New Insights into Radio-Resistance Mechanism Revealed by (Phospho)Proteome Analysis of Deinococcus Radiodurans after Heavy Ion Irradiation
Abstract
1. Introduction
2. Results
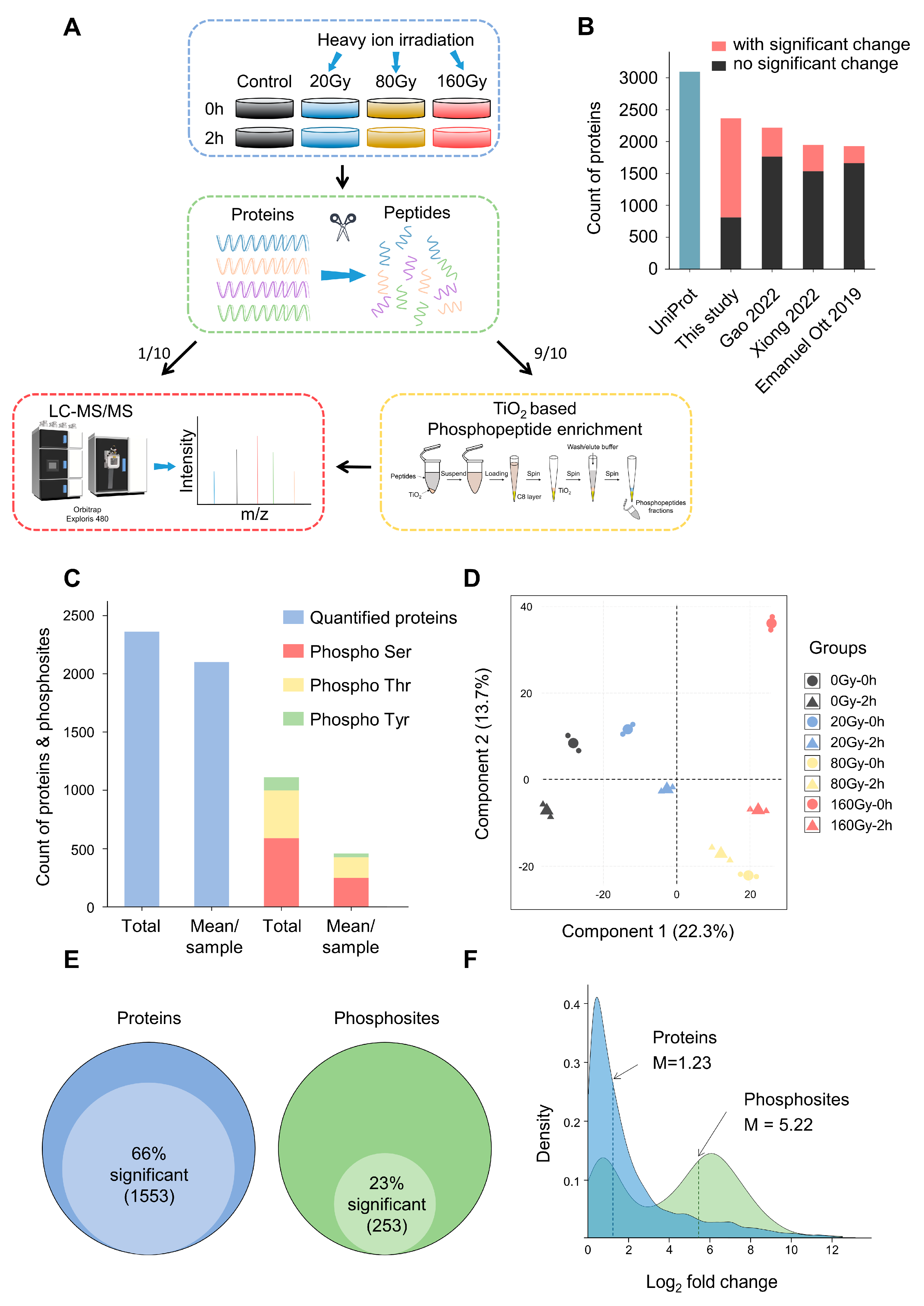
2.1. Global Analysis for (Phospho)Proteome of D. radiodurans after Heavy Ion Irradiation
2.2. Dose and Stage Dependence of D. radiodurans Irradiation Response
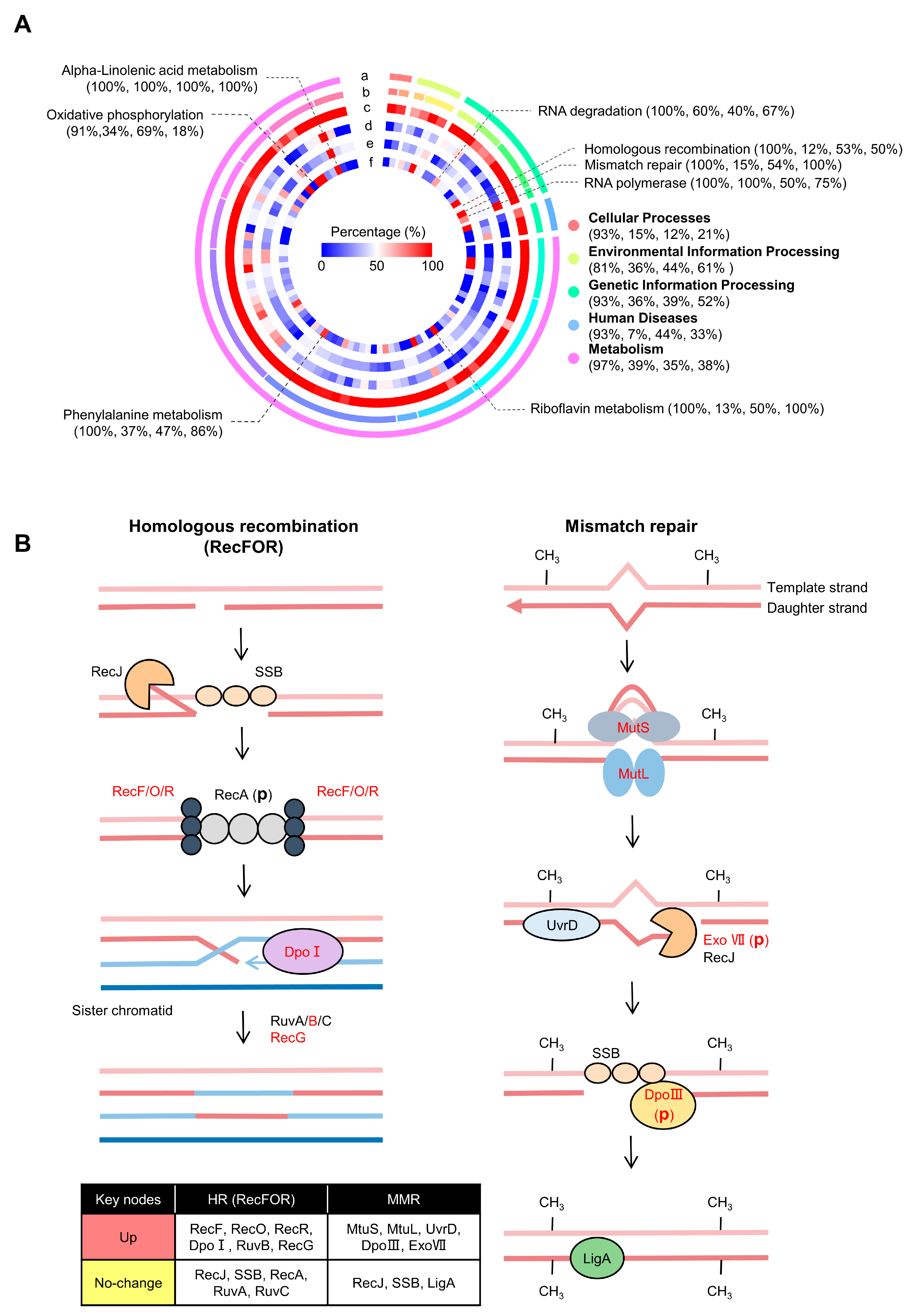
2.3. KEGG Pathway Analysis of Differentially Expressed (Phospho)Proteins
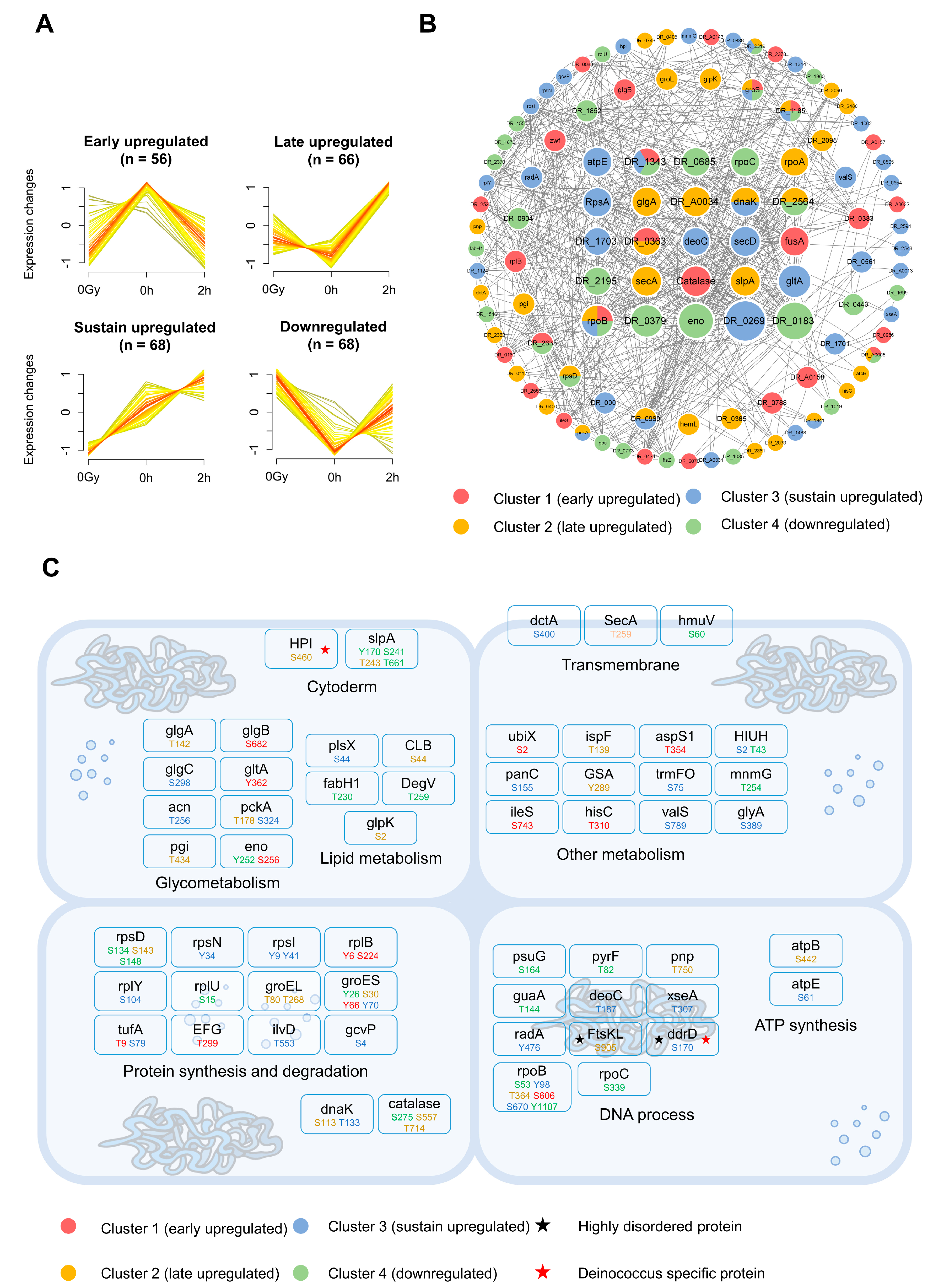
2.4. Dynamic Patterns of Phosphorylation and Functional Interpretation
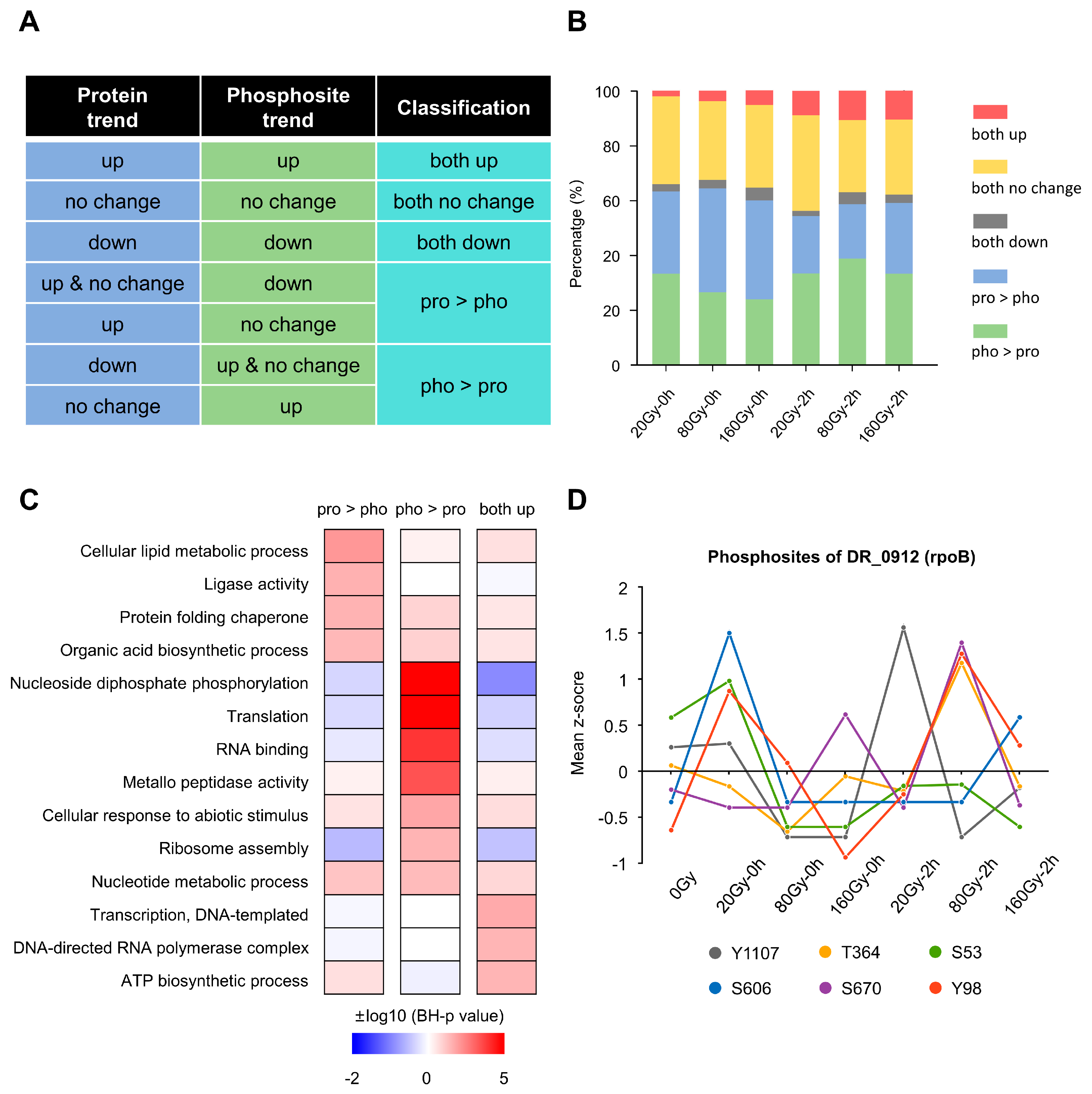
2.5. Comparative Analysis of Phosphosites and Protein Expression
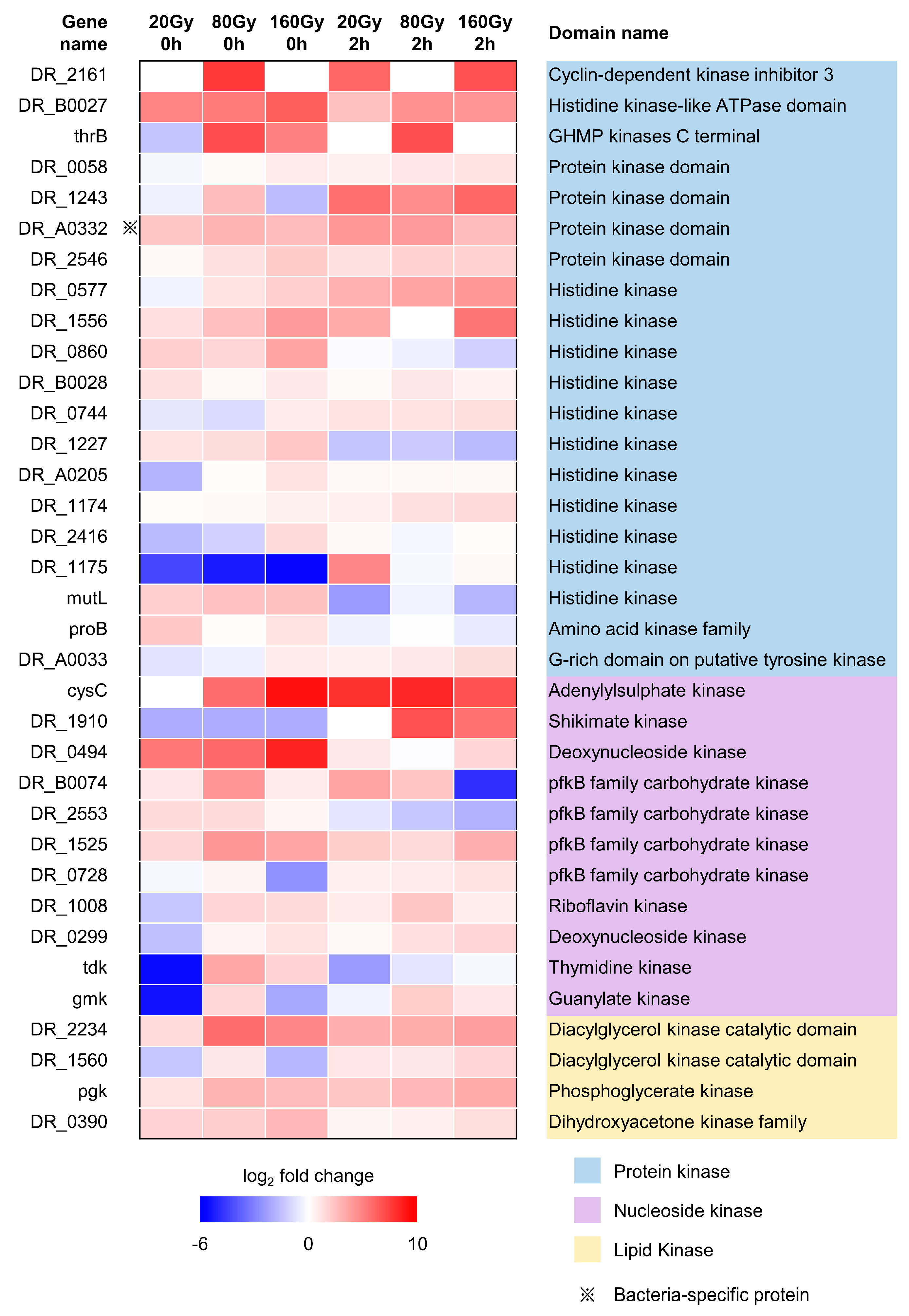
2.6. Irradiation-Sensitive Proteins Containing Kinase Domains
3. Discussion
4. Materials and Methods
4.1. Strain and Culture Conditions
4.2. Pre-Irradiation Preparation, Irradiation, and Post-Irradiation Treatment
4.3. Protein Lysis and Digestion for (Phospho)Proteomics Analysis
4.4. Chromatography and Mass Spectrometry
4.5. Database Searching, Quality Control and Data Normalization
4.6. GO and KEGG Annotation
4.7. PPI Network Analysis
4.8. Protein Structure and Evolutionary Age Annotation
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Lim, S.; Jung, J.-H.; Blanchard, L.; de Groot, A. Conservation and diversity of radiation and oxidative stress resistance mechanisms in Deinococcus species. FEMS Microbiol. Rev. 2018, 43, 19–52. [Google Scholar] [CrossRef] [PubMed]
- Bauermeister, A.; Hahn, C.; Rettberg, P.; Reitz, G.; Moeller, R. Roles of DNA repair and membrane integrity in heat resistance of Deinococcus radiodurans. Arch. Microbiol. 2012, 194, 959–966. [Google Scholar] [CrossRef] [PubMed]
- Mogul, R.; Bol’Shakov, A.; Chan, S.L.; Stevens, R.M.; Khare, B.N.; Meyyappan, M.; Trent, J.D. Impact of Low-Temperature Plasmas on Deinococcus radiodurans and Biomolecules. Biotechnol. Prog. 2003, 19, 776–783. [Google Scholar] [CrossRef] [PubMed]
- Peltek, S.; Meshcheryakova, I.; Kiseleva, E.; Oshchepkov, D.; Rozanov, A.; Serdyukov, D.; Demidov, E.; Vasiliev, G.; Vinokurov, N.; Bryanskaya, A.; et al. E. coli aggregation and impaired cell division after terahertz irradiation. Sci. Rep. 2021, 11, 20464. [Google Scholar] [CrossRef] [PubMed]
- Shukla, S.K.; Manobala, T.; Rao, T.S. The role of S-layer protein (SlpA) in biofilm-formation of Deinococcus radiodurans. J. Appl. Microbiol. 2022, 133, 796–807. [Google Scholar] [CrossRef] [PubMed]
