Preclinical Models and Technologies in Glioblastoma Research: Evolution, Current State, and Future Avenues
Abstract
1. Introduction
2. Methods
3. Cell Lines Used in GBM Research
3.1. C6 Cell Line
3.2. 9L Cell Line
3.3. F98 Cell Line
3.4. RG2 Cell Line
3.5. GL261 Cell Line
3.6. CT2A Cell Line
3.7. U87 Cell Line
3.8. U251 Cell Line
4. Three-Dimensional In Vitro Models
4.1. GBM Spherical Cancer Models
4.2. GBM–Brain Organotypic Models (GBOMs)
4.3. Scaffolds
4.3.1. 3D Bioprinting
4.3.2. 4D Bioprinting
5. Microfluidics
5.1. Using Microfluidics to Isolate Circulating Tumor Cells
5.2. Microfluidics in Molecular Diagnostics
5.3. Efficacy Screening with Microfluidics
5.4. Microfluidics in Studying GBM Progression and Cell Localization
5.5. Microfluidics for Studying Extracellular Matrix Signaling
5.6. Microfluidic Devices in the Study of Cell–Cell Interactions
5.7. Microfluidics for Modeling Interactions with Vascular Flow, Hypoxia, and Angiogenesis
5.8. Microfluidics for Modeling Immune Cell Interactions
6. Animal Models Used for In Vivo GBM Research
6.1. Murine Models
6.2. Canine Models
6.3. Non-Human Primates
6.4. Other Animals
7. Generation and Applications of Available Animal Models
7.1. Grafting Tumor-Initiating Cells
7.2. Engineering Models with Spontaneously Arising Tumors
7.2.1. Genetically Engineered Models
7.2.2. Viral Vector-Induced Models
8. Conclusions and Future Perspectives
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| 5-ALA | 5-aminolevulenic acid |
| BBB | Blood–brain barrier |
| BNCT | boron neutron capture therapy |
| BRCA1 | breast cancer 1 gene |
| CAR | chimeric antigen receptor |
| CNS | central nervous system |
| CTC | circulating tumor cell |
| ECM | extracellular matrix |
| EGF | epidermal growth factor |
| EGFR | epidermal growth factor receptor |
| ESC | embryonic stem cells |
| EV | extracellular vesicles |
| FGF | fibroblast growth factor |
| GBM | glioblastoma |
| GBO | glioblastoma organoid |
| GBOM | GBM–Brain Organotypic Models |
| GFAP | glial fibrillary acidic protein |
| GliCO | glioma cerebral organoids |
| GSC | glioma stem cells |
| GVHD | graft versus host disease |
| HIF | hypoxia inducible factor |
| HSV-TK | herpes simplex virus–thymidine kinase |
| IDH | isocitrate dehydrogenase |
| IDH | isocitrate dehydrogenase |
| IGF | insulin-like growth factor |
| iPSC | induced pluripotent stem cells |
| MCTS | multicellular tumor spheroids |
| MGMT | O6-methylgunaine DNA methyltransferase |
| MMP | matrix metalloproteases |
| NHP | non-human primate |
| NK | natural killer |
| PBMC | peripheral blood mononuclear cells |
| PDGF | platelet-derived growth factor |
| PDMS | poly-dimethylsiloxane |
| PDX | patient-derived xenograft |
| PTEN | phosphatase and tensin homolog |
| PVN | perivascular niche |
| RCAS | avian leukosis sarcoma virus splice-acceptor system |
| SCID | severe combined immunodeficient |
| SCM | spherical cancer models |
| STING | stimulator of interferon genes |
| TME | tumor microenvironment |
| TMZ | temozolomide |
| VEGF | vascular endothelial growth factor |
References
- Ostrom, Q.T.; Price, M.; Neff, C.; Cioffi, G.; Waite, K.A.; Kruchko, C.; Barnholtz-Sloan, J.S. CBTRUS Statistical Report: Primary Brain and Other Central Nervous System Tumors Diagnosed in the United States in 2015–2019. Neuro Oncol. 2022, 24, v1–v95. [Google Scholar] [CrossRef] [PubMed]
- Schaff, L.R.; Mellinghoff, I.K. Glioblastoma and Other Primary Brain Malignancies in Adults: A Review. JAMA 2023, 329, 574–587. [Google Scholar] [CrossRef] [PubMed]
- McKinnon, C.; Nandhabalan, M.; Murray, S.A.; Plaha, P. Glioblastoma: Clinical Presentation, Diagnosis, and Management. BMJ 2021, 374, n1560. [Google Scholar] [CrossRef]
- McMahon, D.J.; Gleeson, J.P.; O’Reilly, S.; Bambury, R.M. Management of Newly Diagnosed Glioblastoma Multiforme: Current State of the Art and Emerging Therapeutic Approaches. Med. Oncol. 2022, 39, 129. [Google Scholar] [CrossRef] [PubMed]
- Hersh, A.M.; Bhimreddy, M.; Weber-Levine, C.; Jiang, K.; Alomari, S.; Theodore, N.; Manbachi, A.; Tyler, B.M. Applications of Focused Ultrasound for the Treatment of Glioblastoma: A New Frontier. Cancers 2022, 14, 4920. [Google Scholar] [CrossRef] [PubMed]
- Iuchi, T.; Inoue, A.; Hirose, Y.; Morioka, M.; Horiguchi, K.; Natsume, A.; Arakawa, Y.; Iwasaki, K.; Fujiki, M.; Kumabe, T.; et al. Long-Term Effectiveness of Gliadel Implant for Malignant Glioma and Prognostic Factors for Survival: 3-Year Results of a Postmarketing Surveillance in Japan. Neurooncol. Adv. 2022, 4, vdab189. [Google Scholar] [CrossRef]
- Xing, W.K.; Shao, C.; Qi, Z.Y.; Yang, C.; Wang, Z. The Role of Gliadel Wafers in the Treatment of Newly Diagnosed GBM: A Meta-Analysis. Drug Des. Devel. Ther. 2015, 9, 3341–3348. [Google Scholar] [CrossRef]
