The Homeodomain–Leucine Zipper Subfamily I Contributes to Leaf Age- and Time-Dependent Resistance to Pathogens in Arabidopsis thaliana
Abstract
:1. Introduction
2. Results
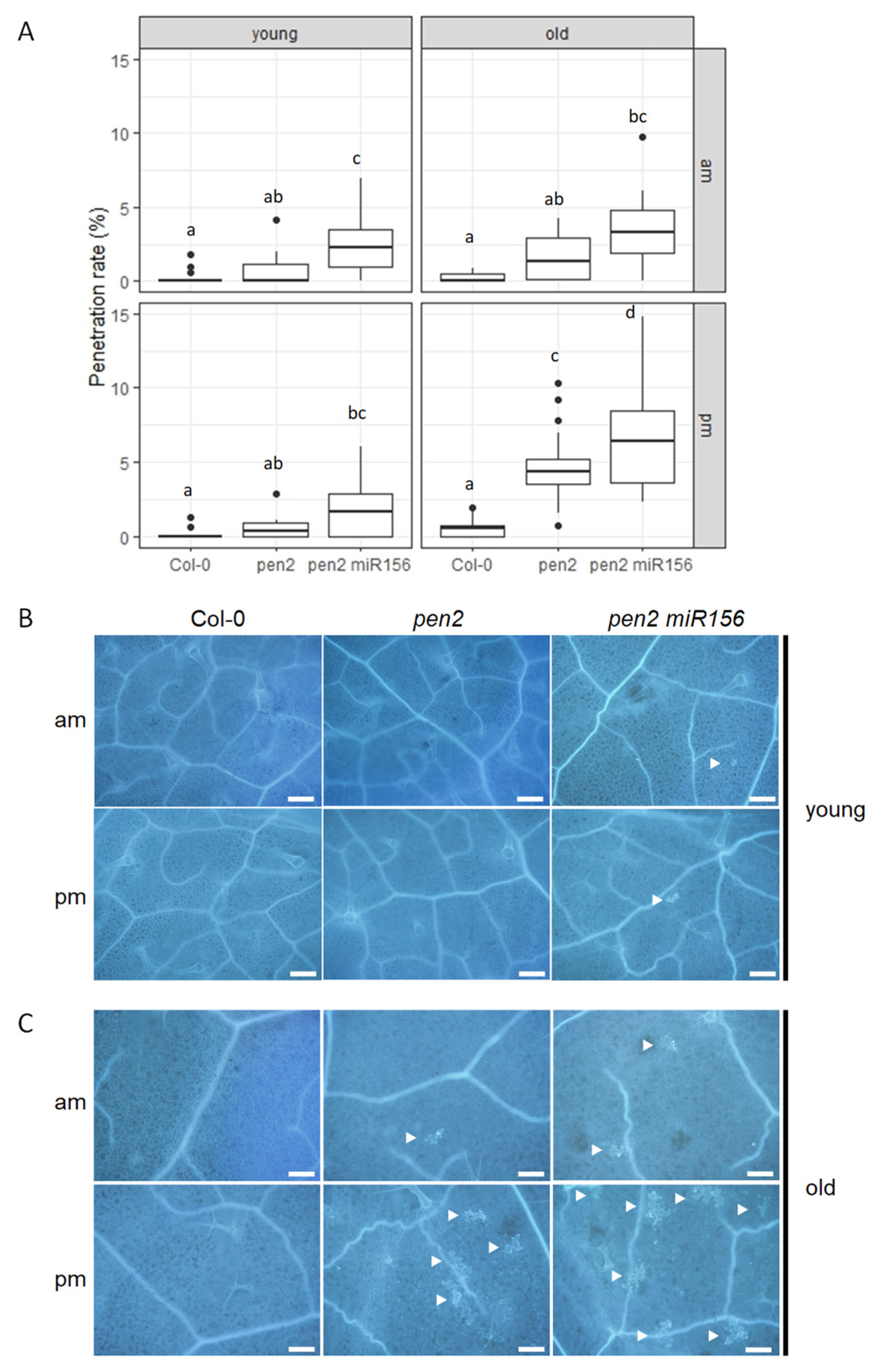
2.1. Nonhost Resistance to Pyricularia oryzae in Arabidopsis thaliana pen2 35S::miR156a Plants
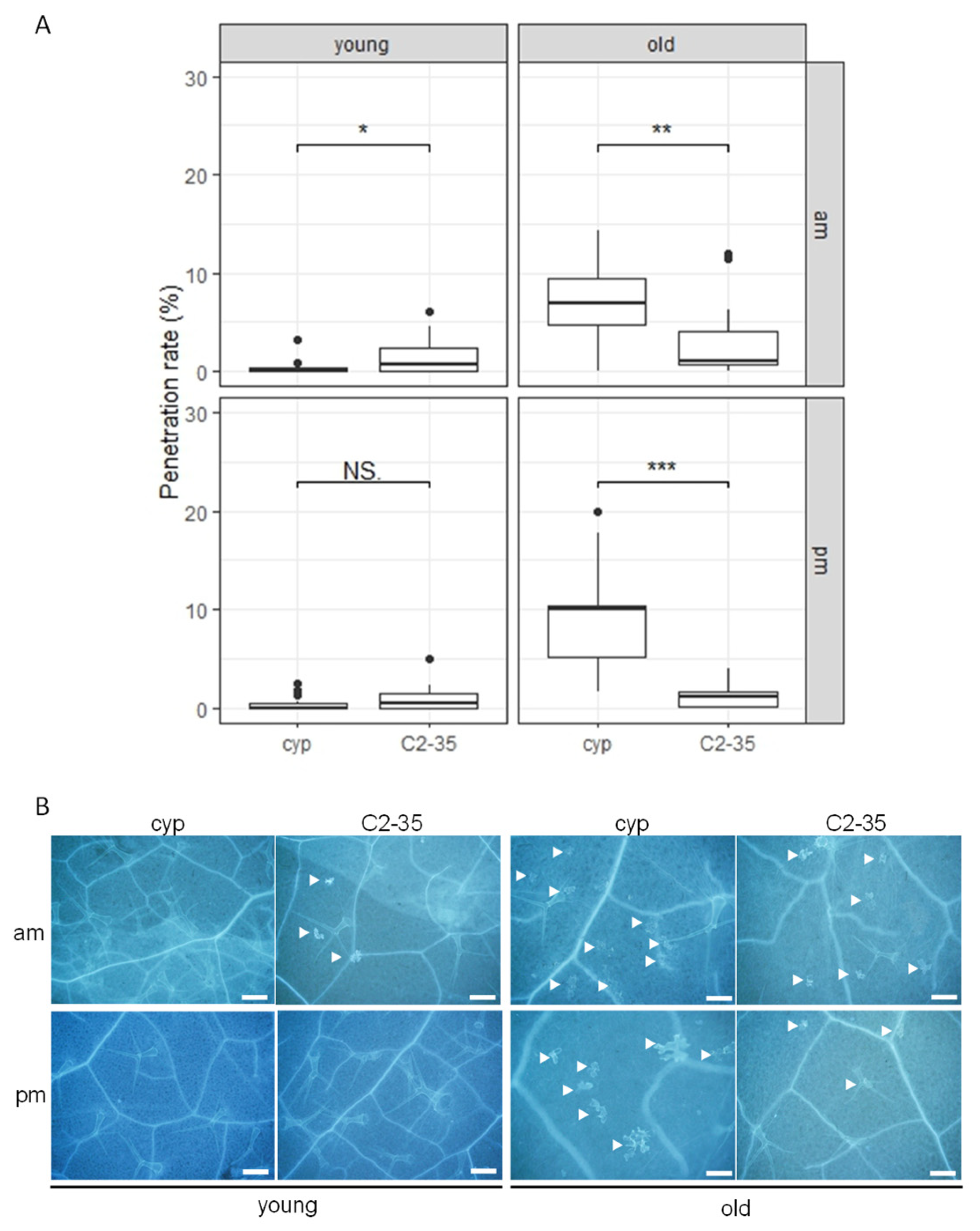
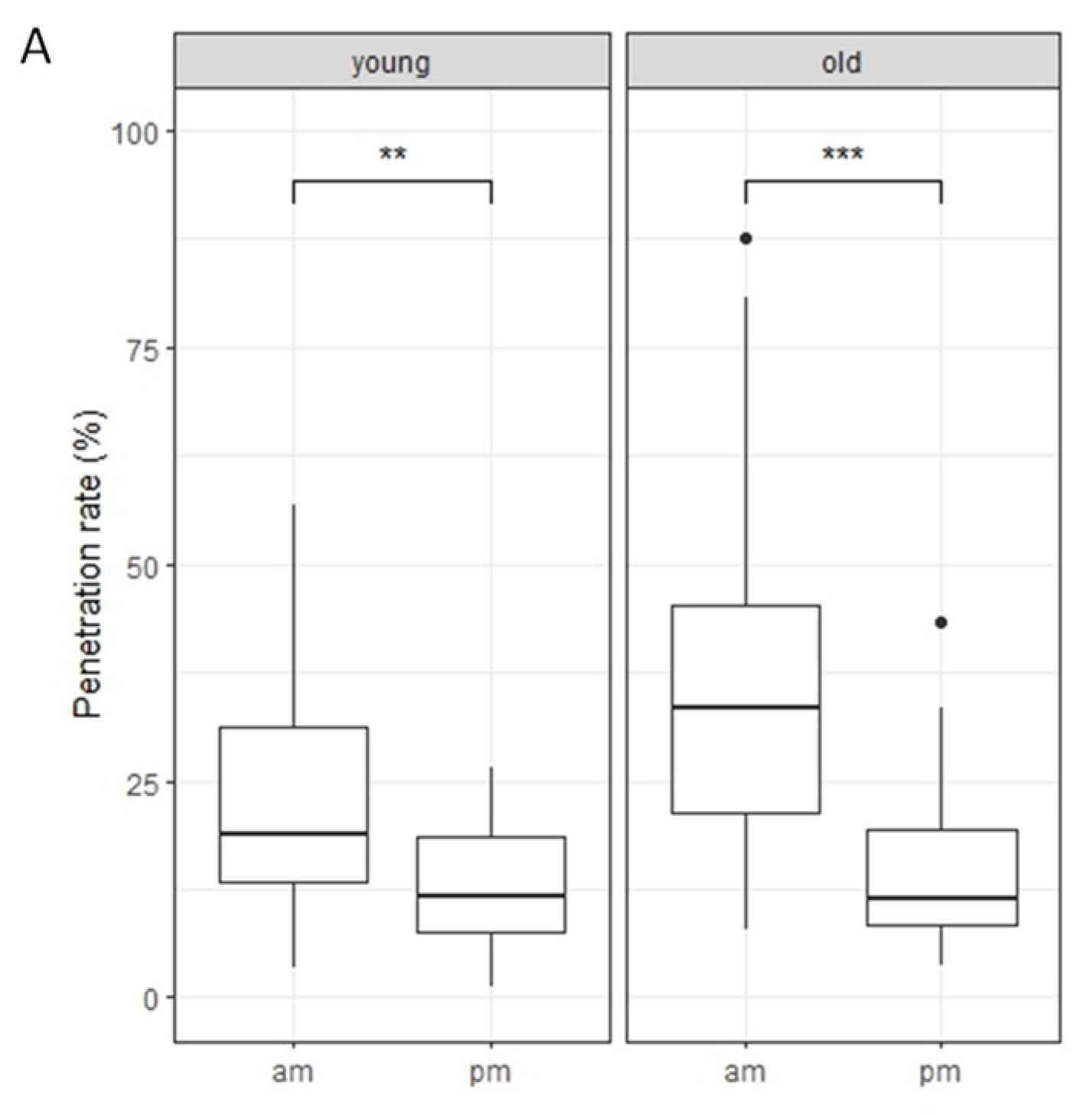
2.2. Nonhost Resistance to Pyricularia oryzae in Rice-FOX Arabidopsis C2-35 Plants
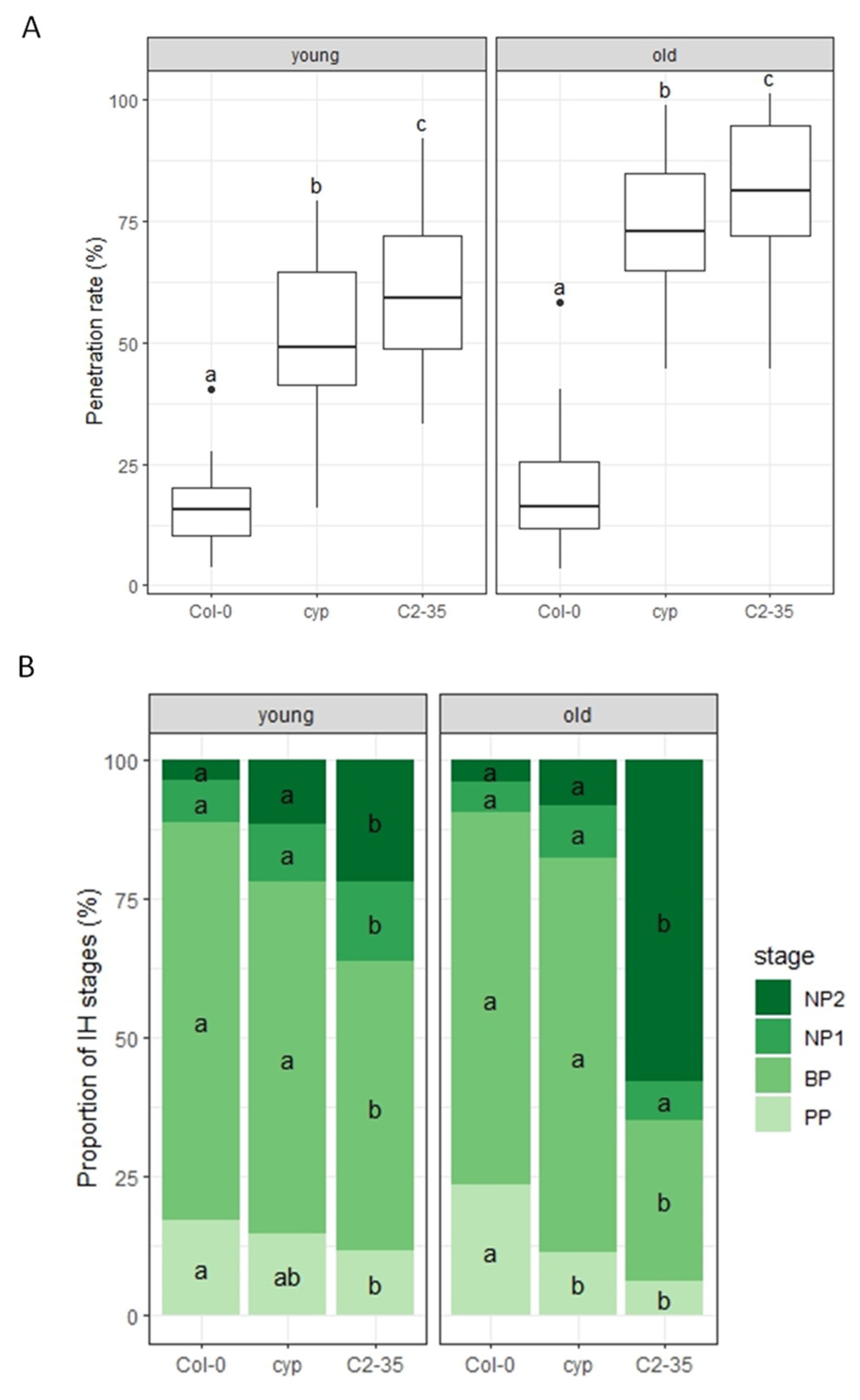
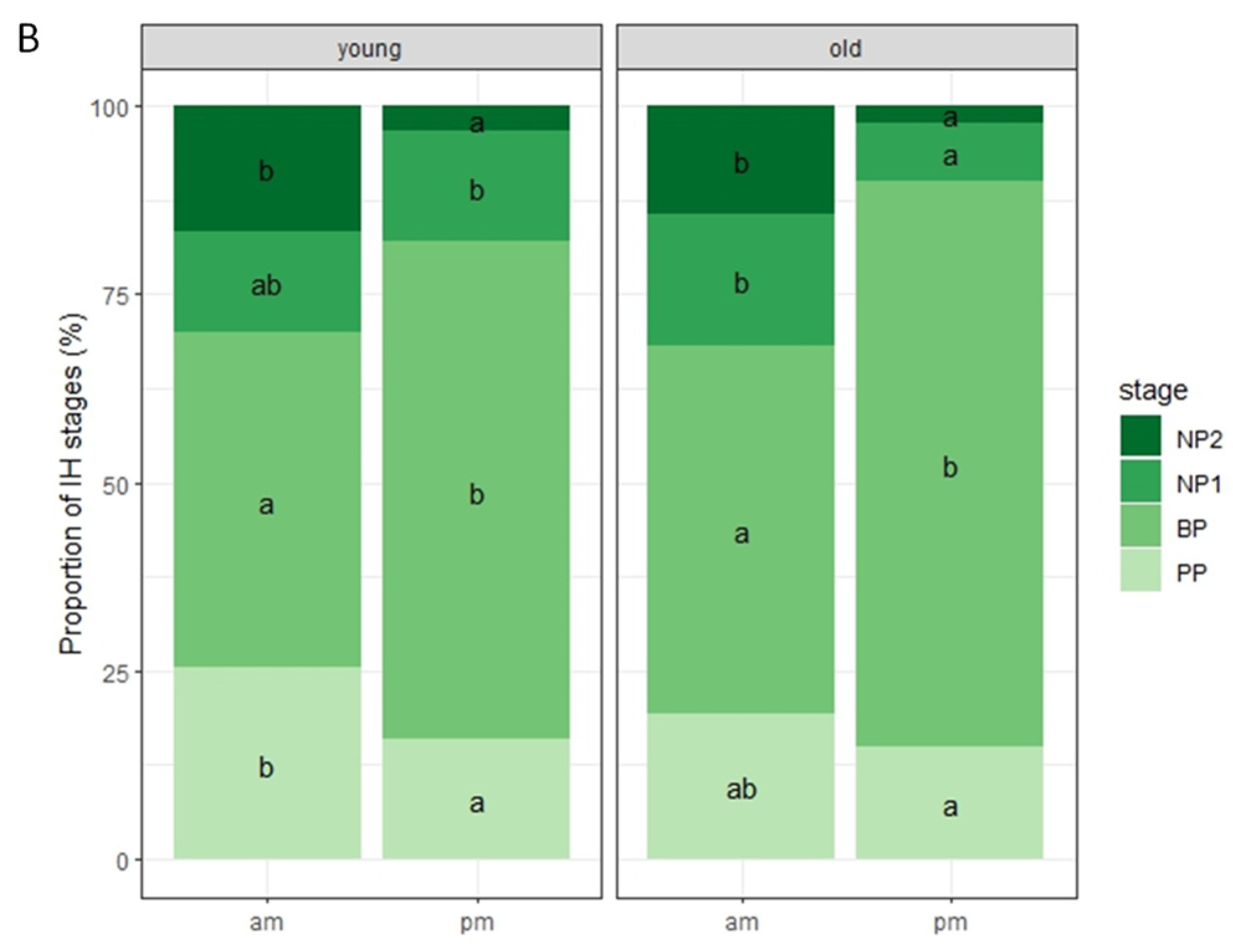
2.3. Nonhost Resistance to Colletotrichum nymphaeae in Rice-FOX Arabidopsis C2-35 Plants
2.4. Host Resistance to Colletotrichum higginsianum in Rice-FOX Arabidopsis C2-35 Plants
2.5. Host Resistance to C. higginsianum in Col-0 Plants
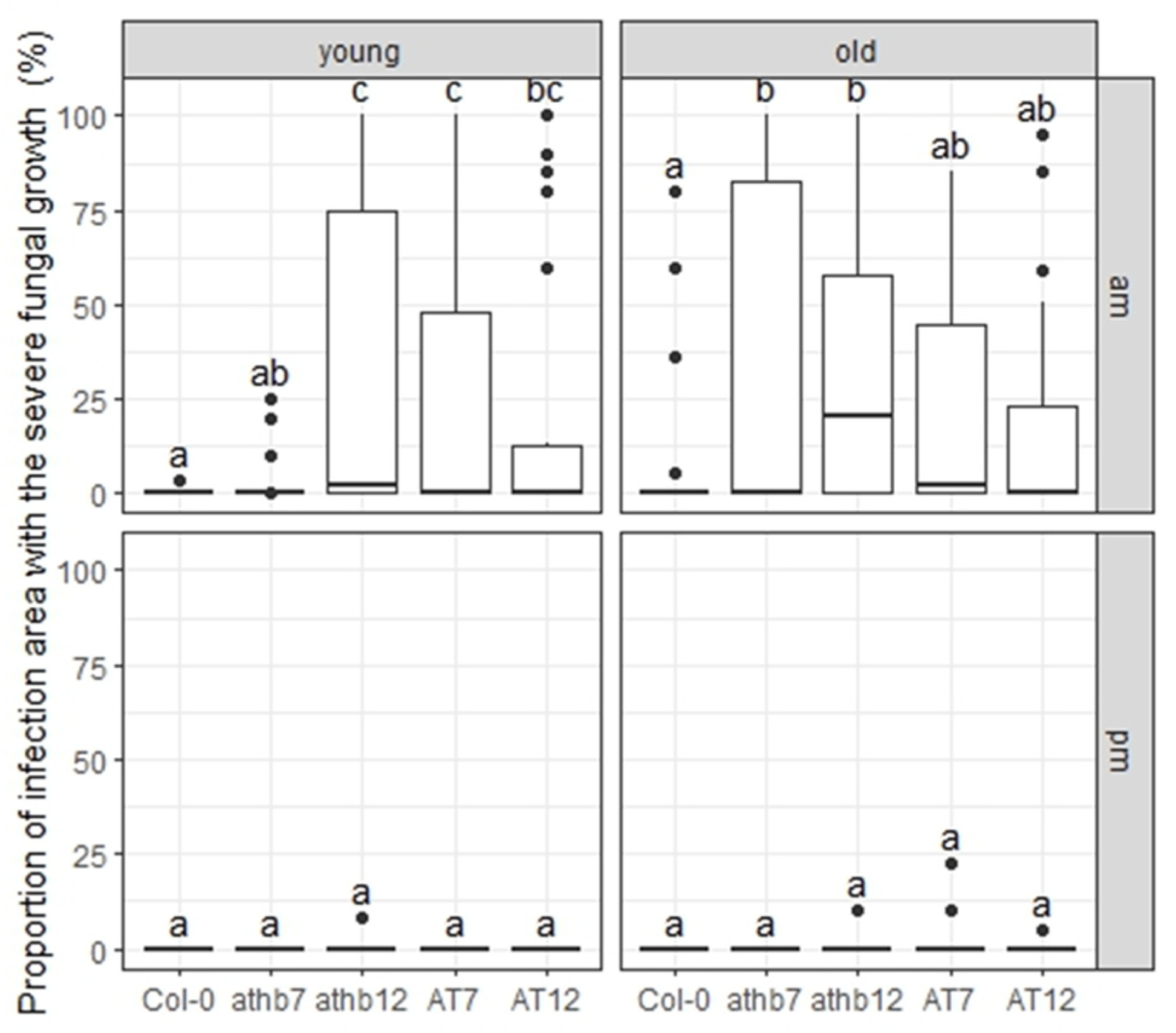
2.6. C. higginsianum Growth in Arabidopsis Mutant Plants
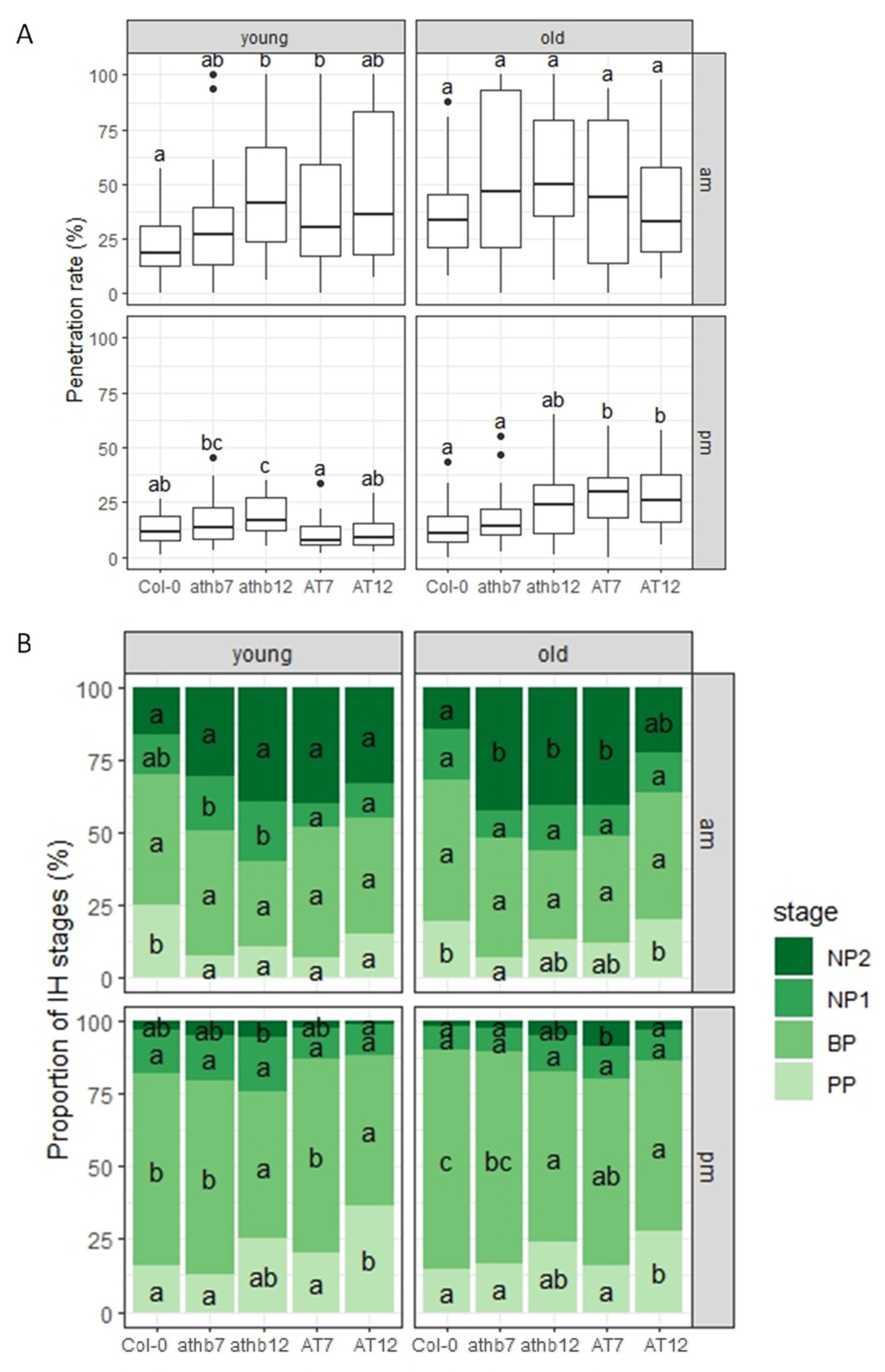
2.7. Host Resistance to C. higginsianum in Arabidopsis Mutant Plants
3. Discussion
4. Materials and Methods
4.1. Plant Materials and Growth Conditions
4.2. Fungal Strains and Media
4.3. Fungal Inoculation
4.4. Rice-FOX Arabidopsis cyp79b2 cyp79b3 Lines and P. oryzae Screening
Supplementary Materials
Author Contributions
Funding
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Scholthof, K.B. The disease triangle: Pathogens, the environment and society. Nat. Rev. Microbiol. 2007, 5, 152–156. [Google Scholar] [CrossRef] [PubMed]
- Roden, L.C.; Ingle, R.A. Lights, rhythms, infection: The role of light and the circadian clock in determining the outcome of plant-pathogen interactions. Plant Cell 2009, 21, 2546–2552. [Google Scholar] [CrossRef] [PubMed]
- Seo, P.J.; Mas, P. STRESSing the role of the plant circadian clock. Trends Plant Sci. 2015, 20, 230–237. [Google Scholar] [CrossRef] [PubMed]
