MicroRNAs in Small Extracellular Vesicles from Amniotic Fluid and Maternal Plasma Associated with Fetal Palate Development in Mice
Abstract
:1. Introduction
2. Results
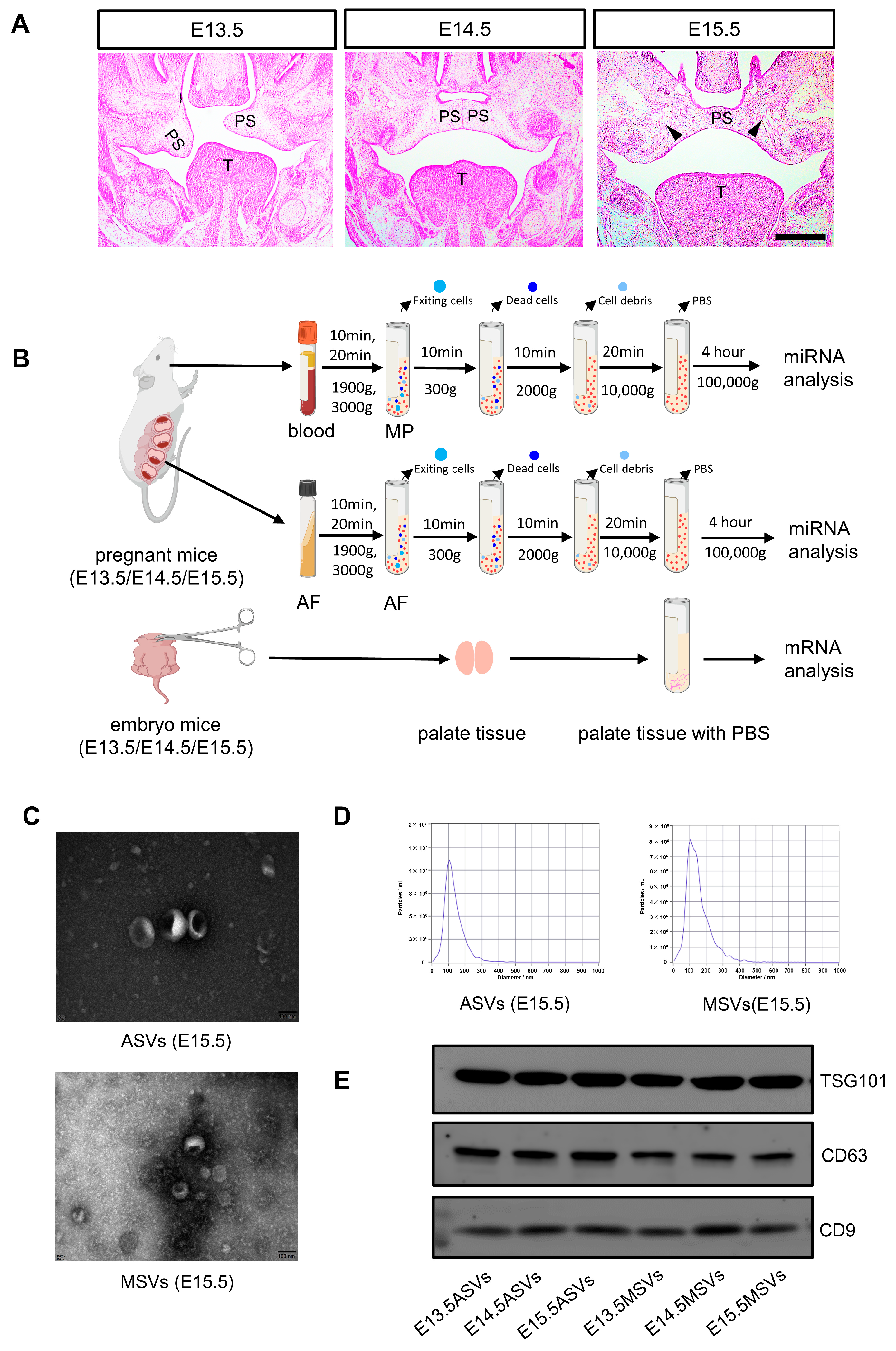
2.1. Morphology and Histology of Palate at E13.5/E14.5/E15.5
2.2. Isolation and Identification of MSVs and ASVs
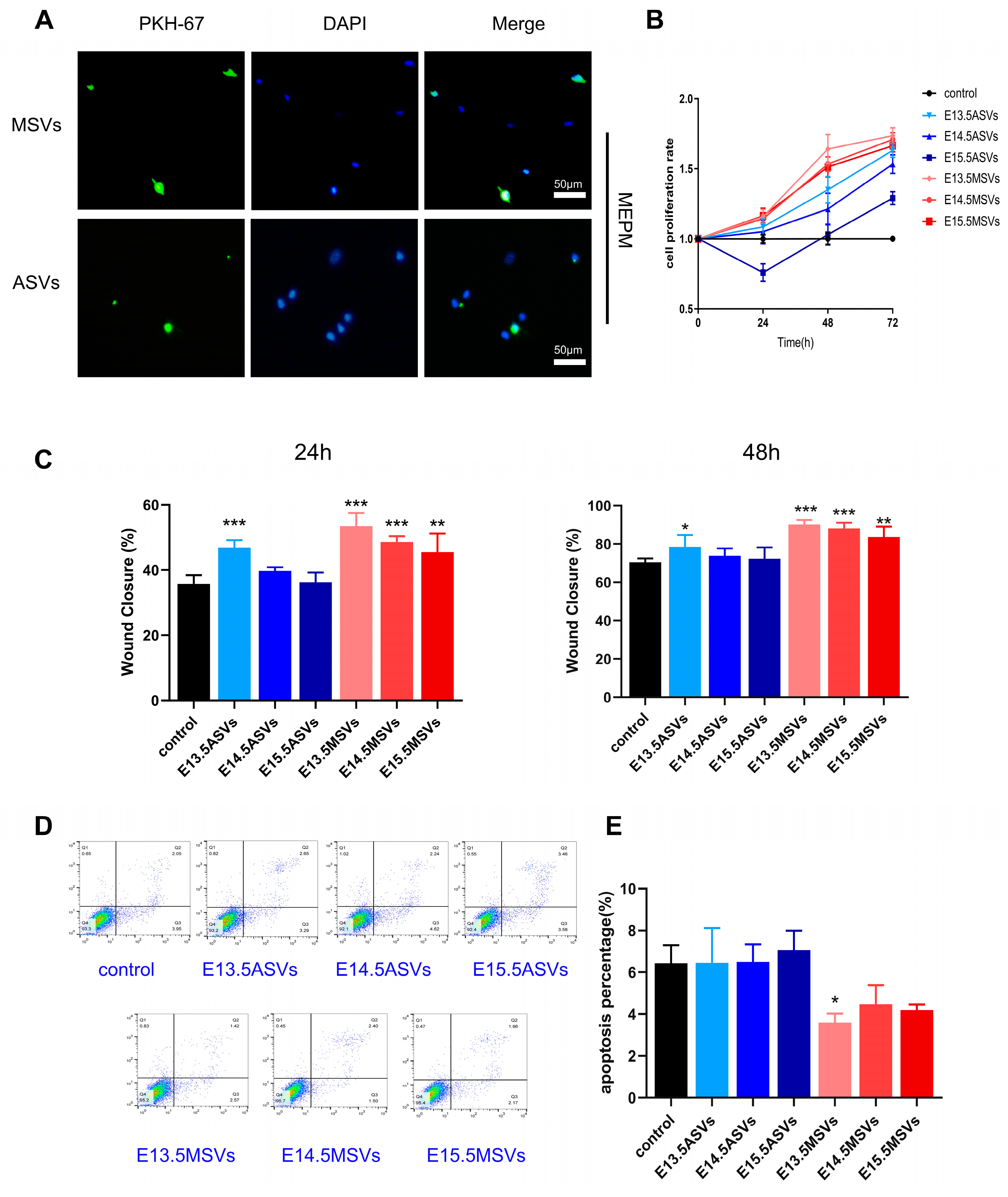
2.3. Different Effects of MSVs and ASVs on the Biological Behaviors of MEPM Cells and MEE Cells
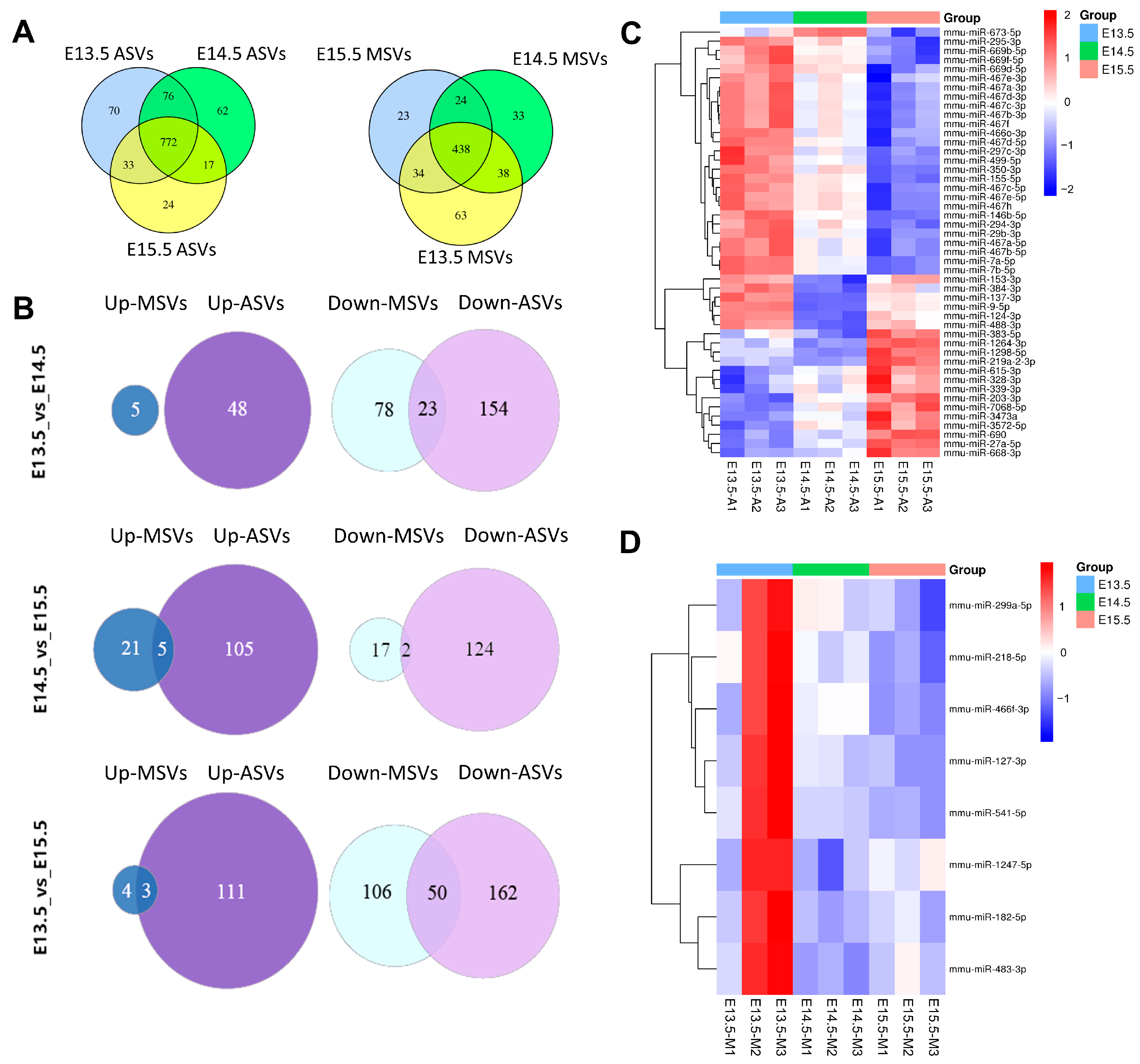
2.4. Analysis of miRNA Sequences in ASVs and MSVs from Different Stages
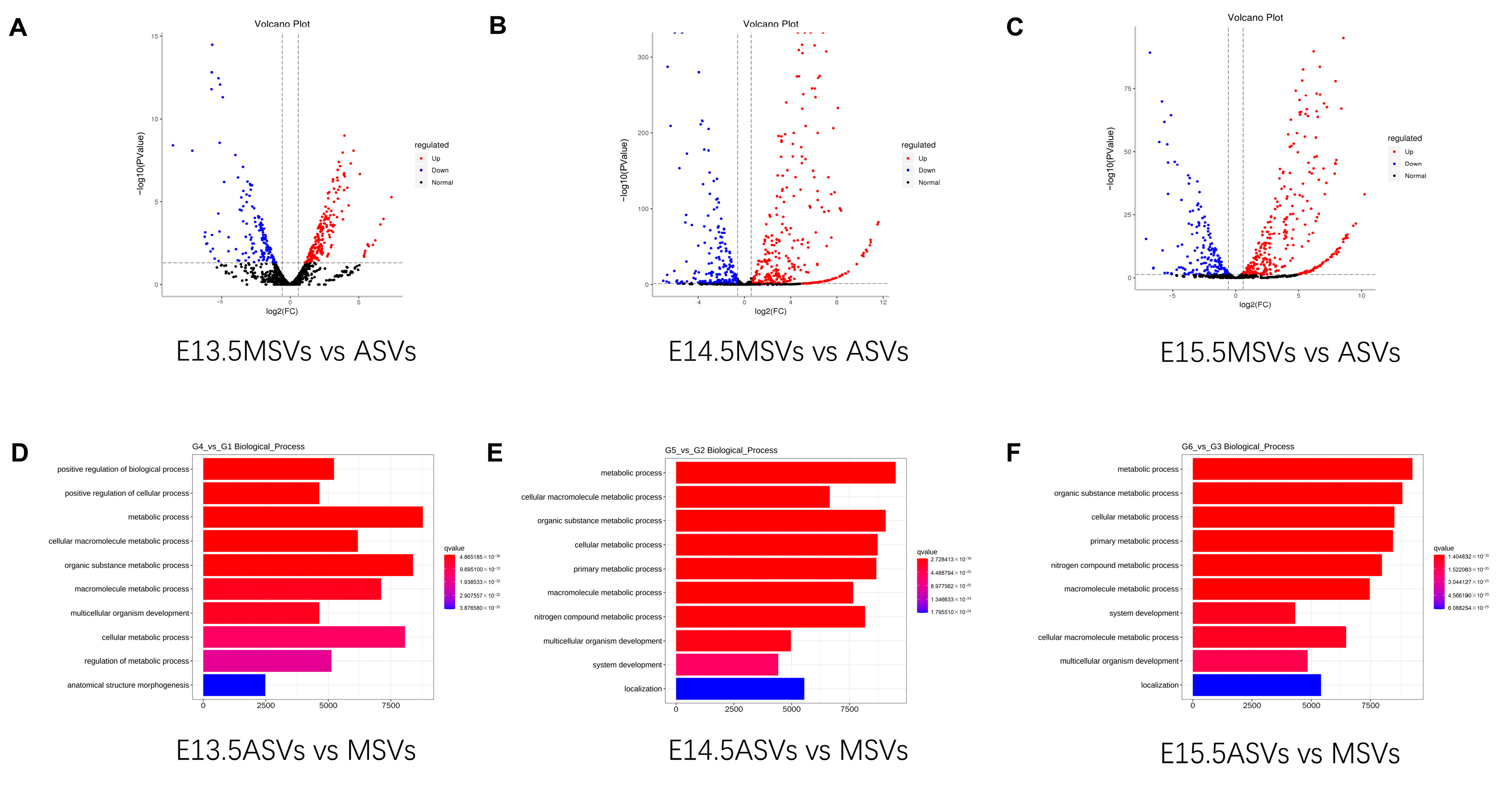
2.5. Analysis of DE miRNAs in ASVs and MSVs
2.6. Regulatory Networks of miRNAs in ASVs and mRNA in Palate Tissue
2.7. Regulatory Networks of miRNAs in MSVs and mRNA in Palate Tissue
2.8. Quantitative Analysis of miRNA Expression
3. Discussion
4. Materials and Methods
4.1. Animals
4.2. Collection of AF and MP
4.3. Hematoxylin and Eosin (HE) Staining
4.4. SEV Isolation and Characterization
4.5. Isolation and Culture of MEPM Cells and MEE Cells
4.6. CCK8 Assay
4.7. Apoptosis Assay
4.8. Scratch Test
4.9. Alizarin Red Staining
4.10. Western Blot
4.11. Library Preparation and Sequencing
4.12. Quantification and DE Analysis of miRNAs
4.13. Transcriptome-Seq Analysis
4.14. GO and KEGG Enrichment Analysis
4.15. Curation of miRNA in SEVs and mRNAs in Palate Tissue Regulation Pairs
4.16. Validation of miRNA Expression
4.17. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Abbreviations
| Abbreviation | Full Name |
| CP | cleft palate |
| miRNA | microRNA |
| GO | gene ontology |
| SEV | small extracellular vesicle |
| MP | maternal plasma |
| AF | amniotic fluid |
| ASV | SEVs derived from AF |
| MSV | SEVs derived from MP |
| CL/P | cleft lip with or without palate |
| E | embryonic day |
| MEPM cell | mouse embryonic palatal mesenchyme cell |
| MEE cell | medial edge epithelial cell |
| DE | differential expression |
| MES | midpalate epithelial suture |
| CCK8 | cell counting kit 8 |
| KEGG | Kyoto Encyclopedia of Genes and Genomes |
| EMT | epithelial–mesenchymal transition |
| VEGFA | vascular endothelial growth factor A |
| HE | hematoxylin and eosin |
| RA | retinoic acid |
| Smpd3 | sphingomyelin phosphodiesterase 3 |
| Agt | angiotensinogen |
| Phex | phosphate-regulating gene with homologies to endopeptidases on the X chromosome |
| Rflna | refilin A |
| Foxc2 | forkhead box C2 |
| Matn4 | matrilin-4 |