- Rothfuss, H.; Lara, J.C.; Schmid, A.K.; Lidstrom, M.E. Involvement of the S-layer proteins Hpi and SlpA in the maintenance of cell envelope integrity in Deinococcus radiodurans R1. Microbiology 2006, 152, 2779–2787. [Google Scholar] [CrossRef]
- Levin-Zaidman, S.; Englander, J.; Shimoni, E.; Sharma, A.K.; Minton, K.W.; Minsky, A. Ringlike Structure of the Deinococcus radiodurans Genome: A Key to Radioresistance? Science 2003, 299, 254–256. [Google Scholar] [CrossRef]
- Winter, M.; Dokic, I.; Schlegel, J.; Warnken, U.; Debus, J.; Abdollahi, A.; Schnölzer, M. Deciphering the Acute Cellular Phosphoproteome Response to Irradiation with X-rays, Protons and Carbon Ions. Mol. Cell. Proteom. 2017, 16, 855–872. [Google Scholar] [CrossRef]
- Mehnati, P.; Morimoto, S.; Yatagai, F.; Furusawa, Y.; Kobayashi, Y.; Wada, S.; Kanai, T.; Hanaoka, F.; Sasaki, H. Exploration of ‘Over Kill Effect’ of High-LET Ar- and Fe-ions by Evaluating the Fraction of Non-hit Cell and Interphase Death. J. Radiat. Res. 2005, 46, 343–350. [Google Scholar] [CrossRef][Green Version]
- Gao, Y.; Li, N.; Zhou, Y.; Zhang, Z.; Zhang, Y.; Fan, P.; Zhou, H.; Zhang, T.; Chang, L.; Gao, H.; et al. iTRAQ-based proteomic analysis of Deinococcus radiodurans in response to 12C6+ heavy ion irradiation. BMC Microbiol. 2022, 22, 264. [Google Scholar] [CrossRef]
- Slade, D.; Lindner, A.B.; Paul, G.; Radman, M. Recombination and Replication in DNA Repair of Heavily Irradiated Deinococcus radiodurans. Cell 2009, 136, 1044–1055. [Google Scholar] [CrossRef] [PubMed]
- Pagès, V. Single-strand gap repair involves both RecF and RecBCD pathways. Curr. Genet. 2016, 62, 519–521. [Google Scholar] [CrossRef] [PubMed]
- Morimatsu, K.; Kowalczykowski, S.C. RecQ helicase and RecJ nuclease provide complementary functions to resect DNA for homologous recombination. Proc. Natl. Acad. Sci. USA 2014, 111, E5133–E5142. [Google Scholar] [CrossRef]
- Zahradka, K.; Slade, D.; Bailone, A.; Sommer, S.; Averbeck, D.; Petranovic, M.; Lindner, A.B.; Radman, M. Reassembly of shattered chromosomes in Deinococcus radiodurans. Nature 2006, 443, 569–573. [Google Scholar] [CrossRef] [PubMed]
- Seck, A.; De Bonis, S.; Stelter, M.; Ökvist, M.; Senarisoy, M.; Hayek, M.R.; Le Roy, A.; Martin, L.; Saint-Pierre, C.; Silveira, C.M.; et al. Structural and functional insights into the activation of the dual incision activity of UvrC, a key player in bacterial NER. Nucleic Acids Res. 2023, 51, 2931–2949. [Google Scholar] [CrossRef] [PubMed]