- Bhardwaj, V.; Meier, S.; Petersen, L.N.; Ingle, R.A.; Roden, L.C. Defence responses of Arabidopsis thaliana to infection by Pseudomonas syringae are regulated by the circadian clock. PLoS ONE 2011, 6, e26968. [Google Scholar] [CrossRef]
- Wang, W.; Barnaby, J.Y.; Tada, Y.; Li, H.; Tor, M.; Caldelari, D.; Lee, D.U.; Fu, X.D.; Dong, X. Timing of plant immune responses by a central circadian regulator. Nature 2011, 470, 110–114. [Google Scholar] [CrossRef]
- Zhang, C.; Xie, Q.; Anderson, R.G.; Ng, G.; Seitz, N.C.; Peterson, T.; McClung, C.R.; McDowell, J.M.; Kong, D.; Kwak, J.M.; et al. Crosstalk between the circadian clock and innate immunity in Arabidopsis. PLoS Pathog. 2013, 9, e1003370. [Google Scholar] [CrossRef]
- Develey-Riviere, M.P.; Galiana, E. Resistance to pathogens and host developmental stage: A multifaceted relationship within the plant kingdom. New Phytol. 2007, 175, 405–416. [Google Scholar] [CrossRef]
- Thomas, H.; Ougham, H.J.; Wagstaff, C.; Stead, A.D. Defining senescence and death. J. Exp. Bot. 2003, 54, 1127–1132. [Google Scholar] [CrossRef]
- Rankenberg, T.; Geldhof, B.; van Veen, H.; Holsteens, K.; Van de Poel, B.; Sasidharan, R. Age-Dependent Abiotic Stress Resilience in Plants. Trends Plant Sci. 2021, 26, 692–705. [Google Scholar] [CrossRef]
- Hu, L.; Yang, L. Time to Fight: Molecular Mechanisms of Age-Related Resistance. Phytopathology 2019, 109, 1500–1508. [Google Scholar] [CrossRef]
- Wu, G.; Poethig, R.S. Temporal regulation of shoot development in Arabidopsis thaliana by miR156 and its target SPL3. Development 2006, 133, 3539–3547. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.W.; Czech, B.; Weigel, D. miR156-regulated SPL transcription factors define an endogenous flowering pathway in Arabidopsis thaliana. Cell 2009, 138, 738–749. [Google Scholar] [CrossRef] [PubMed]
- Li, P.; Lu, Y.J.; Chen, H.; Day, B. The Lifecycle of the Plant Immune System. CRC Crit. Rev. Plant Sci. 2020, 39, 72–100. [Google Scholar] [CrossRef]