| P2rx4 | P2X purinoceptor 4 |
| Dcn | decorin |
| Rbp4 | retinol-binding protein 4 |
| Lox | lysyl oxidase |
| Sgms2 | sphingomyelin synthase 2 |
| Dsg3 | desmoglein 3 |
| Omd | osteomodulin |
| Dnah10 | dynein axonemal heavy chain 10 |
| Col22a1 | collagen XXII |
| Dnali1 | dynein axonemal light intermediate chain 1 |
| Cd9 | cluster of differentiation 9 |
References
- Lewis, C.W.; Jacob, L.S.; Lehmann, C.U. The Primary Care Pediatrician and the Care of Children with Cleft Lip and/or Cleft Palate. Pediatrics 2017, 139, e20170628. [Google Scholar] [CrossRef] [PubMed]
- Dixon, M.J.; Marazita, M.L.; Beaty, T.H.; Murray, J.C. Cleft lip and palate: Understanding genetic and environmental influences. Nat. Rev. Genet. 2011, 12, 167–178. [Google Scholar] [CrossRef] [PubMed]
- van der Hoek-Snieders, H.E.M.; van den Heuvel, A.; van Os-Medendorp, H.; Kamalski, D.M.A. Diagnostic accuracy of fetal MRI to detect cleft palate: A meta-analysis. Eur. J. Pediatr. 2020, 179, 29–38. [Google Scholar] [CrossRef] [PubMed]
- Gross, N.; Kropp, J.; Khatib, H. MicroRNA Signaling in Embryo Development. Biology 2017, 6, 34. [Google Scholar] [CrossRef] [PubMed]
- Babenko, O.; Kovalchuk, I.; Metz, G.A. Stress-induced perinatal and transgenerational epigenetic programming of brain development and mental health. Neurosci. Biobehav. Rev. 2015, 48, 70–91. [Google Scholar] [CrossRef] [PubMed]
- Yoshioka, H.; Suzuki, A.; Iwaya, C.; Iwata, J. Suppression of microRNA 124-3p and microRNA 340-5p ameliorates retinoic acid-induced cleft palate in mice. Development 2022, 149, dev200476. [Google Scholar] [CrossRef]
- Yan, F.; Jia, P.; Yoshioka, H.; Suzuki, A.; Iwata, J.; Zhao, Z. A developmental stage-specific network approach for studying dynamic co-regulation of transcription factors and microRNAs during craniofacial development. Development 2020, 147, dev192948. [Google Scholar] [CrossRef]
- Iwaya, C.; Suzuki, A.; Iwata, J. MicroRNAs and Gene Regulatory Networks Related to Cleft Lip and Palate. Int. J. Mol. Sci. 2023, 24, 3552. [Google Scholar] [CrossRef]
- Xu, Y.; Yuan, D.; Fan, Z.; Wang, S.; Du, J. Identification and profiles of microRNAs in different development stages of miniature pig secondary palate. Genomics 2021, 113, 2634–2644. [Google Scholar] [CrossRef]
- Nazdikbin Yamchi, N.; Ahmadian, S.; Mobarak, H.; Amjadi, F.; Beheshti, R.; Tamadon, A.; Rahbarghazi, R.; Mahdipour, M. Amniotic fluid-derived exosomes attenuated fibrotic changes in POI rats through modulation of the TGF-β/Smads signaling pathway. J. Ovarian Res. 2023, 16, 118. [Google Scholar] [CrossRef]