- Hicks, W.H.; Bird, C.E.; Pernik, M.N.; Haider, A.S.; Dobariya, A.; Abdullah, K.G.; Aoun, S.G.; Bentley, R.T.; Cohen-Gadol, A.A.; Bachoo, R.M.; et al. Large Animal Models of Glioma: Current Status and Future Prospects. Anticancer. Res. 2021, 41, 5343–5353. [Google Scholar] [CrossRef] [PubMed]
- Mandel, J.J.; Yust-Katz, S.; Patel, A.J.; Cachia, D.; Liu, D.; Park, M.; Yuan, Y.; Kent, T.A.; de Groot, J.F. Inability of Positive Phase II Clinical Trials of Investigational Treatments to Subsequently Predict Positive Phase III Clinical Trials in Glioblastoma. Neuro Oncol. 2018, 20, 113–122. [Google Scholar] [CrossRef]
- Mak, I.W.; Evaniew, N.; Ghert, M. Lost in Translation: Animal Models and Clinical Trials in Cancer Treatment. Am. J. Transl. Res. 2014, 6, 114–118. [Google Scholar]
- Cihoric, N.; Tsikkinis, A.; Minniti, G.; Lagerwaard, F.J.; Herrlinger, U.; Mathier, E.; Soldatovic, I.; Jeremic, B.; Ghadjar, P.; Elicin, O.; et al. Current Status and Perspectives of Interventional Clinical Trials for Glioblastoma—Analysis of ClinicalTrials.Gov. Radiat. Oncol. 2017, 12, 1. [Google Scholar] [CrossRef] [PubMed]
- Gunjur, A.; Balasubramanian, A.; Hafeez, U.; Menon, S.; Cher, L.; Parakh, S.; Gan, H.K. Poor Correlation between Preclinical and Patient Efficacy Data for Tumor Targeted Monotherapies in Glioblastoma: The Results of a Systematic Review. J. Neurooncol. 2022, 159, 539–549. [Google Scholar] [CrossRef] [PubMed]
- Benda, P.; Lightbody, J.; Sato, G.; Levine, L.; Sweet, W. Differentiated Rat Glial Cell Strain in Tissue Culture. Science 1968, 161, 370–371. [Google Scholar] [CrossRef] [PubMed]
- Giakoumettis, D.; Kritis, A.; Foroglou, N. C6 Cell Line: The Gold Standard in Glioma Research. Hippokratia 2018, 22, 105. [Google Scholar] [PubMed]
- Sahu, U.; Barth, R.F.; Otani, Y.; McCormack, R.; Kaur, B. Rat and Mouse Brain Tumor Models for Experimental Neuro-Oncology Research. J. Neuropathol. Exp. Neurol. 2022, 81, 312–329. [Google Scholar] [CrossRef] [PubMed]
- Schlegel, J.; Piontek, G.; Kersting, M.; Schuermann, M.; Kappler, R.; Scherthan, H.; Weghorst, C.; Buzard, G.; Mennel, H.D. The P16/Cdkn2a/Ink4a Gene Is Frequently Deleted in Nitrosourea-Induced Rat Glial Tumors. Pathobiology 1999, 67, 202–206. [Google Scholar] [CrossRef] [PubMed]
- Sibenaller, Z.A.; Etame, A.B.; Ali, M.M.; Barua, M.; Braun, T.A.; Casavant, T.L.; Ryken, T.C. Genetic Characterization of Commonly Used Glioma Cell Lines in the Rat Animal Model System. Neurosurg. Focus. 2005, 19, 1–9. [Google Scholar] [CrossRef]
- Fu, Y.; Zheng, Y.; Li, K.; Huang, R.; Zheng, S.; An, N.; Liang, A. Mutations in Isocitrate Dehydrogenase 2 Accelerate Glioma Cell Migration via Matrix Metalloproteinase-2 and 9. Biotechnol. Lett. 2012, 34, 441–446. [Google Scholar] [CrossRef]
- Barth, R.F. Rat Brain Tumor Models in Experimental Neuro-Oncology: The 9L, C6, T9, F98, RG2 (D74), RT-2 and CNS-1 Gliomas. J. Neurooncol. 1998, 36, 91–102. [Google Scholar] [CrossRef]
- Barth, R.F.; Kaur, B. Rat Brain Tumor Models in Experimental Neuro-Oncology: The C6, 9L, T9, RG2, F98, BT4C, RT-2 and CNS-1 Gliomas. J. Neurooncol. 2009, 94, 299–312. [Google Scholar] [CrossRef] [PubMed]
- Parsa, A.T.; Chakrabarti, I.; Hurley, P.T.; Chi, J.H.; Hall, J.S.; Kaiser, M.G.; Bruce, J.N. Limitations of the C6/Wistar Rat Intracerebral Glioma Model: Implications for Evaluating Immunotherapy. Neurosurgery 2000, 47, 993–1000. [Google Scholar] [CrossRef]
- Plate, K.H.; Breier, G.; Millauer, B.; Ullrich, A.; Risau, W. Up-Regulation of Vascular Endothelial Growth Factor and Its Cognate Receptors in a Rat Glioma Model of Tumor Angiogenesis. Cancer Res. 1993, 53, 582. [Google Scholar]
- Schmidek, H.H.; Nielsen, S.L.; Schiller, A.L.; Messer, J. Morphological Studies of Rat Brain Tumors Induced by N-Nitrosomethylurea. J. Neurosurg. 1971, 34, 335–340. [Google Scholar] [CrossRef]
- Benda, P.; Someda, K.; Messer, J.; Sweet, W.H. Morphological and Immunochemical Studies of Rat Glial Tumors and Clonal Strains Propagated in Culture. J. Neurosurg. 1971, 34, 310–323. [Google Scholar] [CrossRef]
- Mathieu, D.; Lecomte, R.; Tsanaclis, A.M.; Larouche, A.; Fortin, D. Standardization and Detailed Characterization of the Syngeneic Fischer/F98 Glioma Model. Can. J. Neurol. Sci. 2007, 34, 296–306. [Google Scholar] [CrossRef] [PubMed]
- Szatmari, T.; Lumniczky, K.; Desaknai, S.; Trajcevski, S.; Hidvegi, E.J.; Hamada, H.; Safrany, G. Detailed Characterization of the Mouse Glioma 261 Tumor Model for Experimental Glioblastoma Therapy. Cancer Sci. 2006, 97, 546–553. [Google Scholar] [CrossRef]
- Oh, T.; Fakurnejad, S.; Sayegh, E.T.; Clark, A.J.; Ivan, M.E.; Sun, M.Z.; Safaee, M.; Bloch, O.; James, C.D.; Parsa, A.T. Immunocompetent Murine Models for the Study of Glioblastoma Immunotherapy. J. Transl. Med. 2014, 12, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Semenkow, S.; Li, S.; Kahlert, U.D.; Raabe, E.H.; Xu, J.; Arnold, A.; Janowski, M.; Oh, B.C.; Brandacher, G.; Bulte, J.W.M.; et al. An Immunocompetent Mouse Model of Human Glioblastoma. Oncotarget 2017, 8, 61072–61082. [Google Scholar] [CrossRef][Green Version]