- Banneville, A.-S.; de la Tour, C.B.; De Bonis, S.; Hognon, C.; Colletier, J.-P.; Teulon, J.-M.; Le Roy, A.; Pellequer, J.-L.; Monari, A.; Dehez, F.; et al. Structural and functional characterization of DdrC, a novel DNA damage-induced nucleoid associated protein involved in DNA compaction. Nucleic Acids Res. 2022, 50, 7680–7696. [Google Scholar] [CrossRef] [PubMed]
- Agapov, A.A.; Kulbachinskiy, A.V. Mechanisms of stress resistance and gene regulation in the radioresistant bacterium Deinococcus radiodurans. Biochemistry 2015, 80, 1201–1216. [Google Scholar] [CrossRef]
- Selvam, K.; Duncan, J.R.; Tanaka, M.; Battista, J.R. DdrA, DdrD, and PprA: Components of UV and Mitomycin C Resistance in Deinococcus radiodurans R1. PLoS ONE 2013, 8, e69007. [Google Scholar] [CrossRef]
- Santos, S.P.; Mitchell, E.P.; Franquelim, H.G.; Castanho, M.A.R.B.; Abreu, I.A.; Romão, C.V. Dps from Deinococcus radiodurans: Oligomeric forms of Dps1 with distinct cellular functions and Dps2 involved in metal storage. FEBS J. 2015, 282, 4307–4327. [Google Scholar] [CrossRef]
- Reon, B.J.; Nguyen, K.H.; Bhattacharyya, G.; Grove, A. Functional comparison of Deinococcus radiodurans Dps proteins suggests distinct in vivo roles. Biochem. J. 2012, 447, 381–391. [Google Scholar] [CrossRef]
- Grove, A.; Wilkinson, S.P. Differential DNA Binding and Protection by Dimeric and Dodecameric forms of the Ferritin Homolog Dps from Deinococcus radiodurans. J. Mol. Biol. 2005, 347, 495–508. [Google Scholar] [CrossRef] [PubMed]
- Gaidamakova, E.K.; Sharma, A.; Matrosova, V.Y.; Grichenko, O.; Volpe, R.P.; Tkavc, R.; Conze, I.H.; Klimenkova, P.; Balygina, I.; Horne, W.H.; et al. Small-Molecule Mn Antioxidants in Caenorhabditis elegans and Deinococcus radiodurans Supplant MnSOD Enzymes during Aging and Irradiation. mBio 2022, 13, e0339421. [Google Scholar] [CrossRef]
- Zhou, C.; Dai, J.; Lu, H.; Chen, Z.; Guo, M.; He, Y.; Gao, K.; Ge, T.; Jin, J.; Wang, L.; et al. Succinylome Analysis Reveals the Involvement of Lysine Succinylation in the Extreme Resistance of Deinococcus radiodurans. Proteomics 2019, 19, e1900158. [Google Scholar] [CrossRef]
- Li, W.; Han, X.; Lan, P. Emerging roles of protein phosphorylation in plant iron homeostasis. Trends Plant Sci. 2022, 27, 908–921. [Google Scholar] [CrossRef] [PubMed]
- Sharma, K.; D’Souza, R.C.J.; Tyanova, S.; Schaab, C.; Wiśniewski, J.R.; Cox, J.; Mann, M. Ultradeep Human Phosphoproteome Reveals a Distinct Regulatory Nature of Tyr and Ser/Thr-Based Signaling. Cell Rep. 2014, 8, 1583–1594. [Google Scholar] [CrossRef] [PubMed]