- Jia, S.; Zhang, Q.; Wang, Y.; Wang, Y.; Liu, D.; He, Y.; Wei, X.; Gu, H.; Ma, W.; Luo, W.; et al. PIWI-interacting RNA sequencing profiles in maternal plasma-derived exosomes reveal novel non-invasive prenatal biomarkers for the early diagnosis of nonsyndromic cleft lip and palate. EBioMedicine 2021, 65, 103253. [Google Scholar] [CrossRef] [PubMed]
- Fame, R.M.; Shannon, M.L.; Chau, K.F.; Head, J.P.; Lehtinen, M.K. A concerted metabolic shift in early forebrain alters the CSF proteome and depends on MYC downregulation for mitochondrial maturation. Development 2019, 146, dev182857. [Google Scholar] [CrossRef] [PubMed]
- Guo, T.; Tang, X.H.; Gao, X.Y.; Zhou, Y.; Jin, B.; Deng, Z.Q.; Hu, Y.; Xing, X.F.; Li, Z.Y.; Ji, J.F. A liquid biopsy signature of circulating exosome-derived mRNAs, miRNAs and lncRNAs predict therapeutic efficacy to neoadjuvant chemotherapy in patients with advanced gastric cancer. Mol. Cancer 2022, 21, 216. [Google Scholar] [CrossRef] [PubMed]
- Aharon, A.; Rebibo-Sabbah, A.; Ahmad, R.S.; Dangot, A.; Bar-Lev, T.H.; Brenner, B.; Cohen, A.H.; David, C.B.; Weiner, Z.; Solt, I. Associations of maternal and placental extracellular vesicle miRNA with preeclampsia. Front. Cell Dev. Biol. 2023, 11, 1080419. [Google Scholar] [CrossRef] [PubMed]
- Arenaccio, C.; Chiozzini, C.; Ferrantelli, F.; Leone, P.; Olivetta, E.; Federico, M. Exosomes in Therapy: Engineering, Pharmacokinetics and Future Applications. Curr. Drug Targets 2019, 20, 87–95. [Google Scholar] [CrossRef] [PubMed]
- Jia, S.; Zhang, Q.; Wang, Y.; Wei, X.; Gu, H.; Liu, D.; Ma, W.; He, Y.; Luo, W.; Yuan, Z. Identification by RNA-Seq of let-7 clusters as prenatal biomarkers for nonsyndromic cleft lip with palate. Ann. N. Y. Acad. Sci. 2022, 1516, 234–246. [Google Scholar] [CrossRef]
- Chen, Q.; Xie, Y.; Dong, X.; Zhang, X.; Zhang, Y.; Yuan, X.; Ding, X.; Qiu, L. TCDD induces cleft palate through exosomes derived from mesenchymal cells. Toxicol. Res. 2022, 11, 901–910. [Google Scholar] [CrossRef]
- Fabietti, I.; Nardi, T.; Favero, C.; Dioni, L.; Cantone, L.; Pergoli, L.; Hoxha, M.; Pinatel, E.; Mosca, F.; Bollati, V.; et al. Extracellular Vesicles and Their miRNA Content in Amniotic and Tracheal Fluids of Fetuses with Severe Congenital Diaphragmatic Hernia Undergoing Fetal Intervention. Cells 2021, 10, 1493. [Google Scholar] [CrossRef]
