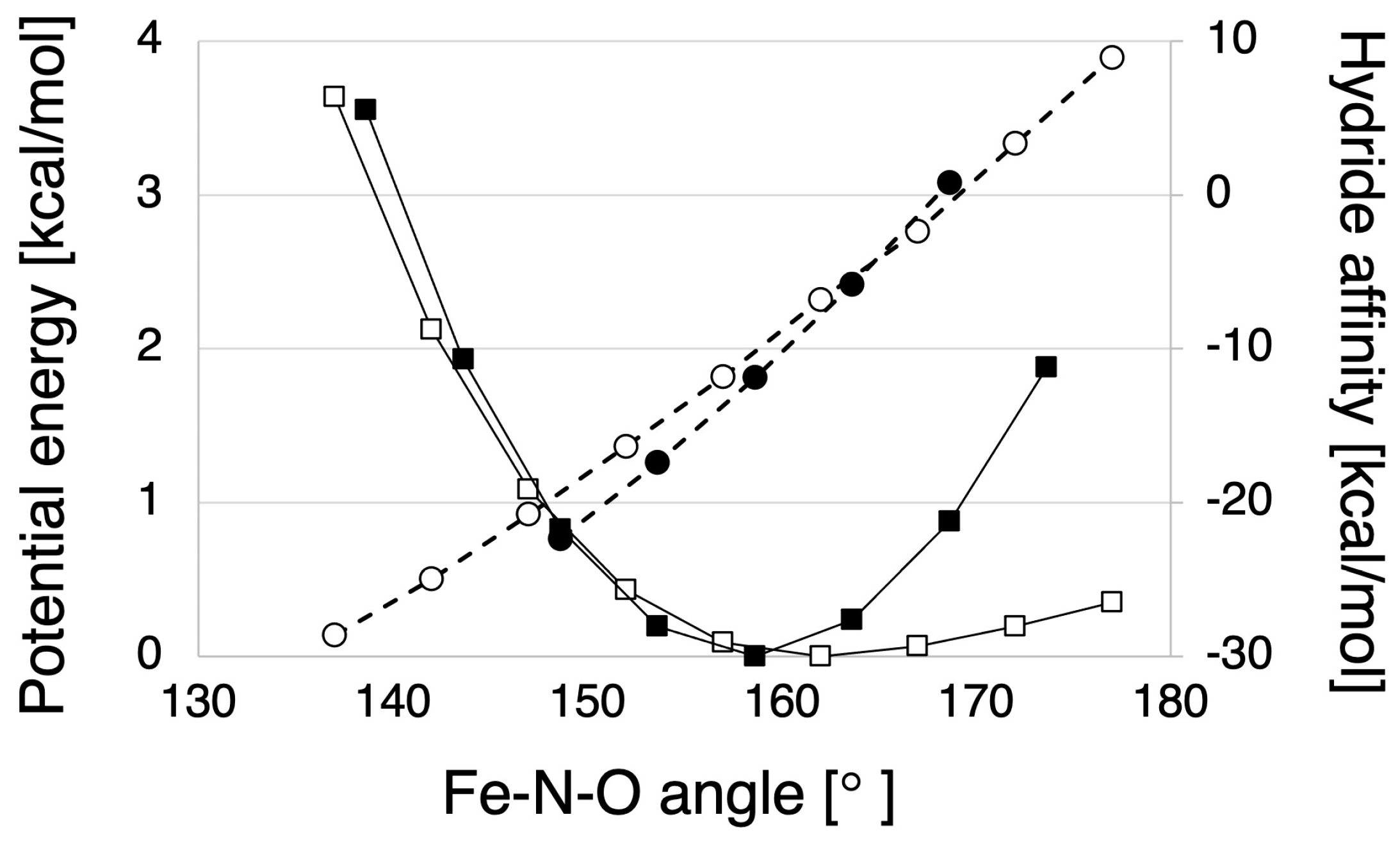
2.1.2. Hydride Affinity of the NO-Bound State with or without the Protein Environment of P450nor
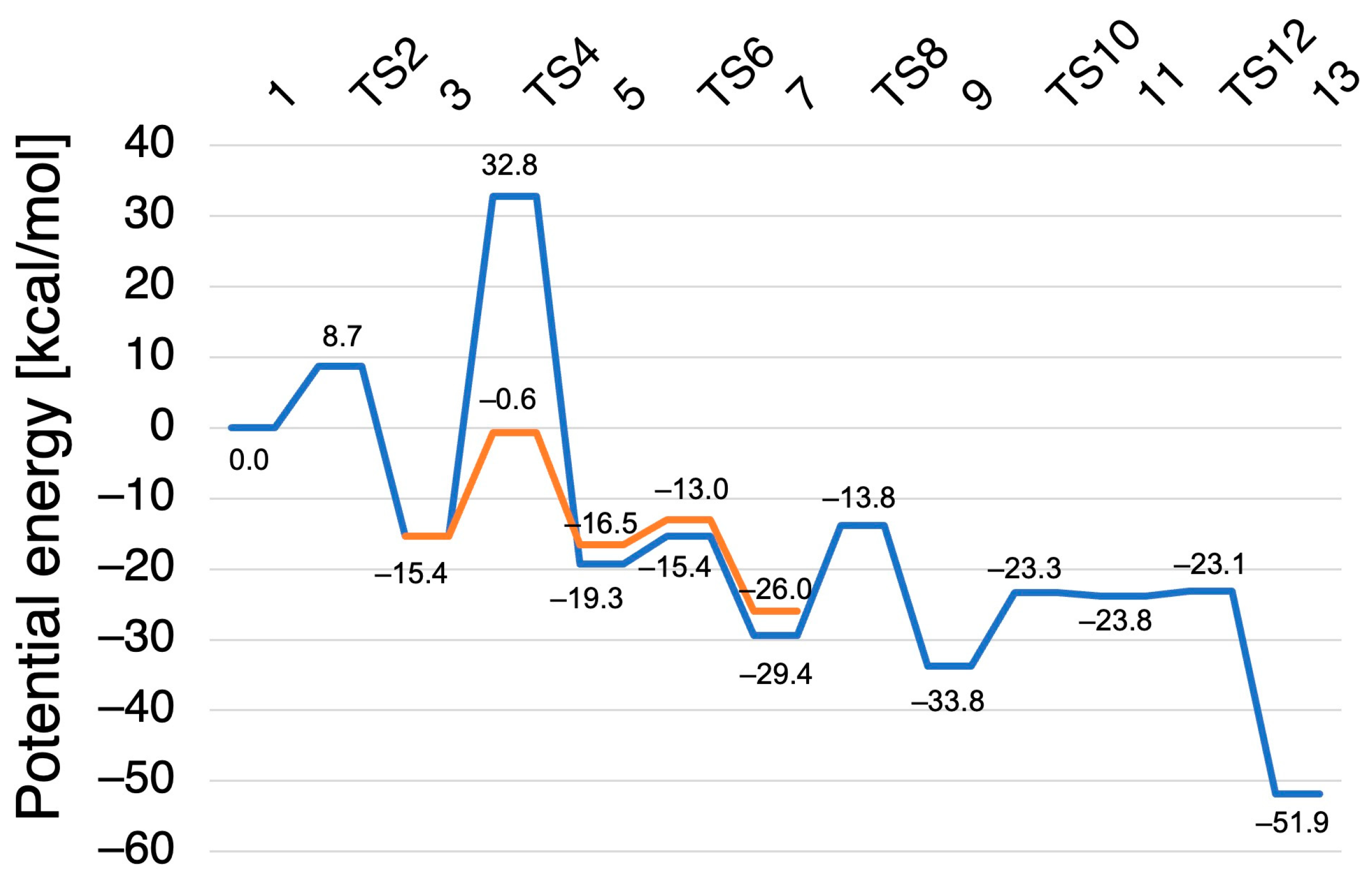
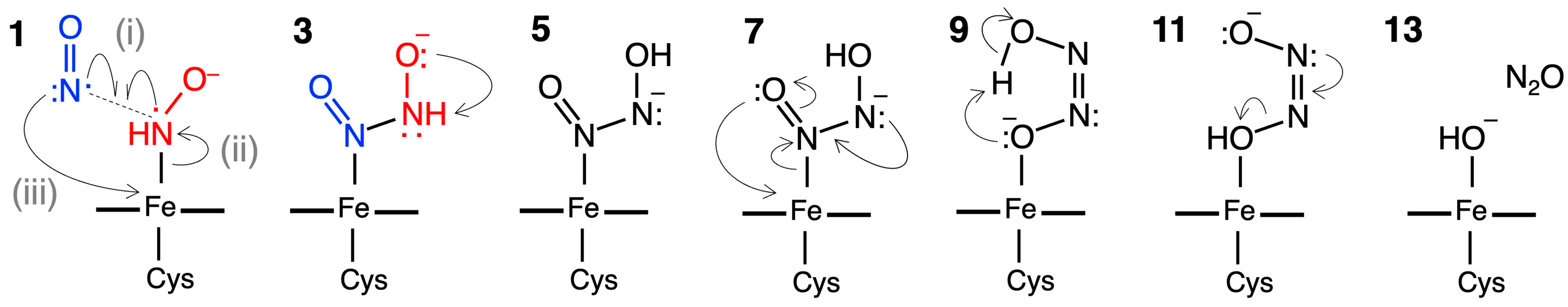
2.2. Computed Reaction Mechanism of the N2O Formation from the Intermediate I of P450nor
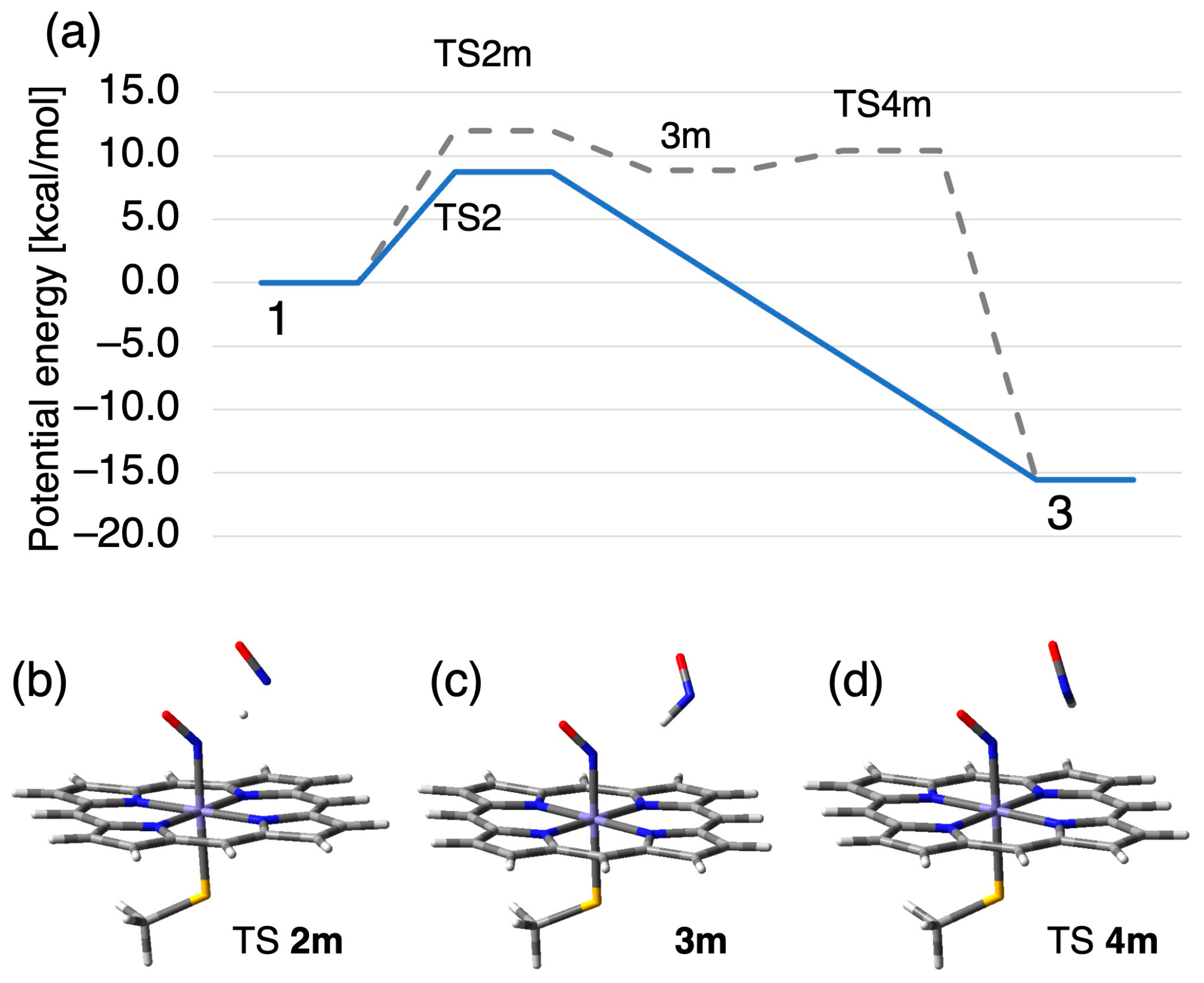
2.2.1. N–N Bond Formation Followed by Barrierless Rearrangement Step
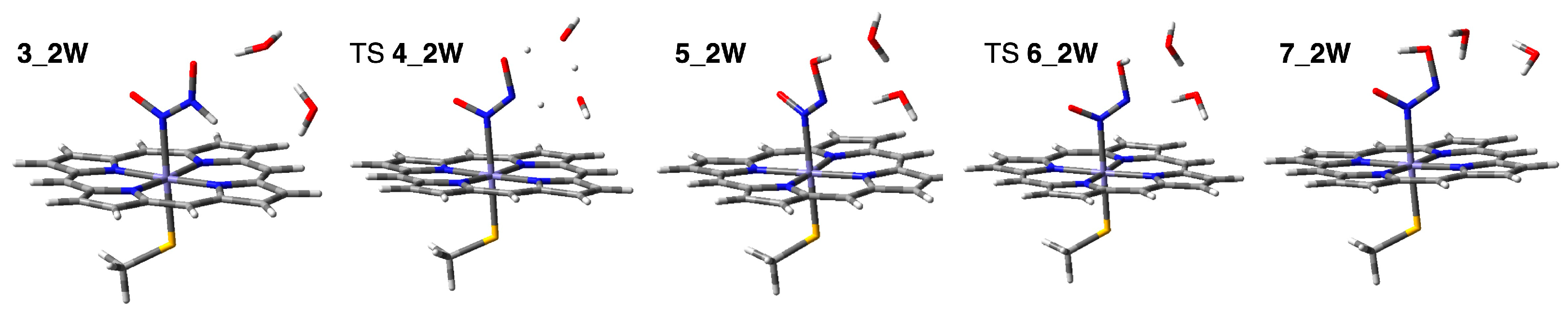
2.2.2. Water-Mediated Proton Transfer Step
2.2.3. Rearrangement of the Fe-ONNHO Species
2.2.4. O–N Bond Dissociation Step
3. Materials and Methods
3.1. Comparison of the Active Site Structure of P450nor to Those of the CYP Family with Heme Protein Database, PyDISH
3.2. QM/MM Calculation for Hydride Affinity Evaluation
3.3. Quantum Chemical Exploration of a Reaction Path of the N2O Generation
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Zumft, W.G. Cell Biology and Molecular Basis of Denitrification. Microbiol. Mol. Biol. Rev. 1997, 61, 533–616. [Google Scholar] [CrossRef]
- Joyner, M.J.; Dietz, N.M. Nitric Oxide and Vasodilation in Human Limbs. J. Appl. Physiol. 1997, 83, 1785–1796. [Google Scholar] [CrossRef] [PubMed]
- Traylor, T.G.; Sharma, V.S. Why Nitric Oxide? Biochemistry 1992, 31, 2847–2849. [Google Scholar] [CrossRef] [PubMed]
- Stamler, J.S.; Singel, D.J.; Loscalzo, J. Biochemistry of Nitric Oxide and Its Redox-Activated Forms. Science 1992, 258, 1898–1902. [Google Scholar] [CrossRef]
- Koshland, D.E. The Molecule of the Year. Science 1992, 258, 1861. [Google Scholar] [CrossRef] [PubMed]
- Shiro, Y. Structure and Function of Bacterial Nitric Oxide Reductases. Biochim. Biophys. Acta BBA-Bioenerg. 2012, 1817, 1907–1913. [Google Scholar] [CrossRef] [PubMed]