- Haddad, A.F.; Young, J.S.; Amara, D.; Berger, M.S.; Raleigh, D.R.; Aghi, M.K.; Butowski, N.A. Mouse Models of Glioblastoma for the Evaluation of Novel Therapeutic Strategies. Neurooncol. Adv. 2021, 3, vdab100. [Google Scholar] [CrossRef] [PubMed]
- Genoud, V.; Marinari, E.; Nikolaev, S.I.; Castle, J.C.; Bukur, V.; Dietrich, P.Y.; Okada, H.; Walker, P.R. Responsiveness to Anti-PD-1 and Anti-CTLA-4 Immune Checkpoint Blockade in SB28 and GL261 Mouse Glioma Models. Oncoimmunology 2018, 7, e1501137. [Google Scholar] [CrossRef] [PubMed]
- Maes, W.; Van Gool, S.W. Experimental Immunotherapy for Malignant Glioma: Lessons from Two Decades of Research in the GL261 Model. Cancer Immunol. Immunother. 2011, 60, 153–160. [Google Scholar] [CrossRef]
- Renner, D.N.; Jin, F.; Litterman, A.J.; Balgeman, A.J.; Hanson, L.M.; Gamez, J.D.; Chae, M.; Carlson, B.L.; Sarkaria, J.N.; Parney, I.F.; et al. Effective Treatment of Established GL261 Murine Gliomas through Picornavirus Vaccination-Enhanced Tumor Antigen-Specific CD8+ T Cell Responses. PLoS ONE 2015, 10, e0125565. [Google Scholar] [CrossRef] [PubMed]
- Zagzag, D.; Amirnovin, R.; Greco, M.A.; Yee, H.; Holash, J.; Wiegand, S.J.; Zabski, S.; Yancopoulos, G.D.; Grumet, M. Vascular Apoptosis and Involution in Gliomas Precede Neovascularization: A Novel Concept for Glioma Growth and Angiogenesis. Lab. Investig. 2000, 80, 837–849. [Google Scholar] [CrossRef] [PubMed]
- Shelton, L.M.; Mukherjee, P.; Huysentruyt, L.C.; Urits, I.; Rosenberg, J.A.; Seyfried, T.N. A Novel Pre-Clinical In Vivo Mouse Model for Malignant Brain Tumor Growth and Invasion. J. Neurooncol. 2010, 99, 165–176. [Google Scholar] [CrossRef] [PubMed]
- Binello, E.; Qadeer, Z.A.; Kothari, H.P.; Emdad, L.; Germano, I.M. Stemness of the CT-2A Immunocompetent Mouse Brain Tumor Model: Characterization In Vitro. J. Cancer 2012, 3, 166–174. [Google Scholar] [CrossRef]
- Martinez-Murillo, R.; Martinez, A. Standardization of an Orthotopic Mouse Brain Tumor Model Following Transplantation of CT-2A Astrocytoma Cells. Histol. Histopathol. 2007, 22, 1309–1326. [Google Scholar] [CrossRef] [PubMed]
- Camphausen, K.; Purow, B.; Sproull, M.; Scott, T.; Ozawa, T.; Deen, D.F.; Tofilon, P.J. Influence of In Vivo Growth on Human Glioma Cell Line Gene Expression: Convergent Profiles under Orthotopic Conditions. Proc. Natl. Acad. Sci. USA 2005, 102, 8287–8292. [Google Scholar] [CrossRef] [PubMed]
- Diao, W.; Tong, X.; Yang, C.; Zhang, F.; Bao, C.; Chen, H.; Liu, L.; Li, M.; Ye, F.; Fan, Q.; et al. Behaviors of Glioblastoma Cells in In Vitro Microenvironments. Sci. Rep. 2019, 9, 85. [Google Scholar] [CrossRef]
- Mesti, T.; Savarin, P.; Triba, M.N.; Le Moyec, L.; Ocvirk, J.; Banissi, C.; Carpentier, A.F. Metabolic Impact of Anti-Angiogenic Agents on U87 Glioma Cells. PLoS ONE 2014, 9, e99198. [Google Scholar] [CrossRef]
- Tamargo, R.J.; Epstein, J.I.; Brem, H. Heterotransplantation of Malignant Human Gliomas in Neonatal Rats. J. Neurosurg. 1988, 69, 928–933. [Google Scholar] [CrossRef]
- Strojnik, T.; Kavalar, R.; Lah, T.T. Experimental Model and Immunohistochemical Analyses of U87 Human Glioblastoma Cell Xenografts in Immunosuppressed Rat Brains. Anticancer. Res. 2006, 26, 2887–2900. [Google Scholar]
- Schulz, J.A.; Rodgers, L.T.; Kryscio, R.J.; Hartz, A.M.S.; Bauer, B. Characterization and Comparison of Human Glioblastoma Models. BMC Cancer 2022, 22, 844. [Google Scholar] [CrossRef] [PubMed]
- Demircan, T.; Yavuz, M.; Kaya, E.; Akgül, S.; Altuntaş, E. Cellular and Molecular Comparison of Glioblastoma Multiform Cell Lines. Cureus 2021, 13, e16043. [Google Scholar] [CrossRef]
- Ishii, N.; Maier, D.; Merlo, A.; Tada, M.; Sawamura, Y.; Diserens, A.C.; Van Meir, E.G. Frequent Co-Alterations of TP53, P16/CDKN2A, P14, PTEN Tumor Suppressor Genes in Human Glioma Cell Lines. Brain Pathol. 1999, 9, 469. [Google Scholar] [CrossRef] [PubMed]
- Heimberger, A.B.; Suki, D.; Yang, D.; Shi, W.; Aldape, K. The Natural History of EGFR and EGFRvIII in Glioblastoma Patients. J. Transl. Med. 2005, 3, 38. [Google Scholar] [CrossRef]
- Guo, P.; Hu, B.; Gu, W.; Xu, L.; Wang, D.; Huang, H.J.S.; Cavenee, W.K.; Cheng, S.Y. Platelet-Derived Growth Factor-B Enhances Glioma Angiogenesis by Stimulating Vascular Endothelial Growth Factor Expression in Tumor Endothelia and by Promoting Pericyte Recruitment. Am. J. Pathol. 2003, 162, 1083–1093. [Google Scholar] [CrossRef]
- Nakada, M.; Niska, J.A.; Tran, N.L.; McDonough, W.S.; Berens, M.E. EphB2/R-Ras Signaling Regulates Glioma Cell Adhesion, Growth, and Invasion. Am. J. Pathol. 2005, 167, 565–576. [Google Scholar] [CrossRef] [PubMed]
- Guha, A.; Feldkamp, M.M.; Lau, N.; Boss, G.; Pawson, A. Proliferation of Human Malignant Astrocytomas Is Dependent on Ras Activation. Oncogene 1997, 15, 2755–2765. [Google Scholar] [CrossRef]