- Tavanasefat, H.; Li, F.; Koyano, K.; Gourtani, B.K.; Marty, V.; Mulpuri, Y.; Lee, S.H.; Shin, K.H.; Wong, D.T.W.; Xiao, X.; et al. Molecular consequences of fetal alcohol exposure on amniotic exosomal miRNAs with functional implications for stem cell potency and differentiation. PLoS ONE 2020, 15, e0242276. [Google Scholar] [CrossRef]
- Bush, J.O.; Jiang, R. Palatogenesis: Morphogenetic and molecular mechanisms of secondary palate development. Development 2012, 139, 231–243. [Google Scholar] [CrossRef]
- Hammond, N.L.; Dixon, M.J. Revisiting the embryogenesis of lip and palate development. Oral Dis. 2022, 28, 1306–1326. [Google Scholar] [CrossRef] [PubMed]
- Peng, X.; Chen, J.; Wang, Y.; Wang, X.; Zhao, X.; Zheng, X.; Wang, Z.; Yuan, D.; Du, J. Osteogenic microenvironment affects palatal development through glycolysis. Differ. Res. Biol. Divers. 2023, 133, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Rodriguez, G.; Han, X.; Janečková, E.; Kahng, S.; Song, B.; Chai, Y. Regulatory Mechanisms of Soft Palate Development and Malformations. J. Dent. Res. 2019, 98, 959–967. [Google Scholar] [CrossRef] [PubMed]
- Ozekin, Y.H.; O’Rourke, R.; Bates, E.A. Single cell sequencing of the mouse anterior palate reveals mesenchymal heterogeneity. Dev. Dyn. Off. Publ. Am. Assoc. Anat. 2023, 252, 713–727. [Google Scholar] [CrossRef] [PubMed]
- Cesario, J.M.; Landin Malt, A.; Deacon, L.J.; Sandberg, M.; Vogt, D.; Tang, Z.; Zhao, Y.; Brown, S.; Rubenstein, J.L.; Jeong, J. Lhx6 and Lhx8 promote palate development through negative regulation of a cell cycle inhibitor gene, p57Kip2. Hum. Mol. Genet. 2015, 24, 5024–5039. [Google Scholar] [CrossRef] [PubMed]
- Li, N.; Liu, J.; Liu, H.; Wang, S.; Hu, P.; Zhou, H.; Xiao, J.; Liu, C. Altered BMP-Smad4 signaling causes complete cleft palate by disturbing osteogenesis in palatal mesenchyme. J. Mol. Histol. 2021, 52, 45–61. [Google Scholar] [CrossRef]
- Cuervo, R.; Covarrubias, L. Death is the major fate of medial edge epithelial cells and the cause of basal lamina degradation during palatogenesis. Development 2004, 131, 15–24. [Google Scholar] [CrossRef] [PubMed]
- Li, A.; Jia, P.; Mallik, S.; Fei, R.; Yoshioka, H.; Suzuki, A.; Iwata, J.; Zhao, Z. Critical microRNAs and regulatory motifs in cleft palate identified by a conserved miRNA-TF-gene network approach in humans and mice. Brief. Bioinform. 2020, 21, 1465–1478. [Google Scholar] [CrossRef]