- Tosha, T.; Yamagiwa, R.; Sawai, H.; Shiro, Y. NO Dynamics in Microbial Denitrification System. Chem. Lett. 2021, 50, 280–288. [Google Scholar] [CrossRef]
- Ravishankara, A.R.; Daniel, J.S.; Portmann, R.W. Nitrous Oxide (N2O): The Dominant Ozone-Depleting Substance Emitted in the 21st Century. Science 2009, 326, 123–125. [Google Scholar] [CrossRef]
- Takeda, H.; Kimura, T.; Nomura, T.; Horitani, M.; Yokota, A.; Matsubayashi, A.; Ishii, S.; Shiro, Y.; Kubo, M.; Tosha, T. Timing of NO Binding and Protonation in the Catalytic Reaction of Bacterial Nitric Oxide Reductase as Established by Time-Resolved Spectroscopy. Bull. Chem. Soc. Jpn. 2020, 93, 825–833. [Google Scholar] [CrossRef]
- Girsch, P.; De Vries, S. Purification and Initial Kinetic and Spectroscopic Characterization of NO Reductase from Paracoccus Denitrificans. Biochim. Biophys. Acta BBA-Bioenerg. 1997, 1318, 202–216. [Google Scholar] [CrossRef]
- Cheesman, M.R.; Zumft, W.G.; Thomson, A.J. The MCD and EPR of the Heme Centers of Nitric Oxide Reductase from Pseudomonas Stutzeri: Evidence That the Enzyme Is Structurally Related to the Heme-Copper Oxidases. Biochemistry 1998, 37, 3994–4000. [Google Scholar] [CrossRef] [PubMed]
- Shoun, H.; Sudo, Y.; Seto, Y.; Beppu, T. Purification and Properties of a Cyotchrome P-450 of a Fungus, Fusarium Oxysporum. J. Biochem. 1983, 94, 1219–1229. [Google Scholar] [CrossRef] [PubMed]
- Nakahara, K.; Tanimoto, T.; Hatano, K.; Usuda, K.; Shoun, H. Cytochrome P-450 55A1 (P-450dNIR) Acts as Nitric Oxide Reductase Employing NADH as the Direct Electron Donor. J. Biol. Chem. 1993, 268, 8350–8355. [Google Scholar] [CrossRef]