- Sharma, D.K.; Siddiqui, M.Q.; Gadewal, N.; Choudhary, R.K.; Varma, A.K.; Misra, H.S.; Rajpurohit, Y.S. Phosphorylation of deinococcal RecA affects its structural and functional dynamics implicated for its roles in radioresistance of Deinococcus radiodurans. J. Biomol. Struct. Dyn. 2019, 38, 114–123. [Google Scholar] [CrossRef]
- Maurya, G.K.; Modi, K.; Banerjee, M.; Chaudhary, R.; Rajpurohit, Y.S.; Misra, H.S. Phosphorylation of FtsZ and FtsA by a DNA Damage-Responsive Ser/Thr Protein Kinase Affects Their Functional Interactions in Deinococcus radiodurans. mSphere 2018, 3, e00325–e18. [Google Scholar] [CrossRef]
- Rajpurohit, Y.S.; Bihani, S.C.; Waldor, M.K.; Misra, H.S. Phosphorylation of Deinococcus radiodurans RecA Regulates Its Activity and May Contribute to Radioresistance. J. Biol. Chem. 2016, 291, 16672–16685. [Google Scholar] [CrossRef]
- Chaudhary, R.; Mishra, S.; Maurya, G.K.; Rajpurohit, Y.S.; Misra, H.S. FtsZ phosphorylation brings about growth arrest upon DNA damage in Deinococcus radiodurans. FASEB BioAdvances 2022, 5, 27–42. [Google Scholar] [CrossRef]
- Potel, C.M.; Lin, M.-H.; Heck, A.J.R.; Lemeer, S. Widespread bacterial protein histidine phosphorylation revealed by mass spectrometry-based proteomics. Nat. Methods 2018, 15, 187–190. [Google Scholar] [CrossRef]
- Garcia-Garcia, T.; Poncet, S.; Derouiche, A.; Shi, L.; Mijakovic, I.; Noirot-Gros, M.-F. Role of Protein Phosphorylation in the Regulation of Cell Cycle and DNA-Related Processes in Bacteria. Front. Microbiol. 2016, 7, 184. [Google Scholar] [CrossRef]
- Xiong, Y.; Wei, L.; Xin, S.; Min, R.; Liu, F.; Li, N.; Zhang, Y. Comprehensive Temporal Protein Dynamics during Postirradiation Recovery in Deinococcus radiodurans. Oxidative Med. Cell. Longev. 2022, 2022, 1622829. [Google Scholar] [CrossRef] [PubMed]
- Ott, E.; Kawaguchi, Y.; Özgen, N.; Yamagishi, A.; Rabbow, E.; Rettberg, P.; Weckwerth, W.; Milojevic, T. Proteomic and Metabolomic Profiling of Deinococcus radiodurans Recovering After Exposure to Simulated Low Earth Orbit Vacuum Conditions. Front. Microbiol. 2019, 10, 909. [Google Scholar] [CrossRef] [PubMed]
- Stracke, C.; Meyer, B.H.; Hagemann, A.; Jo, E.; Lee, A.; Albers, S.-V.; Cha, J.; Bräsen, C.; Siebers, B. Salt Stress Response of Sulfolobus acidocaldarius Involves Complex Trehalose Metabolism Utilizing a Novel Trehalose-6-Phosphate Synthase (TPS)/Trehalose-6-Phosphate Phosphatase (TPP) Pathway. Appl. Environ. Microbiol. 2020, 86, 24. [Google Scholar] [CrossRef] [PubMed]
- Sakaguchi, M. Diverse and common features of trehalases and their contributions to microbial trehalose metabolism. Appl. Microbiol. Biotechnol. 2020, 104, 1837–1847. [Google Scholar] [CrossRef]