- Yan, F.; Simon, L.M.; Suzuki, A.; Iwaya, C.; Jia, P.; Iwata, J.; Zhao, Z. Spatiotemporal MicroRNA-Gene Expression Network Related to Orofacial Clefts. J. Dent. Res. 2022, 101, 1398–1407. [Google Scholar] [CrossRef]
- Yu, D.; Lim, J.; Wang, X.; Liang, F.; Xiao, G. Enhanced construction of gene regulatory networks using hub gene information. BMC Bioinform. 2017, 18, 186. [Google Scholar] [CrossRef]
- Tatullo, M.; Marrelli, B.; Zullo, M.J.; Codispoti, B.; Paduano, F.; Benincasa, C.; Fortunato, F.; Scacco, S.; Zavan, B.; Cocco, T. Exosomes from Human Periapical Cyst-MSCs: Theranostic Application in Parkinson’s Disease. Int. J. Med. Sci. 2020, 17, 657–663. [Google Scholar] [CrossRef]
- Li, R.; Sun, Y.; Chen, Z.; Zheng, M.; Shan, Y.; Ying, X.; Weng, M.; Chen, Z. The Fibroblast Growth Factor 9 (Fgf9) Participates in Palatogenesis by Promoting Palatal Growth and Elevation. Front. Physiol. 2021, 12, 653040. [Google Scholar] [CrossRef] [PubMed]
- Nakajima, A.; Tanaka, E.; Ito, Y.; Maeno, M.; Iwata, K.; Shimizu, N.; Shuler, C.F. The expression of TGF-β3 for epithelial-mesenchyme transdifferentiated MEE in palatogenesis. J. Mol. Histol. 2010, 41, 343–355. [Google Scholar] [CrossRef] [PubMed]
- Tanahashi, H.; Tian, Q.B.; Hara, Y.; Sakagami, H.; Endo, S.; Suzuki, T. Polyhydramnios in Lrp4 knockout mice with bilateral kidney agenesis: Defects in the pathways of amniotic fluid clearance. Sci. Rep. 2016, 6, 20241. [Google Scholar] [CrossRef] [PubMed]
- Yang, H.; Ma, Q.; Wang, Y.; Tang, Z. Clinical application of exosomes and circulating microRNAs in the diagnosis of pregnancy complications and foetal abnormalities. J. Transl. Med. 2020, 18, 32. [Google Scholar] [CrossRef] [PubMed]
- Logan, S.M.; Ruest, L.B.; Benson, M.D.; Svoboda, K.K.H. Extracellular Matrix in Secondary Palate Development. Anat. Rec. 2020, 303, 1543–1556. [Google Scholar] [CrossRef]
- Ozturk, F.; Li, Y.; Zhu, X.; Guda, C.; Nawshad, A. Systematic analysis of palatal transcriptome to identify cleft palate genes within TGFβ3-knockout mice alleles: RNA-Seq analysis of TGFβ3 Mice. BMC Genom. 2013, 14, 113. [Google Scholar] [CrossRef]
- Takenoshita, M.; Takechi, M.; Vu Hoang, T.; Furutera, T.; Akagawa, C.; Namangkalakul, W.; Aoto, K.; Kume, T.; Miyashin, M.; Iwamoto, T.; et al. Cell lineage- and expression-based inference of the roles of forkhead box transcription factor Foxc2 in craniofacial development. Dev. Dyn. Off. Publ. Am. Assoc. Anat. 2021, 250, 1125–1139. [Google Scholar] [CrossRef]