- Hino, T.; Matsumoto, Y.; Nagano, S.; Sugimoto, H.; Fukumori, Y.; Murata, T.; Iwata, S.; Shiro, Y. Structural Basis of Biological N2O Generation by Bacterial Nitric Oxide Reductase. Science 2010, 330, 1666–1670. [Google Scholar] [CrossRef] [PubMed]
- Kumita, H.; Matsuura, K.; Hino, T.; Takahashi, S.; Hori, H.; Fukumori, Y.; Morishima, I.; Shiro, Y. NO Reduction by Nitric-Oxide Reductase from Denitrifying Bacterium Pseudomonas Aeruginosa. J. Biol. Chem. 2004, 279, 55247–55254. [Google Scholar] [CrossRef]
- Shoji, M.; Hanaoka, K.; Kondo, D.; Sato, A.; Umeda, H.; Kamiya, K.; Shiraishi, K. A QM/MM Study of Nitric Oxide Reductase-Catalysed N2O Formation. Mol. Phys. 2014, 112, 393–397. [Google Scholar] [CrossRef]
- Kizawa, H.; Tomura, D.; Oda, M.; Fukamizu, A.; Hoshino, T.; Gotoh, O.; Yasui, T.; Shoun, H. Nucleotide Sequence of the Unique Nitrate/Nitrite-Inducible Cytochrome P-450 cDNA from Fusarium Oxysporum. J. Biol. Chem. 1991, 266, 10632–10637. [Google Scholar] [CrossRef] [PubMed]
- Shoun, H.; Fushinobu, S.; Jiang, L.; Kim, S.-W.; Wakagi, T. Fungal Denitrification and Nitric Oxide Reductase Cytochrome P450nor. Philos. Trans. R. Soc. B Biol. Sci. 2012, 367, 1186–1194. [Google Scholar] [CrossRef]
- Shiro, Y.; Kato, M.; Iizuka, T.; Nakahara, K.; Shoun, H. Kinetics and Thermodynamics of CO Binding to Cytochrome P450nor. Biochemistry 1994, 33, 8673–8677. [Google Scholar] [CrossRef]
- Ortiz de Montellano, P.R. (Ed.) Cytochrome P450: Structure, Mechanism, and Biochemistry, 2nd ed.; Springer: New York, NY, USA, 1995; ISBN 978-1-4757-2391-5. [Google Scholar]