- Lu, R.-S.; Li, P.; Qiu, Y.-X. The Complete Chloroplast Genomes of Three Cardiocrinum (Liliaceae) Species: Comparative Genomic and Phylogenetic Analyses. Front. Plant Sci. 2017, 7, 2054. [Google Scholar] [CrossRef]
- Caserta, E.; Liu, L.-C.; Grundy, F.J.; Henkin, T.M. Codon-Anticodon Recognition in the Bacillus subtilis glyQS T Box Riboswitch. J. Biol. Chem. 2015, 290, 23336–23347. [Google Scholar] [CrossRef]
- Chakravarty, A.K.; Subbotin, R.; Chait, B.T.; Shuman, S. RNA ligase RtcB splices 3′-phosphate and 5′-OH ends via covalent RtcB-(histidinyl)-GMP and polynucleotide-(3′)pp(5′)G intermediates. Proc. Natl. Acad. Sci. USA 2012, 109, 6072–6077. [Google Scholar] [CrossRef]
- Zhang, Y.; Morar, M.; Ealick, S.E. Structural biology of the purine biosynthetic pathway. Cell. Mol. Life Sci. 2008, 65, 3699–3724. [Google Scholar] [CrossRef]
- Fujimura, S.; Mio, K.; Ohkubo, T.; Arai, T.; Kuramochi, M.; Sekiguchi, H.; Sasaki, Y.C. Diffracted X-ray Tracking Method for Measuring Intramolecular Dynamics of Membrane Proteins. Int. J. Mol. Sci. 2022, 23, 2343. [Google Scholar] [CrossRef]
- Cai, L.; Zhao, D.; Hou, J.; Wu, J.; Cai, S.; Dassarma, P.; Xiang, H. Cellular and organellar membrane-associated proteins in haloarchaea: Perspectives on the physiological significance and biotechnological applications. Sci. China Life Sci. 2012, 55, 404–414. [Google Scholar] [CrossRef] [PubMed]
- Yang, Y.; Jin, H.; Chen, Y.; Lin, W.; Wang, C.; Chen, Z.; Han, N.; Bian, H.; Zhu, M.; Wang, J. A chloroplast envelope membrane protein containing a putative LrgB domain related to the control of bacterial death and lysis is required for chloroplast development in Arabidopsis thaliana. New Phytol. 2011, 193, 81–95. [Google Scholar] [CrossRef] [PubMed]
- Kanehisa, M.; Furumichi, M.; Sato, Y.; Ishiguro-Watanabe, M.; Tanabe, M. KEGG: Integrating viruses and cellular organisms. Nucleic Acids Res. 2020, 49, D545–D551. [Google Scholar] [CrossRef] [PubMed]
- Xu, Q.; Cao, Y.; Zhong, X.; Qin, X.; Feng, J.; Peng, H.; Su, Y.; Ma, Z.; Zhou, S. Riboflavin protects against heart failure via SCAD-dependent DJ-1–Keap1–Nrf2 signalling pathway. Br. J. Pharmacol. 2023, 1–23. [Google Scholar] [CrossRef]
- Alam, S.-I.; Kim, M.-W.; Shah, F.A.; Saeed, K.; Ullah, R.; Kim, M.-O. Alpha-Linolenic Acid Impedes Cadmium-Induced Oxidative Stress, Neuroinflammation, and Neurodegeneration in Mouse Brain. Cells 2021, 10, 2274. [Google Scholar] [CrossRef]