- Nandadasa, S.; Kraft, C.M.; Wang, L.W.; O’Donnell, A.; Patel, R.; Gee, H.Y.; Grobe, K.; Cox, T.C.; Hildebrandt, F.; Apte, S.S. Secreted metalloproteases ADAMTS9 and ADAMTS20 have a non-canonical role in ciliary vesicle growth during ciliogenesis. Nat. Commun. 2019, 10, 953. [Google Scholar] [CrossRef]
- Yuan, G.; Singh, G.; Chen, S.; Perez, K.C.; Wu, Y.; Liu, B.; Helms, J.A. Cleft Palate and Aglossia Result From Perturbations in Wnt and Hedgehog Signaling. Cleft Palate-Craniofacial J. Off. Publ. Am. Cleft Palate-Craniofacial Assoc. 2017, 54, 269–280. [Google Scholar] [CrossRef]
- Moore, E.R. Primary Cilia: The New Face of Craniofacial Research. Biomolecules 2022, 12, 1724. [Google Scholar] [CrossRef] [PubMed]
- Masyuk, A.I.; Huang, B.Q.; Ward, C.J.; Gradilone, S.A.; Banales, J.M.; Masyuk, T.V.; Radtke, B.; Splinter, P.L.; LaRusso, N.F. Biliary exosomes influence cholangiocyte regulatory mechanisms and proliferation through interaction with primary cilia. Am. J. Physiol. Gastrointest. Liver Physiol. 2010, 299, G990–G999. [Google Scholar] [CrossRef] [PubMed]
- Sun, S.; Fisher, R.L.; Bowser, S.S.; Pentecost, B.T.; Sui, H. Three-dimensional architecture of epithelial primary cilia. Proc. Natl. Acad. Sci. USA 2019, 116, 9370–9379. [Google Scholar] [CrossRef] [PubMed]
- Wood, C.R.; Huang, K.; Diener, D.R.; Rosenbaum, J.L. The cilium secretes bioactive ectosomes. Curr. Biol. 2013, 23, 906–911. [Google Scholar] [CrossRef] [PubMed]
- Ikegami, K.; Ijaz, F. Current understandings of the relationship between extracellular vesicles and cilia. J. Biochem. 2021, 169, 139–145. [Google Scholar] [CrossRef] [PubMed]
- Suttorp, C.M.; Cremers, N.A.; van Rheden, R.; Regan, R.F.; Helmich, P.; van Kempen, S.; Kuijpers-Jagtman, A.M.; Wagener, F. Chemokine Signaling during Midline Epithelial Seam Disintegration Facilitates Palatal Fusion. Front. Cell Dev. Biol. 2017, 5, 94. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.; Chen, J.; Yuan, D.; Sun, L.; Fan, Z.; Wang, S.; Du, J. Dynamic mRNA Expression Analysis of the Secondary Palatal Morphogenesis in Miniature Pigs. Int. J. Mol. Sci. 2019, 20, 4284. [Google Scholar] [CrossRef] [PubMed]
- Cavallucci, V.; Bisicchia, E.; Cencioni, M.T.; Ferri, A.; Latini, L.; Nobili, A.; Biamonte, F.; Nazio, F.; Fanelli, F.; Moreno, S.; et al. Acute focal brain damage alters mitochondrial dynamics and autophagy in axotomized neurons. Cell Death Dis. 2014, 5, e1545. [Google Scholar] [CrossRef]