- Shiro, Y.; Fujii, M.; Iizuka, T.; Adachi, S.; Tsukamoto, K.; Nakahara, K.; Shoun, H. Spectroscopic and Kinetic Studies on Reaction of Cytochrome P450nor with Nitric Oxide. J. Biol. Chem. 1995, 270, 1617–1623. [Google Scholar] [CrossRef]
- Obayashi, E.; Tsukamoto, K.; Adachi, S.; Takahashi, S.; Nomura, M.; Iizuka, T.; Shoun, H.; Shiro, Y. Unique Binding of Nitric Oxide to Ferric Nitric Oxide Reductase from Fusarium Oxysporum Elucidated with Infrared, Resonance Raman, and X-Ray Absorption Spectroscopies. J. Am. Chem. Soc. 1997, 119, 7807–7816. [Google Scholar] [CrossRef]
- Tosha, T.; Nomura, T.; Nishida, T.; Saeki, N.; Okubayashi, K.; Yamagiwa, R.; Sugahara, M.; Nakane, T.; Yamashita, K.; Hirata, K.; et al. Capturing an Initial Intermediate during the P450nor Enzymatic Reaction Using Time-Resolved XFEL Crystallography and Caged-Substrate. Nat. Commun. 2017, 8, 1585. [Google Scholar] [CrossRef] [PubMed]
- Daiber, A.; Nauser, T.; Takaya, N.; Kudo, T.; Weber, P.; Hultschig, C.; Shoun, H.; Ullrich, V. Isotope Effects and Intermediates in the Reduction of NO by P450NOR. J. Inorg. Biochem. 2002, 88, 343–352. [Google Scholar] [CrossRef] [PubMed]
- Lehnert, N.; Praneeth, V.K.K.; Paulat, F. Electronic Structure of Iron(II)–Porphyrin Nitroxyl Complexes: Molecular Mechanism of Fungal Nitric Oxide Reductase (P450nor). J. Comput. Chem. 2006, 27, 1338–1351. [Google Scholar] [CrossRef] [PubMed]
- Riplinger, C.; Bill, E.; Daiber, A.; Ullrich, V.; Shoun, H.; Neese, F. New Insights into the Nature of Observable Reaction Intermediates in Cytochrome P450 NO Reductase by Using a Combination of Spectroscopy and Quantum Mechanics/Molecular Mechanics Calculations. Chem.-Eur. J. 2014, 20, 1602–1614. [Google Scholar] [CrossRef]