- Xu, Y.P.; Lv, L.H.; Xu, Y.J.; Yang, J.; Cao, J.Y.; Cai, X.Z. Leaf stage-associated resistance is correlated with phytohormones in a pathosystem-dependent manner. J. Integr. Plant Biol. 2018, 60, 703–722. [Google Scholar] [CrossRef] [PubMed]
- Hu, L.; Qi, P.; Peper, A.; Kong, F.; Yao, Y.; Yang, L. Distinct function of SPL genes in age-related resistance in Arabidopsis. PLoS Pathog. 2023, 19, e1011218. [Google Scholar] [CrossRef] [PubMed]
- Maeda, K.; Houjyou, Y.; Komatsu, T.; Hori, H.; Kodaira, T.; Ishikawa, A. AGB1 and PMR5 contribute to PEN2-mediated preinvasion resistance to Magnaporthe oryzae in Arabidopsis thaliana. Mol. Plant-Microbe Interact. 2009, 22, 1331–1340. [Google Scholar] [CrossRef]
- Nakao, M.; Nakamura, R.; Kita, K.; Inukai, R.; Ishikawa, A. Non-host resistance to penetration and hyphal growth of Magnaporthe oryzae in Arabidopsis. Sci. Rep. 2011, 1, 171. [Google Scholar] [CrossRef]
- Lipka, V.; Dittgen, J.; Bednarek, P.; Bhat, R.; Wiermer, M.; Stein, M.; Landtag, J.; Brandt, W.; Rosahl, S.; Scheel, D.; et al. Pre- and postinvasion defenses both contribute to nonhost resistance in Arabidopsis. Science 2005, 310, 1180–1183. [Google Scholar] [CrossRef]
- Pastorczyk, M.; Bednarek, P. The Function of Glucosinolates and Related Metabolites in Plant Innate Immunity. Glucosinolates 2016, 80, 171–198. [Google Scholar] [CrossRef]
- Zhao, Y.D.; Hull, A.K.; Gupta, N.R.; Goss, K.A.; Alonso, J.; Ecker, J.R.; Normanly, J.; Chory, J.; Celenza, J.L. Trp-dependent auxin biosynthesis in Arabidopsis: Involvement of cytochrome P450s CYP79B2 and CYP79B3. Genes Dev. 2002, 16, 3100–3112. [Google Scholar] [CrossRef]
- Sanchez-Vallet, A.; Ramos, B.; Bednarek, P.; Lopez, G.; Pislewska-Bednarek, M.; Schulze-Lefert, P.; Molina, A. Tryptophan-derived secondary metabolites in Arabidopsis thaliana confer non-host resistance to necrotrophic Plectosphaerella cucumerina fungi. Plant J. 2010, 63, 115–127. [Google Scholar] [CrossRef]