- Ljubojevic, S.; Radulovic, S.; Leitinger, G.; Sedej, S.; Sacherer, M.; Holzer, M.; Winkler, C.; Pritz, E.; Mittler, T.; Schmidt, A.; et al. Early remodeling of perinuclear Ca2+ stores and nucleoplasmic Ca2+ signaling during the development of hypertrophy and heart failure. Circulation 2014, 130, 244–255. [Google Scholar] [CrossRef]
- Sun, B.; Liu, Y.; Huang, W.; Zhang, Q.; Lin, J.; Li, W.; Zhang, J.; Chen, F. Functional identification of a rare vascular endothelial growth factor a (VEGFA) variant associating with the nonsyndromic cleft lip with/without cleft palate. Bioengineered 2021, 12, 1471–1483. [Google Scholar] [CrossRef]
- Wang, Y.; Chen, J.; Wang, X.; Guo, C.; Peng, X.; Liu, Y.; Li, T.; Du, J. Novel investigations in retinoic-acid-induced cleft palate about the gut microbiome of pregnant mice. Front. Cell. Infect. Microbiol. 2022, 12, 1042779. [Google Scholar] [CrossRef] [PubMed]
- Dong, S.; Zhang, Y.; Huang, H. Involvement of RBP4 in all-trans retinoic acid induced cleft palate. Mol. Med. Rep. 2017, 16, 5915–5923. [Google Scholar] [CrossRef]
- Rot, I.; Kablar, B. Role of skeletal muscle in palate development. Histol. Histopathol. 2013, 28, 1–13. [Google Scholar] [PubMed]
- Vaiman, A.; Fritz, S.; Beauvallet, C.; Boussaha, M.; Grohs, C.; Daniel-Carlier, N.; Relun, A.; Boichard, D.; Vilotte, J.L.; Duchesne, A. Mutation of the MYH3 gene causes recessive cleft palate in Limousine cattle. Genet. Sel. Evol. 2022, 54, 71. [Google Scholar] [CrossRef] [PubMed]
- Dudas, M.; Kim, J.; Li, W.Y.; Nagy, A.; Larsson, J.; Karlsson, S.; Chai, Y.; Kaartinen, V. Epithelial and ectomesenchymal role of the type I TGF-beta receptor ALK5 during facial morphogenesis and palatal fusion. Dev. Biol. 2006, 296, 298–314. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Y.; Zhao, R.; Lyu, Y.; Shi, H.; Ye, W.; Tan, Y.; Li, R.; Xu, Y. Serum and Amniotic Fluid Metabolic Profile Changes in Response to Gestational Diabetes Mellitus and the Association with Maternal-Fetal Outcomes. Nutrients 2021, 13, 3644. [Google Scholar] [CrossRef] [PubMed]
- Lee, A.M.; Hu, J.; Xu, Y.; Abraham, A.G.; Xiao, R.; Coresh, J.; Rebholz, C.; Chen, J.; Rhee, E.P.; Feldman, H.I.; et al. Using Machine Learning to Identify Metabolomic Signatures of Pediatric Chronic Kidney Disease Etiology. J. Am. Soc. Nephrol. 2022, 33, 375–386. [Google Scholar] [CrossRef] [PubMed]
- Tatullo, M.; Codispoti, B.; Paduano, F.; Nuzzolese, M.; Makeeva, I. Strategic Tools in Regenerative and Translational Dentistry. Int. J. Mol. Sci. 2019, 20, 1879. [Google Scholar] [CrossRef]