- Feng, L.; Li, W.; Liu, Y.; Jiang, W.-D.; Kuang, S.-Y.; Wu, P.; Jiang, J.; Tang, L.; Tang, W.-N.; Zhang, Y.-A.; et al. Protective role of phenylalanine on the ROS-induced oxidative damage, apoptosis and tight junction damage via Nrf2, TOR and NF-κB signalling molecules in the gill of fish. Fish Shellfish. Immunol. 2016, 60, 185–196. [Google Scholar] [CrossRef]
- Bentchikou, E.; Servant, P.; Coste, G.; Sommer, S. A Major Role of the RecFOR Pathway in DNA Double-Strand-Break Repair through ESDSA in Deinococcus radiodurans. PLOS Genet. 2010, 6, e1000774. [Google Scholar] [CrossRef]
- Szklarczyk, D.; Kirsch, R.; Koutrouli, M.; Nastou, K.; Mehryary, F.; Hachilif, R.; Gable, A.L.; Fang, T.; Doncheva, N.T.; Pyysalo, S.; et al. The STRING database in 2023: Protein–protein association networks and functional enrichment analyses for any sequenced genome of interest. Nucleic Acids Res. 2022, 51, D638–D646. [Google Scholar] [CrossRef]
- Jiao, C.; Gu, Z. iTRAQ-based proteomic analysis reveals changes in response to UV-B treatment in soybean sprouts. Food Chem. 2018, 275, 467–473. [Google Scholar] [CrossRef]
- Prasad, S.M.; Kumar, S.; Parihar, P.; Singh, A.; Singh, R. Evaluating the combined effects of pretilachlor and UV-B on two Azolla species. Pestic. Biochem. Physiol. 2016, 128, 45–56. [Google Scholar] [CrossRef]
- Huang, G.; Wang, L.; Zhou, Q. Lanthanum (III) Regulates the Nitrogen Assimilation in Soybean Seedlings under Ultraviolet-B Radiation. Biol. Trace Element Res. 2012, 151, 105–112. [Google Scholar] [CrossRef] [PubMed]
- Boothby, T.C.; Tapia, H.; Brozena, A.H.; Piszkiewicz, S.; Smith, A.E.; Giovannini, I.; Rebecchi, L.; Pielak, G.J.; Koshland, D.; Goldstein, B. Tardigrades Use Intrinsically Disordered Proteins to Survive Desiccation. Mol. Cell 2017, 65, 975–984. [Google Scholar] [CrossRef] [PubMed]
- Ramasamy, P.; Vandermarliere, E.; Vranken, W.F.; Martens, L. Panoramic Perspective on Human Phosphosites. J. Proteome Res. 2022, 21, 1894–1915. [Google Scholar] [CrossRef]
- Giansanti, P.; Strating, J.R.P.M.; Defourny, K.A.Y.; Cesonyte, I.; Bottino, A.M.S.; Post, H.; Viktorova, E.G.; Ho, V.Q.T.; Langereis, M.A.; Belov, G.A.; et al. Dynamic remodelling of the human host cell proteome and phosphoproteome upon enterovirus infection. Nat. Commun. 2020, 11, 4332. [Google Scholar] [CrossRef]
- Fujiwara, D.; Kawaguchi, Y.; Kinoshita, I.; Yatabe, J.; Narumi, I.; Hashimoto, H.; Yokobori, S.-I.; Yamagishi, A. Mutation Analysis of the rpoB Gene in the Radiation-Resistant Bacterium Deinococcus radiodurans R1 Exposed to Space during the Tanpopo Experiment at the International Space Station. Astrobiology 2021, 21, 1494–1504. [Google Scholar] [CrossRef] [PubMed]