- Hiruma, K.; Fukunaga, S.; Bednarek, P.; Pislewska-Bednarek, M.; Watanabe, S.; Narusaka, Y.; Shirasu, K.; Takano, Y. Glutathione and tryptophan metabolism are required for Arabidopsis immunity during the hypersensitive response to hemibiotrophs. Proc. Natl. Acad. Sci. USA 2013, 110, 9589–9594. [Google Scholar] [CrossRef]
- Kosaka, A.; Pastorczyk, M.; Pislewska-Bednarek, M.; Nishiuchi, T.; Ono, E.; Suemoto, H.; Ishikawa, A.; Frerigmann, H.; Kaido, M.; Mise, K.; et al. Tryptophan-derived metabolites and BAK1 separately contribute to Arabidopsis postinvasive immunity against Alternaria brassicicola. Sci. Rep. 2021, 11, 1488. [Google Scholar] [CrossRef]
- Shimizu, S.; Yamauchi, Y.; Ishikawa, A. Photoperiod Following Inoculation of Arabidopsis with Pyricularia oryzae (syn. Magnaporthe oryzae) Influences on the Plant-Pathogen Interaction. Int. J. Mol. Sci. 2021, 22, 5004. [Google Scholar] [CrossRef]
- Yamauchi, Y.; Makihara, M.; Ishikawa, A. Leaf age and time of inoculation contribute to nonhost resistance to Pyricularia oryzae in Arabidopsis thaliana. Plant Biotechnol. 2017, 34, 207–210. [Google Scholar] [CrossRef] [PubMed]
- Yamaura, S.; Yamauchi, Y.; Makihara, M.; Yamashino, T.; Ishikawa, A. CCA1 and LHY contribute to nonhost resistance to Pyricularia oryzae (syn. Magnaporthe oryzae) in Arabidopsis thaliana. Biosci. Biotechnol. Biochem. 2020, 84, 76–84. [Google Scholar] [CrossRef] [PubMed]
- Maeda, N.; Noguchi, T.; Nakamichi, N.; Suzuki, T.; Ishikawa, A. Epidermal CCA1 and PMR5 contribute to nonhost resistance in Arabidopsis. Biosci. Biotechnol. Biochem. 2022, 86, 1623–1630. [Google Scholar] [CrossRef] [PubMed]
- Yan, Y.Q.; Yuan, Q.F.; Tang, J.T.; Huang, J.B.; Hsiang, T.; Wei, Y.D.; Zheng, L. Colletotrichum higginsianum as a Model for Understanding Host-Pathogen Interactions: A Review. Int. J. Mol. Sci. 2018, 19, 2142. [Google Scholar] [CrossRef]