- Biggs, L.C.; Goudy, S.L.; Dunnwald, M. Palatogenesis and cutaneous repair: A two-headed coin. Dev. Dyn. Off. Publ. Am. Assoc. Anat. 2015, 244, 289–310. [Google Scholar] [CrossRef]
- Chen, M.; Xie, Y.; Luo, Y.; Xie, Y.; Wu, N.; Peng, S.; Chen, Q. Exosomes-a potential indicator and mediator of cleft lip and palate: A narrative review. Ann. Transl. Med. 2021, 9, 1485. [Google Scholar] [CrossRef]
- Khan, N.Z.; Cao, T.; He, J.; Ritzel, R.M.; Li, Y.; Henry, R.J.; Colson, C.; Stoica, B.A.; Faden, A.I.; Wu, J. Spinal cord injury alters microRNA and CD81+ exosome levels in plasma extracellular nanoparticles with neuroinflammatory potential. Brain Behav. Immun. 2021, 92, 165–183. [Google Scholar] [CrossRef] [PubMed]
- Chen, J.; Yao, Y.; Wang, Y.; Wang, X.; Peng, X.; Li, T.; Liu, Y.; Du, J. Autophagy triggered by the ROS/ERK signaling pathway protects mouse embryonic palatal cells from apoptosis induced by nicotine. Environ. Sci. Pollut. Res. Int. 2022, 29, 81909–81922. [Google Scholar] [CrossRef] [PubMed]




Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhao, X.; Peng, X.; Wang, Z.; Zheng, X.; Wang, X.; Wang, Y.; Chen, J.; Yuan, D.; Liu, Y.; Du, J. MicroRNAs in Small Extracellular Vesicles from Amniotic Fluid and Maternal Plasma Associated with Fetal Palate Development in Mice. Int. J. Mol. Sci. 2023, 24, 17173. https://doi.org/10.3390/ijms242417173
Zhao X, Peng X, Wang Z, Zheng X, Wang X, Wang Y, Chen J, Yuan D, Liu Y, Du J. MicroRNAs in Small Extracellular Vesicles from Amniotic Fluid and Maternal Plasma Associated with Fetal Palate Development in Mice. International Journal of Molecular Sciences. 2023; 24(24):17173. https://doi.org/10.3390/ijms242417173
Chicago/Turabian StyleZhao, Xige, Xia Peng, Zhiwei Wang, Xiaoyu Zheng, Xiaotong Wang, Yijia Wang, Jing Chen, Dong Yuan, Ying Liu, and Juan Du. 2023. "MicroRNAs in Small Extracellular Vesicles from Amniotic Fluid and Maternal Plasma Associated with Fetal Palate Development in Mice" International Journal of Molecular Sciences 24, no. 24: 17173. https://doi.org/10.3390/ijms242417173
APA StyleZhao, X., Peng, X., Wang, Z., Zheng, X., Wang, X., Wang, Y., Chen, J., Yuan, D., Liu, Y., & Du, J. (2023). MicroRNAs in Small Extracellular Vesicles from Amniotic Fluid and Maternal Plasma Associated with Fetal Palate Development in Mice. International Journal of Molecular Sciences, 24(24), 17173. https://doi.org/10.3390/ijms242417173