- Riplinger, C.; Neese, F. The Reaction Mechanism of Cytochrome P450 NO Reductase: A Detailed Quantum Mechanics/Molecular Mechanics Study. ChemPhysChem 2011, 12, 3192–3203. [Google Scholar] [CrossRef] [PubMed]
- Nomura, T.; Kimura, T.; Kanematsu, Y.; Yamada, D.; Yamashita, K.; Hirata, K.; Ueno, G.; Murakami, H.; Hisano, T.; Yamagiwa, R.; et al. Short-Lived Intermediate in N 2 O Generation by P450 NO Reductase Captured by Time-Resolved IR Spectroscopy and XFEL Crystallography. Proc. Natl. Acad. Sci. USA 2021, 118, e2101481118. [Google Scholar] [CrossRef] [PubMed]
- Obayashi, E.; Takahashi, S.; Shiro, Y. Electronic Structure of Reaction Intermediate of Cytochrome P450nor in Its Nitric Oxide Reduction. J. Am. Chem. Soc. 1998, 120, 12964–12965. [Google Scholar] [CrossRef]
- Kondo, H.X.; Kanematsu, Y.; Masumoto, G.; Takano, Y. PyDISH: Database and Analysis Tools for Heme Porphyrin Distortion in Heme Proteins. Database 2020, baaa066. [Google Scholar] [CrossRef]
- Shimizu, H.; Park, S.-Y.; Lee, D.-S.; Shoun, H.; Shiro, Y. Crystal Structures of Cytochrome P450nor and Its Mutants (Ser286→Val, Thr) in the Ferric Resting State at Cryogenic Temperature: A Comparative Analysis with Monooxygenase Cytochrome P450s. J. Inorg. Biochem. 2000, 81, 191–205. [Google Scholar] [CrossRef]
- Hasemann, C.A.; Kurumbail, R.G.; Boddupalli, S.S.; Peterson, J.A.; Deisenhofer, J. Structure and Function of Cytochromes P450:A Comparative Analysis of Three Crystal Structures. Structure 1995, 3, 41–62. [Google Scholar] [CrossRef] [PubMed]
- Imada, Y.; Nakamura, H.; Takano, Y. Density Functional Study of Porphyrin Distortion Effects on Redox Potential of Heme. J. Comput. Chem. 2018, 39, 143–150. [Google Scholar] [CrossRef] [PubMed]