- Jin, J.P.; Tian, F.; Yang, D.C.; Meng, Y.Q.; Kong, L.; Luo, J.C.; Gao, G. PlantTFDB 4.0: Toward a central hub for transcription factors and regulatory interactions in plants. Nucleic Acids Res. 2017, 45, D1040–D1045. [Google Scholar] [CrossRef]
- Ariel, F.D.; Manavella, P.A.; Dezar, C.A.; Chan, R.L. The true story of the HD-Zip family. Trends Plant Sci. 2007, 12, 419–426. [Google Scholar] [CrossRef] [PubMed]
- Perotti, M.F.; Ribone, P.A.; Chan, R.L. Plant transcription factors from the homeodomain-leucine zipper family I. Role in development and stress responses. IUBMB Life 2017, 69, 280–289. [Google Scholar] [CrossRef]
- Liu, Y.; Xu, J.; Guo, S.; Yuan, X.; Zhao, S.; Tian, H.; Dai, S.; Kong, X.; Ding, Z. AtHB7/12 Regulate Root Growth in Response to Aluminum Stress. Int. J. Mol. Sci. 2020, 21, 4080. [Google Scholar] [CrossRef]
- Agalou, A.; Purwantomo, S.; Overnas, E.; Johannesson, H.; Zhu, X.; Estiati, A.; de Kam, R.J.; Engstrom, P.; Slamet-Loedin, I.H.; Zhu, Z.; et al. A genome-wide survey of HD-Zip genes in rice and analysis of drought-responsive family members. Plant Mol. Biol. 2008, 66, 87–103. [Google Scholar] [CrossRef]
- Zhang, S.; Haider, I.; Kohlen, W.; Jiang, L.; Bouwmeester, H.; Meijer, A.H.; Schluepmann, H.; Liu, C.M.; Ouwerkerk, P.B. Function of the HD-Zip I gene Oshox22 in ABA-mediated drought and salt tolerances in rice. Plant Mol. Biol. 2012, 80, 571–585. [Google Scholar] [CrossRef]
- Bhattacharjee, A.; Sharma, R.; Jain, M. Over-Expression of OsHOX24 Confers Enhanced Susceptibility to Abiotic Stresses in Transgenic Rice via Modulating Stress-Responsive Gene Expression. Front. Plant Sci. 2017, 8, 628. [Google Scholar] [CrossRef] [PubMed]