- Glick, R.P.; Lichtor, T.; Unterman, T.G. Insulin-like Growth Factors in Central Nervous System Tumors. J. Neurooncol. 1997, 35, 315–325. [Google Scholar] [CrossRef]
- Yan, H.; Parsons, D.W.; Jin, G.; McLendon, R.; Rasheed, B.A.; Yuan, W.; Kos, I.; Batinic-Haberle, I.; Jones, S.; Riggins, G.J.; et al. IDH1 and IDH2 Mutations in Gliomas. N. Engl. J. Med. 2009, 360, 765. [Google Scholar] [CrossRef] [PubMed]
- Beiko, J.; Suki, D.; Hess, K.R.; Fox, B.D.; Cheung, V.; Cabral, M.; Shonka, N.; Gilbert, M.R.; Sawaya, R.; Prabhu, S.S.; et al. IDH1 Mutant Malignant Astrocytomas Are More Amenable to Surgical Resection and Have a Survival Benefit Associated with Maximal Surgical Resection. Neuro Oncol. 2014, 16, 81. [Google Scholar] [CrossRef] [PubMed]
- Fu, Y.; Huang, R.; Zheng, Y.; Zhang, Z.; Liang, A. Glioma-Derived Mutations in Isocitrate Dehydrogenase 2 Beneficial to Traditional Chemotherapy. Biochem. Biophys. Res. Commun. 2011, 410, 218–223. [Google Scholar] [CrossRef] [PubMed]
- Jung, C.S.; Foerch, C.; Schänzer, A.; Heck, A.; Plate, K.H.; Seifert, V.; Steinmetz, H.; Raabe, A.; Sitzer, M. Serum GFAP Is a Diagnostic Marker for Glioblastoma Multiforme. Brain 2007, 130, 3336–3341. [Google Scholar] [CrossRef]
- Huhndorf, M.; Moussavi, A.; Kramann, N.; Will, O.; Hattermann, K.; Stadelmann, C.; Jansen, O.; Boretius, S. Alterations of the Blood-Brain Barrier and Regional Perfusion in Tumor Development: MRI Insights from a Rat C6 Glioma Model. PLoS ONE 2016, 11, e0168174. [Google Scholar] [CrossRef] [PubMed]
- Le, T.N.T.; Lim, H.; Hamilton, A.M.; Parkins, K.M.; Chen, Y.; Scholl, T.J.; Ronald, J.A. Characterization of an Orthotopic Rat Model of Glioblastoma Using Multiparametric Magnetic Resonance Imaging and Bioluminescence Imaging. Tomography 2018, 4, 55. [Google Scholar] [CrossRef]
- San-Galli, F.; Vrignaud, P.; Robert, J.; Coindre, J.M.; Cohadon, F. Assessment of the Experimental Model of Transplanted C6 Glioblastoma in Wistar Rats. J. Neurooncol. 1989, 7, 299–304. [Google Scholar] [CrossRef]
- Farrell, C.L.; Stewart, P.A.; Del Maestro, R.F. A New Glioma Model in Rat: The C6 Spheroid Implantation Technique Permeability and Vascular Characterization. J. Neurooncol. 1987, 4, 403–415. [Google Scholar] [CrossRef]
- Gross, J.L.; Morrison, R.S.; Eidsvoog, K.; Herblin, W.F.; Kornblith, P.L.; Dexter, D.L. Basic Fibroblast Growth Factor: A Potential Autocrine Regulator of Human Glioma Cell Growth. J. Neurosci. Res. 1990, 27, 689–696. [Google Scholar] [CrossRef]
- Solly, F.; Fish, R.; Simard, B.; Bolle, N.; Kruithof, E.; Polack, B.; Pernod, G. Tissue-Type Plasminogen Activator Has Antiangiogenic Properties without Effect on Tumor Growth in a Rat C6 Glioma Model. Cancer Gene Ther. 2008, 15, 685–692. [Google Scholar] [CrossRef] [PubMed]
- Doblas, S.; Saunders, D.; Kshirsagar, P.; Pye, Q.; Oblander, J.; Gordon, B.; Kosanke, S.; Floyd, R.A.; Towner, R.A. Phenyl-Tert-Butylnitrone Induces Tumor Regression and Decreases Angiogenesis in a C6 Rat Glioma Model. Free Radic. Biol. Med. 2008, 44, 63–72. [Google Scholar] [CrossRef]
- Sheehan, J.; Ionescu, A.; Pouratian, N.; Hamilton, D.K.; Schlesinger, D.; Oskouian, R.J.; Sansur, C. Use of Trans Sodium Crocetinate for Sensitizing Glioblastoma Multiforme to Radiation: Laboratory Investigation. J. Neurosurg. 2008, 108, 972–978. [Google Scholar] [CrossRef]
- Yang, W.Q.; Lun, X.; Palmer, C.A.; Wilcox, M.E.; Muzik, H.; Shi, Z.Q.; Dyck, R.; Coffey, M.; Thompson, B.; Hamilton, M.; et al. Efficacy and Safety Evaluation of Human Reovirus Type 3 in Immunocompetent Animals: Racine and Nonhuman Primates. Clin. Cancer Res. 2004, 10, 8561–8576. [Google Scholar] [CrossRef]
- Tanriover, N.; Ulu, M.O.; Sanus, G.Z.; Bilir, A.; Canbeyli, R.; Oz, B.; Akar, Z.; Kuday, C. The Effects of Systemic and Intratumoral Interleukin-12 Treatment in C6 Rat Glioma Model. Neurol. Res. 2008, 30, 511–517. [Google Scholar] [CrossRef] [PubMed]
- Mannino, S.; Molinari, A.; Sabatino, G.; Ciafrè, S.A.; Colone, M.; Maira, G.; Anile, C.; Arancia, G.; Mangiola, A. Intratumoral vs Systemic Administration of Meta-Tetrahydroxyphenylchlorin for Photodynamic Therapy of Malignant Gliomas: Assessment of Uptake and Spatial Distribution in C6 Rat Glioma Model. Int. J. Immunopathol. Pharmacol. 2008, 21, 227–231. [Google Scholar] [CrossRef] [PubMed]
- Zhao, S.; Zhang, X.; Zhang, J.; Zhang, J.; Zou, H.; Liu, Y.; Dong, X.; Sun, X. Intravenous Administration of Arsenic Trioxide Encapsulated in Liposomes Inhibits the Growth of C6 Gliomas in Rat Brains. J. Chemother. 2008, 20, 253–262. [Google Scholar] [CrossRef]
- Ahmed, A.E.; Jacob, S.; Nagy, A.A.; Abdel-Naim, A.B. Dibromoacetonitrile-Induced Protein Oxidation and Inhibition of Proteasomal Activity in Rat Glioma Cells. Toxicol. Lett. 2008, 179, 29–33. [Google Scholar] [CrossRef] [PubMed]