- Kanematsu, Y.; Kondo, H.X.; Imada, Y.; Takano, Y. Statistical and Quantum-Chemical Analysis of the Effect of Heme Porphyrin Distortion in Heme Proteins: Differences between Oxidoreductases and Oxygen Carrier Proteins. Chem. Phys. Lett. 2018, 710, 108–112. [Google Scholar] [CrossRef]
- Kondo, H.X.; Iizuka, H.; Masumoto, G.; Kabaya, Y.; Kanematsu, Y.; Takano, Y. Prediction of Protein Function from Tertiary Structure of the Active Site in Heme Proteins by Convolutional Neural Network. Biomolecules 2023, 13, 137. [Google Scholar] [CrossRef] [PubMed]
- Bikiel, D.E.; Forti, F.; Boechi, L.; Nardini, M.; Luque, F.J.; Martí, M.A.; Estrin, D.A. Role of Heme Distortion on Oxygen Affinity in Heme Proteins: The Protoglobin Case. J. Phys. Chem. B 2010, 114, 8536–8543. [Google Scholar] [CrossRef] [PubMed]
- Jentzen, W.; Ma, J.-G.; Shelnutt, J.A. Conservation of the Conformation of the Porphyrin Macrocycle in Hemoproteins. Biophys. J. 1998, 74, 753–763. [Google Scholar] [CrossRef]
- Breunig, M.M.; Kriegel, H.-P.; Ng, R.T.; Sander, J. LOF: Identifying Density-Based Local Outliers. ACM SIGMOD Rec. 2000, 29, 93–104. [Google Scholar] [CrossRef]
- Cossi, M.; Rega, N.; Scalmani, G.; Barone, V. Energies, Structures, and Electronic Properties of Molecules in Solution with the C-PCM Solvation Model. J. Comput. Chem. 2003, 24, 669–681. [Google Scholar] [CrossRef]
- Becke, A.D. Density-Functional Thermochemistry. III. The Role of Exact Exchange. J. Chem. Phys. 1993, 98, 5648–5652. [Google Scholar] [CrossRef]
- McLean, A.D.; Chandler, G.S. Contracted Gaussian Basis Sets for Molecular Calculations. I. Second Row Atoms, Z = 11–18. J. Chem. Phys. 1980, 72, 5639–5648. [Google Scholar] [CrossRef]