- Re, D.A.; Capella, M.; Bonaventure, G.; Chan, R.L. Arabidopsis AtHB7 and AtHB12 evolved divergently to fine tune processes associated with growth and responses to water stress. BMC Plant Biol. 2014, 14, 150. [Google Scholar] [CrossRef]
- Ichikawa, T.; Nakazawa, M.; Kawashima, M.; Iizumi, H.; Kuroda, H.; Kondou, Y.; Tsuhara, Y.; Suzuki, K.; Ishikawa, A.; Seki, M.; et al. The FOX hunting system: An alternative gain-of-function gene hunting technique. Plant J. 2006, 48, 974–985. [Google Scholar] [CrossRef]
- Kondou, Y.; Higuchi, M.; Takahashi, S.; Sakurai, T.; Ichikawa, T.; Kuroda, H.; Yoshizumi, T.; Tsumoto, Y.; Horii, Y.; Kawashima, M.; et al. Systematic approaches to using the FOX hunting system to identify useful rice genes. Plant J. 2009, 57, 883–894. [Google Scholar] [CrossRef] [PubMed]
- Yokotani, N.; Ichikawa, T.; Kondou, Y.; Matsui, M.; Hirochika, H.; Iwabuchi, M.; Oda, K. Expression of rice heat stress transcription factor OsHsfA2e enhances tolerance to environmental stresses in transgenic Arabidopsis. Planta 2008, 227, 957–967. [Google Scholar] [CrossRef] [PubMed]
- Yokotani, N.; Ichikawa, T.; Kondou, Y.; Maeda, S.; Iwabuchi, M.; Mori, M.; Hirochika, H.; Matsui, M.; Oda, K. Overexpression of a rice gene encoding a small C2 domain protein OsSMCP1 increases tolerance to abiotic and biotic stresses in transgenic Arabidopsis. Plant Mol. Biol. 2009, 71, 391–402. [Google Scholar] [CrossRef]
- Yokotani, N.; Ichikawa, T.; Kondou, Y.; Iwabuchi, M.; Matsui, M.; Hirochika, H.; Oda, K. Role of the rice transcription factor JAmyb in abiotic stress response. J. Plant Res. 2013, 126, 131–139. [Google Scholar] [CrossRef] [PubMed]
- Dubouzet, J.G.; Maeda, S.; Sugano, S.; Ohtake, M.; Hayashi, N.; Ichikawa, T.; Kondou, Y.; Kuroda, H.; Horii, Y.; Matsui, M.; et al. Screening for resistance against Pseudomonas syringae in rice-FOX Arabidopsis lines identified a putative receptor-like cytoplasmic kinase gene that confers resistance to major bacterial and fungal pathogens in Arabidopsis and rice. Plant Biotechnol. J. 2011, 9, 466–485. [Google Scholar] [CrossRef]