- Valable, S.; Lemasson, B.; Farion, R.; Beaumont, M.; Segebarth, C.; Remy, C.; Barbier, E.L. Assessment of Blood Volume, Vessel Size, and the Expression of Angiogenic Factors in Two Rat Glioma Models: A Longitudinal In Vivo and Ex Vivo Study. NMR Biomed. 2008, 21, 1043–1056. [Google Scholar] [CrossRef]
- Asai, A.; Miyagi, Y.; Sugiyama, A.; Gamanuma, M.; Hong, S.I.; Takamoto, S.; Nomura, K.; Matsutani, M.; Takakura, K.; Kuchino, Y. Negative Effects of Wild-Type P53 and s-Myc on Cellular Growth and Tumorigenicity of Glioma Cells. Implication of the Tumor Suppressor Genes for Gene Therapy. J. Neurooncol. 1994, 19, 259–268. [Google Scholar] [CrossRef]
- Wolff, J.E.A.; Mölenkamp, G.; Hotfilder, M.; Laterra, J. Dexamethasone Inhibits Glioma-Induced Formation of Capillary like Structures In Vitro and Angiogenesis In Vivo. Klin. Padiatr. 1997, 209, 275–277. [Google Scholar] [CrossRef] [PubMed]
- Yang, H.; Chopp, M.; Zhang, X.; Jiang, F.; Zhang, Z.; Kalkanis, S.; Schallert, T. Using Behavioral Measurement to Assess Tumor Progression and Functional Outcome after Antiangiogenic Treatment in Mouse Glioma Models. Behav. Brain Res. 2007, 182, 42–50. [Google Scholar] [CrossRef]
- Bencokova, Z.; Pauron, L.; Devic, C.; Joubert, A.; Gastaldo, J.; Massart, C.; Balosso, J.; Foray, N. Molecular and Cellular Response of the Most Extensively Used Rodent Glioma Models to Radiation and/or Cisplatin. J. Neurooncol 2008, 86, 13–21. [Google Scholar] [CrossRef]
- Bouchet, A.; Bidart, M.; Miladi, I.; Le Clec’h, C.; Serduc, R.; Coutton, C.; Regnard, P.; Khalil, E.; Dufort, S.; Lemasson, B.; et al. Characterization of the 9L Gliosarcoma Implanted in the Fischer Rat: An Orthotopic Model for a Grade IV Brain Tumor. Tumour Biol. 2014, 35, 6221–6233. [Google Scholar] [CrossRef]
- Iwadate, Y.; Inoue, M.; Saegusa, T.; Tokusumi, Y.; Kinoh, H.; Hasegawa, M.; Tagawa, M.; Yamaura, A.; Shimada, H. Recombinant Sendai Virus Vector Induces Complete Remission of Established Brain Tumors through Efficient Interleukin-2 Gene Transfer in Vaccinated Rats. Clin. Cancer Res. 2005, 11, 3821–3827. [Google Scholar] [CrossRef] [PubMed]
- Kumar, S.; Brown, S.L.; Kolozsvary, A.; Freytag, S.O.; Kim, J.H. Efficacy of Suicide Gene Therapy in Hypoxic Rat 9L Glioma Cells. J. Neurooncol. 2008, 90, 19. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Madara, J.; Krewet, J.A.; Shah, M. Heat Shock Protein 72 Expression Allows Permissive Replication of Oncolytic Adenovirus Dl1520 (ONYX-015) in Rat Glioblastoma Cells. Mol. Cancer 2005, 4, 12. [Google Scholar] [CrossRef]
- Aghi, M.; Chou, T.; Suling, K.; Breakefield, X.O.; Chiocca, E.A. Multimodal Cancer Treatment Mediated by a Replicating Oncolytic Virus That Delivers the Oxazaphosphorine/Rat Cytochrome P450 2B1 and Ganciclovir/Herpes Simplex Virus Thymidine Kinase Gene Therapies. Cancer Res. 1999, 59, 3861–3865. [Google Scholar]
- Yuan, H.; Schroeder, T.; Bowsher, J.E.; Hedlund, L.W.; Wong, T.; Dewhirst, M.W. Intertumoral Differences in Hypoxia Selectivity of the PET Imaging Agent 64Cu(II)-Diacetyl-Bis(N4-Methylthiosemicarbazone). J. Nucl. Med. 2006, 47, 989–998. [Google Scholar]
- Bansal, A.; Shuyan, W.; Hara, T.; Harris, R.A.; DeGrado, T.R. Biodisposition and Metabolism of [18F]Fluorocholine in 9L Glioma Cells and 9L Glioma-Bearing Fisher Rats. Eur. J. Nucl. Med. Mol. Imaging 2008, 35, 1192. [Google Scholar] [CrossRef]
- Barcellos-Hoff, M.-H.; Linfoot, P.A.; Marton, L.J.; Deen, D.F. Production of Stable Phenotypes from 9L Rat Brain Tumor Multicellular Spheroids Treated with 1,3-Bis(2-Chloroethyl)-1-Nitrosourea. Int. J. Cancer 1992, 52, 409–413. [Google Scholar] [CrossRef]
- Schepkin, V.D.; Lee, K.C.; Kuszpit, K.; Muthuswami, M.; Johnson, T.D.; Chenevert, T.L.; Rehemtulla, A.; Ross, B.D. Proton and Sodium MRI Assessment of Emerging Tumor Chemotherapeutic Resistance. NMR Biomed. 2006, 19, 1035. [Google Scholar] [CrossRef] [PubMed]
- Black, K.L.; Yin, D.; Konda, B.M.; Wang, X.; Hu, J.; Ko, M.H.K.; Bayan, J.A.; Sacapano, M.R.; Espinoza, A.J.; Ong, J.M.; et al. Different Effects of KCa and KATP Agonists on Brain Tumor Permeability between Syngeneic and Allogeneic Rat Models. Brain Res. 2008, 1227, 198–206. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Fross, R.D.; Warnke, P.C.; Groothuis, D.R. Blood Flow and Blood-to-Tissue Transport in 9L Gliosarcomas: The Role of the Brain Tumor Model in Drug Delivery Research. J. Neurooncol. 1991, 11, 185–197. [Google Scholar] [CrossRef]
- Guarnieri, M.; Carson, B.S.; Khan, A.; Penno, M.; Jallo, G.I. Flexible versus Rigid Catheters for Chronic Administration of Exogenous Agents into Central Nervous System Tissues. J. Neurosci. Methods 2005, 144, 147–152. [Google Scholar] [CrossRef] [PubMed]
- Warnke, P.C.; Blasberg, R.G.; Groothuis, D.R. The Effect of Hyperosmotic Blood-Brain Barrier Disruption on Blood-to-Tissue Transport in ENU-Induced Gliomas. Ann. Neurol. 1987, 22, 300–305. [Google Scholar] [CrossRef]