- Weigend, F.; Ahlrichs, R. Balanced Basis Sets of Split Valence, Triple Zeta Valence and Quadruple Zeta Valence Quality for H to Rn: Design and Assessment of Accuracy. Phys. Chem. Chem. Phys. 2005, 7, 3297. [Google Scholar] [CrossRef] [PubMed]
- Frisch, M.J.; Trucks, G.W.; Schlegel, H.B.; Scuseria, G.E.; Robb, M.A.; Cheeseman, J.R.; Scalmani, G.; Barone, V.; Petersson, G.A.; Nakatsuji, H.; et al. Gaussian 09; Rev. D.01; Gaussian Inc.: Wallingford, CT, USA, 2009. [Google Scholar]



Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kanematsu, Y.; Kondo, H.X.; Takano, Y. Computational Exploration of Minimum Energy Reaction Pathway of N2O Formation from Intermediate I of P450nor Using an Active Center Model. Int. J. Mol. Sci. 2023, 24, 17172. https://doi.org/10.3390/ijms242417172
Kanematsu Y, Kondo HX, Takano Y. Computational Exploration of Minimum Energy Reaction Pathway of N2O Formation from Intermediate I of P450nor Using an Active Center Model. International Journal of Molecular Sciences. 2023; 24(24):17172. https://doi.org/10.3390/ijms242417172
Chicago/Turabian StyleKanematsu, Yusuke, Hiroko X. Kondo, and Yu Takano. 2023. "Computational Exploration of Minimum Energy Reaction Pathway of N2O Formation from Intermediate I of P450nor Using an Active Center Model" International Journal of Molecular Sciences 24, no. 24: 17172. https://doi.org/10.3390/ijms242417172
APA StyleKanematsu, Y., Kondo, H. X., & Takano, Y. (2023). Computational Exploration of Minimum Energy Reaction Pathway of N2O Formation from Intermediate I of P450nor Using an Active Center Model. International Journal of Molecular Sciences, 24(24), 17172. https://doi.org/10.3390/ijms242417172