- Irieda, H.; Takano, Y. Epidermal chloroplasts are defense-related motile organelles equipped with plant immune components. Nat. Commun. 2021, 12, 2739. [Google Scholar] [CrossRef] [PubMed]
- Hiruma, K.; Onozawa-Komori, M.; Takahashi, F.; Asakura, M.; Bednarek, P.; Okuno, T.; Schulze-Lefert, P.; Takano, Y. Entry mode-dependent function of an indole glucosinolate pathway in Arabidopsis for nonhost resistance against anthracnose pathogens. Plant Cell 2010, 22, 2429–2443. [Google Scholar] [CrossRef] [PubMed]
- Berens, M.L.; Wolinska, K.W.; Spaepen, S.; Ziegler, J.; Nobori, T.; Nair, A.; Kruler, V.; Winkelmuller, T.M.; Wang, Y.; Mine, A.; et al. Balancing trade-offs between biotic and abiotic stress responses through leaf age-dependent variation in stress hormone cross-talk. Proc. Natl. Acad. Sci. USA 2019, 116, 2364–2373. [Google Scholar] [CrossRef] [PubMed]
- Lee, Y.H.; Chun, J.Y. A new homeodomain-leucine zipper gene from Arabidopsis thaliana induced by water stress and abscisic acid treatment. Plant Mol. Biol. 1998, 37, 377–384. [Google Scholar] [CrossRef]
- Olsson, A.S.; Engstrom, P.; Soderman, E. The homeobox genes ATHB12 and ATHB7 encode potential regulators of growth in response to water deficit in Arabidopsis. Plant Mol. Biol. 2004, 55, 663–677. [Google Scholar] [CrossRef]
- Son, O.; Hur, Y.S.; Kim, Y.K.; Lee, H.J.; Kim, S.; Kim, M.R.; Nam, K.H.; Lee, M.S.; Kim, B.Y.; Park, J.; et al. ATHB12, an ABA-inducible homeodomain-leucine zipper (HD-Zip) protein of Arabidopsis, negatively regulates the growth of the inflorescence stem by decreasing the expression of a gibberellin 20-oxidase gene. Plant Cell Physiol. 2010, 51, 1537–1547. [Google Scholar] [CrossRef]