- Barth, R.F.; Carpenter, D.E. Rodent Brain Tumor Models for Studies Focusing on Boron Neutron Capture Therapy. Cancer Biother. Radiopharm. 2023, 38, 148–151. [Google Scholar] [CrossRef] [PubMed]
- Elleaume, H.; Barth, R.F.; Rousseau, J.; Bobyk, L.; Balosso, J.; Yang, W.; Huo, T.; Nakkula, R. Radiation Therapy Combined with Intracerebral Convection-Enhanced Delivery of Cisplatin or Carboplatin for Treatment of the F98 Rat Glioma. J. Neurooncol. 2020, 149, 193–208. [Google Scholar] [CrossRef]
- Kaneko, S.; Allen, N.J.; Clendenon, N.R.; Kartha, M. Treatment Schedule of Combination Using Radiation and ACNU in the Experimental Brain Tumors. Neurol. Med. Chir. 1983, 23, 849–855. [Google Scholar] [CrossRef][Green Version]
- Barth, R.F.; Yang, W.; Rotaru, J.H.; Moeschberger, M.L.; Joel, D.D.; Nawrocky, M.M.; Goodman, J.H.; Soloway, A.H. Boron Neutron Capture Therapy of Brain Tumors: Enhanced Survival Following Intracarotid Injection of Either Sodium Borocaptate or Boronophenylalanine with or without Blood-Brain Barrier Disruption. Cancer Res. 1997, 57, 1129–1136. [Google Scholar] [CrossRef]
- Cho, J.Y.; Shen, D.H.Y.; Yang, W.; Williams, B.; Buckwalter, T.L.F.; La Perle, K.M.D.; Hinkle, G.; Pozderac, R.; Kloos, R.; Nagaraja, H.N.; et al. In Vivo Imaging and Radioiodine Therapy Following Sodium Iodide Symporter Gene Transfer in Animal Model of Intracerebral Gliomas. Gene Ther. 2002, 9, 1139–1145. [Google Scholar] [CrossRef]
- Adam, J.F.; Joubert, A.; Biston, M.C.; Charvet, A.M.; Peoc’h, M.; Le Bas, J.F.; Balosso, J.; Estève, F.; Elleaume, H. Prolonged Survival of Fischer Rats Bearing F98 Glioma after Iodine-Enhanced Synchrotron Stereotactic Radiotherapy. Int. J. Radiat. Oncol. Biol. Phys. 2006, 64, 603–611. [Google Scholar] [CrossRef] [PubMed]
- Backer, M.V.; Gaynutdinov, T.I.; Patel, V.; Bandyopadhyaya, A.K.; Thirumamagal, B.T.S.; Tjarks, W.; Barth, R.F.; Claffey, K.; Backer, J.M. Vascular Endothelial Growth Factor Selectively Targets Boronated Dendrimers to Tumor Vasculature. Mol. Cancer Ther. 2005, 4, 1423–1429. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Van Zijl, P.C.M.; Laterra, J.; Salhotra, A.; Lal, B.; Mori, S.; Zhou, J. Unique Patterns of Diffusion Directionality in Rat Brain Tumors Revealed by High-Resolution Diffusion Tensor MRI. Magn. Reson. Med. 2007, 58, 454–462. [Google Scholar] [CrossRef]
- Blanchard, J.; Mathieu, D.; Patenaude, Y.; Fortin, D. MR-Pathological Comparison in F98-Fischer Glioma Model Using a Human Gantry. Can. J. Neurol. Sci. 2006, 33, 86–91. [Google Scholar] [CrossRef]
- Towner, R.A.; Gillespie, D.L.; Schwager, A.; Saunders, D.G.; Smith, N.; Njoku, C.E.; Krysiak, R.S.; Larabee, C.; Iqbal, H.; Floyd, R.A.; et al. Regression of Glioma Tumor Growth in F98 and U87 Rat Glioma Models by the Nitrone OKN-007. Neuro Oncol. 2013, 15, 330. [Google Scholar] [CrossRef]
- Yang, W.; Wu, G.; Barth, R.F.; Swindall, M.R.; Bandyopadhyaya, A.K.; Tjarks, W.; Tordoff, K.; Moeschberger, M.; Sferra, T.J.; Binns, P.J.; et al. Molecular Targeting and Treatment of Composite EGFR and EGFRvIII-Positive Gliomas Using Boronated Monoclonal Antibodies. Clin. Cancer Res. 2008, 14, 883–891. [Google Scholar] [CrossRef] [PubMed]
- Huo, T.; Barth, R.F.; Yang, W.; Nakkula, R.J.; Koynova, R.; Tenchov, B.; Chaudhury, A.R.; Agius, L.; Boulikas, T.; Elleaume, H.; et al. Preparation, Biodistribution and Neurotoxicity of Liposomal Cisplatin Following Convection Enhanced Delivery in Normal and F98 Glioma Bearing Rats. PLoS ONE 2012, 7, e48752. [Google Scholar] [CrossRef]
- Von Eckardstein, K.L.; Patt, S.; Zhu, J.; Zhang, L.; Cervós-Navarro, J.; Reszka, R. Short-Term Neuropathological Aspects of In Vivo Suicide Gene Transfer to the F98 Rat Glioblastoma Using Liposomal and Viral Vectors. Histol. Histopathol. 2001, 16, 735–744. [Google Scholar] [CrossRef] [PubMed]
- Weizsäcker, M.; Nagamune, A.; Winkelströter, R.; Vieten, H.; Wechsler, W. Radiation and Drug Response of the Rat Glioma RG2. Eur. J. Cancer Clin. Oncol. 1982, 18, 891–895. [Google Scholar] [CrossRef] [PubMed]
- Miknyoczki, S.; Chang, H.; Grobelny, J.; Pritchard, S.; Worrell, C.; McGann, N.; Ator, M.; Husten, J.; Deibold, J.; Hudkins, R.; et al. The Selective Poly(ADP-Ribose) Polymerase-1(2) Inhibitor, CEP-8983, Increases the Sensitivity of Chemoresistant Tumor Cells to Temozolomide and Irinotecan but Does Not Potentiate Myelotoxicity. Mol. Cancer Ther. 2007, 6, 2290–2302. [Google Scholar] [CrossRef]
- Tsai, N.M.; Lin, S.Z.; Lee, C.C.; Chen, S.P.; Su, H.C.; Chang, W.L.; Harn, H.J. The Antitumor Effects of Angelica Sinensis on Malignant Brain Tumors In Vitro and In Vivo. Clin. Cancer Res. 2005, 11, 3475–3484. [Google Scholar] [CrossRef]
- Shen, D.H.Y.; Marsee, D.K.; Schaap, J.; Yang, W.; Cho, J.Y.; Hinkle, G.; Nagaraja, H.N.; Kloos, R.T.; Barth, R.F.; Jhiang, S.M. Effects of Dose, Intervention Time, and Radionuclide on Sodium Iodide Symporter (NIS)-Targeted Radionuclide Therapy. Gene Ther. 2004, 11, 161–169. [Google Scholar] [CrossRef] [PubMed]