- Yuan, Q.; Xie, F.; Huang, W.; Hu, M.; Yan, Q.; Chen, Z.; Zheng, Y.; Liu, L. The review of alpha-linolenic acid: Sources, metabolism, and pharmacology. Phytotherapy Res. 2021, 36, 164–188. [Google Scholar] [CrossRef] [PubMed]
- Fila, M.; Chojnacki, C.; Chojnacki, J.; Blasiak, J. Nutrients to Improve Mitochondrial Function to Reduce Brain Energy Deficit and Oxidative Stress in Migraine. Nutrients 2021, 13, 4433. [Google Scholar] [CrossRef]
- Stejskal, K.; de Beeck, J.O.; Dürnberger, G.; Jacobs, P.; Mechtler, K. Ultrasensitive NanoLC-MS of Subnanogram Protein Samples Using Second Generation Micropillar Array LC Technology with Orbitrap Exploris 480 and FAIMS PRO. Anal. Chem. 2021, 93, 8704–8710. [Google Scholar] [CrossRef]
- Shannon, P.; Markiel, A.; Ozier, O.; Baliga, N.S.; Wang, J.T.; Ramage, D.; Amin, N.; Schwikowski, B.; Ideker, T. Cytoscape: A software environment for integrated models of Biomolecular Interaction Networks. Genome Res. 2003, 13, 2498–2504. [Google Scholar] [CrossRef]
- Wang, H.; Zhong, H.; Gao, C.; Zang, J.; Yang, D. The Distinct Properties of the Consecutive Disordered Regions Inside or Outside Protein Domains and Their Functional Significance. Int. J. Mol. Sci. 2021, 22, 10677. [Google Scholar] [CrossRef]
- Hanson, J.; Paliwal, K.K.; Zhou, Y. Accurate Single-Sequence Prediction of Protein Intrinsic Disorder by an Ensemble of Deep Recurrent and Convolutional Architectures. J. Chem. Inf. Model. 2018, 58, 2369–2376. [Google Scholar] [CrossRef] [PubMed]
- Gao, C.; Ma, C.; Wang, H.; Zhong, H.; Zang, J.; Zhong, R.; He, F.; Yang, D. Intrinsic disorder in protein domains contributes to both organism complexity and clade-specific functions. Sci. Rep. 2021, 11, 2985. [Google Scholar] [CrossRef] [PubMed]
- Gsponer, J.; Futschik, M.E.; Teichmann, S.A.; Babu, M.M. Tight Regulation of Unstructured Proteins: From Transcript Synthesis to Protein Degradation. Science 2008, 322, 1365–1368. [Google Scholar] [CrossRef] [PubMed]
- Trigos, A.S.; Pearson, R.B.; Papenfuss, A.T.; Goode, D.L. Altered interactions between unicellular and multicellular genes drive hallmarks of transformation in a diverse range of solid tumors. Proc. Natl. Acad. Sci. USA 2017, 114, 6406–6411. [Google Scholar] [CrossRef] [PubMed]
- Domazet-Lošo, T.; Tautz, D. A phylogenetically based transcriptome age index mirrors ontogenetic divergence patterns. Nature 2010, 468, 815–818. [Google Scholar] [CrossRef]