- Mishra, K.B.; Iannacone, R.; Petrozza, A.; Mishra, A.; Armentano, N.; La Vecchia, G.; Trtilek, M.; Cellini, F.; Nedbal, L. Engineered drought tolerance in tomato plants is reflected in chlorophyll fluorescence emission. Plant Sci. 2012, 182, 79–86. [Google Scholar] [CrossRef]
- Valdes, A.E.; Overnas, E.; Johansson, H.; Rada-Iglesias, A.; Engstrom, P. The homeodomain-leucine zipper (HD-Zip) class I transcription factors ATHB7 and ATHB12 modulate abscisic acid signalling by regulating protein phosphatase 2C and abscisic acid receptor gene activities. Plant Mol. Biol. 2012, 80, 405–418. [Google Scholar] [CrossRef]

Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Maeda, N.; Matsuta, F.; Noguchi, T.; Fujii, A.; Ishida, H.; Kitagawa, Y.; Ishikawa, A. The Homeodomain–Leucine Zipper Subfamily I Contributes to Leaf Age- and Time-Dependent Resistance to Pathogens in Arabidopsis thaliana. Int. J. Mol. Sci. 2023, 24, 16356. https://doi.org/10.3390/ijms242216356
Maeda N, Matsuta F, Noguchi T, Fujii A, Ishida H, Kitagawa Y, Ishikawa A. The Homeodomain–Leucine Zipper Subfamily I Contributes to Leaf Age- and Time-Dependent Resistance to Pathogens in Arabidopsis thaliana. International Journal of Molecular Sciences. 2023; 24(22):16356. https://doi.org/10.3390/ijms242216356
Chicago/Turabian StyleMaeda, Nami, Fuko Matsuta, Takaya Noguchi, Ayumu Fujii, Hikaru Ishida, Yudai Kitagawa, and Atsushi Ishikawa. 2023. "The Homeodomain–Leucine Zipper Subfamily I Contributes to Leaf Age- and Time-Dependent Resistance to Pathogens in Arabidopsis thaliana" International Journal of Molecular Sciences 24, no. 22: 16356. https://doi.org/10.3390/ijms242216356
APA StyleMaeda, N., Matsuta, F., Noguchi, T., Fujii, A., Ishida, H., Kitagawa, Y., & Ishikawa, A. (2023). The Homeodomain–Leucine Zipper Subfamily I Contributes to Leaf Age- and Time-Dependent Resistance to Pathogens in Arabidopsis thaliana. International Journal of Molecular Sciences, 24(22), 16356. https://doi.org/10.3390/ijms242216356