- Wang, W.; Tai, C.-K.; Kershaw, A.D.; Solly, S.K.; Klatzmann, D.; Kasahara, N.; Chen, T.C. Use of Replication-Competent Retroviral Vectors in an Immunocompetent Intracranial Glioma Model. Neurosurg. Focus 2006, 20, E25. [Google Scholar] [CrossRef] [PubMed]
- Zagorac, D.; Jakovcevic, D.; Gebremedhin, D.; Harder, D.R. Anti-Angiogenic Effect of Inhibitors of Cytochrome P450 on Rats with Glioblastoma Multiforme. J. Cereb. Blood Flow. Metab. 2008, 28, 1431. [Google Scholar] [CrossRef]
- Hashizume, K.; Black, K.L. Increased Endothelial Vesicular Transport Correlates with Increased Blood-Tumor Barrier Permeability Induced by Bradykinin and Leukotriene C4. J. Neuropathol. Exp. Neurol. 2002, 61, 725–735. [Google Scholar] [CrossRef]
- Ningaraj, N.S.; Rao, M.; Hashizume, K.; Asotra, K.; Black, K.L. Regulation of Blood-Brain Tumor Barrier Permeability by Calcium-Activated Potassium Channels. J. Pharmacol. Exp. Ther. 2002, 301, 838–851. [Google Scholar] [CrossRef]
- Ferrier, M.C.; Sarin, H.; Fung, S.H.; Schatlo, B.; Pluta, R.M.; Gupta, S.N.; Choykeb, P.L.; Oldfield, E.H.; Thomasson, D.; Butman, J.A. Validation of Dynamic Contrast-Enhanced Magnetic Resonance Imaging-Derived Vascular Permeability Measurements Using Quantitative Autoradiography in the RG2 Rat Brain Tumor Model. Neoplasia 2007, 9, 546. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Kurozumi, K.; Hardcastle, J.; Thakur, R.; Yang, M.; Christoforidis, G.; Fulci, G.; Hochberg, F.H.; Weissleder, R.; Carson, W.; Chiocca, E.A.; et al. Effect of Tumor Microenvironment Modulation on the Efficacy of Oncolytic Virus Therapy. J. Natl. Cancer Inst. 2007, 99, 1768–1781. [Google Scholar] [CrossRef]
- Oshiro, S.; Liu, Y.; Fukushima, T.; Asotra, K.; Black, K.L. Modified Immunoregulation Associated with Interferon-Gamma Treatment of Rat Glioma. Neurol. Res. 2001, 23, 359–366. [Google Scholar] [CrossRef]
- Bilmin, K.; Kujawska, T.; Secomski, W.; Nowicki, A.; Grieb, P. 5-Aminolevulinic Acid-Mediated Sonosensitization of Rat RG2 Glioma Cells In Vitro. Folia Neuropathol. 2016, 54, 234–240. [Google Scholar] [CrossRef]
- Lenting, K.; Verhaak, R.; ter Laan, M.; Wesseling, P.; Leenders, W. Glioma: Experimental Models and Reality. Acta Neuropathol. 2017, 133, 263–282. [Google Scholar] [CrossRef]
- Jacobs, V.L.; Valdes, P.A.; Hickey, W.F.; de Leo, J.A. Current Review of In Vivo GBM Rodent Models: Emphasis on the CNS-1 Tumour Model. ASN Neuro 2011, 3, 171–181. [Google Scholar] [CrossRef]
- Desaknai, S.; Lumniczky, K.; Esik, O.; Hamada, H.; Safrany, G. Local Tumour Irradiation Enhances the Anti-Tumour Effect of a Double-Suicide Gene Therapy System in a Murine Glioma Model. J. Gene Med. 2003, 5, 377–385. [Google Scholar] [CrossRef] [PubMed]
- Herrlinger, U.; Kramm, C.M.; Johnston, K.M.; Louis, D.N.; Finkelstein, D.; Reznikoff, G.; Dranoff, G.; Breakefield, X.O.; Yu, J.S. Vaccination for Experimental Gliomas Using GM-CSF-Transduced Glioma Cells. Cancer Gene Ther. 1997, 4, 345–352. [Google Scholar] [PubMed]
- Yu, J.S.; Burwick, J.A.; Dranoff, G.; Breakefield, X.O. Gene Therapy for Metastatic Brain Tumors by Vaccination with Granulocyte-Macrophage Colony-Stimulating Factor-Transduced Tumor Cells. Hum. Gene Ther. 1997, 8, 1065–1072. [Google Scholar] [CrossRef]
- Natsume, A.; Mizuno, M.; Ryuke, Y.; Yoshida, J. Antitumor Effect and Cellular Immunity Activation by Murine Interferon-Beta Gene Transfer against Intracerebral Glioma in Mouse. Gene Ther. 1999, 6, 1626–1633. [Google Scholar] [CrossRef]
- Natsume, A.; Tsujimura, K.; Mizuno, M.; Takahashi, T.; Yoshida, J. IFN-Beta Gene Therapy Induces Systemic Antitumor Immunity against Malignant Glioma. J. Neurooncol. 2000, 47, 117–124. [Google Scholar] [CrossRef] [PubMed]
- Azzalin, A.; Nato, G.; Parmigiani, E.; Garello, F.; Buffo, A.; Magrassi, L. Inhibitors of GLUT/SLC2A Enhance the Action of BCNU and Temozolomide against High-Grade Gliomas. Neoplasia 2017, 19, 364. [Google Scholar] [CrossRef]
- Medikonda, R.; Choi, J.; Pant, A.; Saleh, L.; Routkevitch, D.; Tong, L.; Belcaid, Z.; Kim, Y.H.; Jackson, C.M.; Jackson, C.; et al. Synergy between Glutamate Modulation and Anti-Programmed Cell Death Protein 1 Immunotherapy for Glioblastoma. J. Neurosurg. 2022, 136, 379–388. [Google Scholar] [CrossRef]
- Seyfried, T.N.; el-Abbadi, M.; Roy, M.L. Ganglioside Distribution in Murine Neural Tumors. Mol. Chem. Neuropathol. 1992, 17, 147–167. [Google Scholar] [CrossRef]
- Fan, D.; Yue, Q.; Chen, J.; Wang, C.; Yu, R.; Jin, Z.; Yin, S.; Wang, Q.; Chen, L.; Liao, X.; et al. Reprogramming the Immunosuppressive Microenvironment of IDH1 Wild-Type Glioblastoma by Blocking Wnt Signaling between Microglia and Cancer Cells. Oncoimmunology 2021, 10, 1932061. [Google Scholar] [CrossRef]
- Marsh, J.; Mukherjee, P.; Seyfried, T.N. Akt-Dependent Proapoptotic Effects of Dietary Restriction on Late-Stage Management of a Phosphatase and Tensin Homologue/Tuberous Sclerosis Complex 2-Deficient Mouse Astrocytoma. Clin. Cancer Res. 2008, 14, 7751–7762. [Google Scholar] [CrossRef] [PubMed]