- Domazet-Lošo, T.; Brajković, J.; Tautz, D. A phylostratigraphy approach to uncover the genomic history of major adaptations in metazoan lineages. Trends Genet. 2007, 23, 533–539. [Google Scholar] [CrossRef]
- Buchfink, B.; Reuter, K.; Drost, H.-G. Sensitive protein alignments at tree-of-life scale using DIAMOND. Nat. Methods 2021, 18, 366–368. [Google Scholar] [CrossRef]

| Uniprot ID | Protein Name | The Upregulated Experiment Groups Dose (Gy)-Time (h) | Phosphosites Amino Acid-Cluster |
|---|---|---|---|
| Q9RV39 | A-adding tRNA nucleotidyltransferase | 160-0 | NULL |
| Q9RR60 | Enolase | 20-0, 80-0, 160-0,20-2, 80-2, 160-2 | T252-4, Y256-1 |
| Q9RSR1 | Polyribonucleotide nucleotidyltransferase | NULL | T750-2 |
| Q9RY23 | Chaperone protein DnaK | 20-0, 80-0, 160-0,20-2, 80-2, 160-2 | S113-2, T133-3 |
| Q9RWQ9 | Chaperonin GroEL | 20-0, 80-0, 160-0,20-2, 80-2, 160-2 | T80-2, T268-2 |
| Q9RSJ6 | RNA polymerase subunit alpha | NULL | Y28-2 |
| Q9RVV9 | RNA polymerase subunit beta | NULL | S53-4, Y98-3, T364-2, S606-1, S670-3, Y1107-4 |
| Q9RVW0 | RNA polymerase subunit beta’ | 80-0, 160-0, 20-2, 160-2 | S339-4 |
| Q9RRJ6 | RNA polymerase subunit omega | 80-0, 160-0, 160-2 | NULL |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Liu, S.; Wang, F.; Chen, H.; Yang, Z.; Ning, Y.; Chang, C.; Yang, D. New Insights into Radio-Resistance Mechanism Revealed by (Phospho)Proteome Analysis of Deinococcus Radiodurans after Heavy Ion Irradiation. Int. J. Mol. Sci. 2023, 24, 14817. https://doi.org/10.3390/ijms241914817
Liu S, Wang F, Chen H, Yang Z, Ning Y, Chang C, Yang D. New Insights into Radio-Resistance Mechanism Revealed by (Phospho)Proteome Analysis of Deinococcus Radiodurans after Heavy Ion Irradiation. International Journal of Molecular Sciences. 2023; 24(19):14817. https://doi.org/10.3390/ijms241914817
Chicago/Turabian StyleLiu, Shihao, Fei Wang, Heye Chen, Zhixiang Yang, Yifan Ning, Cheng Chang, and Dong Yang. 2023. "New Insights into Radio-Resistance Mechanism Revealed by (Phospho)Proteome Analysis of Deinococcus Radiodurans after Heavy Ion Irradiation" International Journal of Molecular Sciences 24, no. 19: 14817. https://doi.org/10.3390/ijms241914817
APA StyleLiu, S., Wang, F., Chen, H., Yang, Z., Ning, Y., Chang, C., & Yang, D. (2023). New Insights into Radio-Resistance Mechanism Revealed by (Phospho)Proteome Analysis of Deinococcus Radiodurans after Heavy Ion Irradiation. International Journal of Molecular Sciences, 24(19), 14817. https://doi.org/10.3390/ijms241914817