- Khalsa, J.K.; Cheng, N.; Keegan, J.; Chaudry, A.; Driver, J.; Bi, W.L.; Lederer, J.; Shah, K. Immune Phenotyping of Diverse Syngeneic Murine Brain Tumors Identifies Immunologically Distinct Types. Nat. Commun. 2020, 11, 3912. [Google Scholar] [CrossRef]
- Saha, D.; Martuza, R.L.; Rabkin, S.D. Macrophage Polarization Contributes to Glioblastoma Eradication by Combination Immunovirotherapy and Immune Checkpoint Blockade. Cancer Cell 2017, 32, 253–267.e5. [Google Scholar] [CrossRef] [PubMed]
- Berger, G.; Knelson, E.H.; Jimenez-Macias, J.L.; Nowicki, M.O.; Han, S.; Panagioti, E.; Lizotte, P.H.; Adu-Berchie, K.; Stafford, A.; Dimitrakakis, N.; et al. STING Activation Promotes Robust Immune Response and NK Cell-Mediated Tumor Regression in Glioblastoma Models. Proc. Natl. Acad. Sci. USA 2022, 119, e2111003119. [Google Scholar] [CrossRef] [PubMed]
- Barnard, Z.; Wakimoto, H.; Zaupa, C.; Patel, A.P.; Klehm, J.; Martuza, R.L.; Rabkin, S.D.; Curry, W.T., Jr. Expression of FMS-like Tyrosine Kinase 3 Ligand by Oncolytic Herpes Simplex Virus Type I Prolongs Survival in Mice Bearing Established Syngeneic Intracranial Malignant Glioma. Neurosurgery 2012, 71, 741–748, discussion 748. [Google Scholar] [CrossRef] [PubMed]
- Pontén, J.; Macintyre, E.H. Long term culture of normal and neoplastic human glia. Acta Pathol. Microbiol. Scand. 1968, 74, 465–486. [Google Scholar] [CrossRef]
- Fueyo, J.; Gomez-Manzano, C.; Yung, W.K.A.; Clayman, G.L.; Liu, T.J.; Bruner, J.; Levin, V.A.; Kyritsis, A.P. Adenovirus-Mediated P16/CDKN2 Gene Transfer Induces Growth Arrest and Modifies the Transformed Phenotype of Glioma Cells. Oncogene 1996, 12, 103–110. [Google Scholar]
- Wierzbicki, M.; Sawosz, E.; Strojny, B.; Jaworski, S.; Grodzik, M.; Chwalibog, A. NF-ΚB-Related Decrease of Glioma Angiogenic Potential by Graphite Nanoparticles and Graphene Oxide Nanoplatelets. Sci. Rep. 2018, 8, 1–9. [Google Scholar] [CrossRef]
- Francescone, R.; Scully, S.; Bentley, B.; Yan, W.; Taylor, S.L.; Oh, D.; Moral, L.; Shao, R. Glioblastoma-Derived Tumor Cells Induce Vasculogenic Mimicry through Flk-1 Protein Activation. J. Biol. Chem. 2012, 287, 24821. [Google Scholar] [CrossRef]
- Qiang, L.; Yang, Y.; Ma, Y.J.; Chen, F.H.; Zhang, L.B.; Liu, W.; Qi, Q.; Lu, N.; Tao, L.; Wang, X.T.; et al. Isolation and Characterization of Cancer Stem like Cells in Human Glioblastoma Cell Lines. Cancer Lett. 2009, 279, 13–21. [Google Scholar] [CrossRef]
- Faury, D.; Nantel, A.; Dunn, S.E.; Guiot, M.C.; Haque, T.; Hauser, P.; Garami, M.; Bognár, L.; Hanzély, Z.; Liberski, P.P.; et al. Molecular Profiling Identifies Prognostic Subgroups of Pediatric Glioblastoma and Shows Increased YB-1 Expression in Tumors. J. Clin. Oncol. 2007, 25, 1196–1208. [Google Scholar] [CrossRef]
- Zhu, H.-J.; Ogawa, M.; Magata, Y.; Hirata, M.; Ohmomo, Y.; Namba, H.; Sakahara, H. Assessment of Epidermal Growth Factor Receptor Status in Glioblastomas. Asia Ocean. J. Nucl. Med. Biol. 2013, 1, 47. [Google Scholar]
- Sun, J.; Zhang, Y.; Zhen, Y.; Cui, J.; Hu, G.; Lin, Y. Antitumor Activity of Tetrandrine Citrate in Human Glioma U87 Cells In Vitro and In Vivo. Oncol. Rep. 2019, 42, 2345–2354. [Google Scholar] [CrossRef]
- Hsu, S.P.C.; Kuo, J.S.; Chiang, H.C.; Wang, H.E.; Wang, Y.S.; Huang, C.C.; Huang, Y.C.; Chi, M.S.; Mehta, M.P.; Chi, K.H. Temozolomide, Sirolimus and Chloroquine Is a New Therapeutic Combination That Synergizes to Disrupt Lysosomal Function and Cholesterol Homeostasis in GBM Cells. Oncotarget 2018, 9, 6883–6896. [Google Scholar] [CrossRef] [PubMed]
- Pechman, K.R.; Donohoe, D.L.; Bedekar, D.P.; Kurpad, S.N.; Hoffmann, R.G.; Schmainda, K.M. Characterization of Bevacizumab Dose Response Relationship in U87 Brain Tumors Using Magnetic Resonance Imaging Measures of Enhancing Tumor Volume and Relative Cerebral Blood Volume. J. Neurooncol. 2011, 105, 233–239. [Google Scholar] [CrossRef][Green Version]
- Pan, Q.; Yang, X.J.; Wang, H.M.; Dong, X.T.; Wang, W.; Li, Y.; Li, J.M. Chemoresistance to Temozolomide in Human Glioma Cell Line U251 Is Associated with Increased Activity of O6-Methylguanine-DNA Methyltransferase and Can Be Overcome by Metronomic Temozolomide Regimen. Cell Biochem. Biophys. 2012, 62, 185–191. [Google Scholar] [CrossRef] [PubMed]
- Chu, S.H.; Zhou, Z.M.; Karri, S.; Li, Z.Q.; Zhao, J.M. In Vitro and In Vivo Radiosensitization of Human Glioma U251 Cells Induced by Upregulated Expression of SLC22A18. Cancer Gene Ther. 2014, 21, 103–109. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Pamies, D.; Zurich, M.-G.; Hartung, T. Organotypic Models to Study Human Glioblastoma: Studying the beast in Its Ecosystem. iScience 2020, 23, 101633. [Google Scholar] [CrossRef]
- Ben-David, U.; Siranosian, B.; Ha, G.; Tang, H.; Oren, Y.; Hinohara, K.; Strathdee, C.A.; Dempster, J.; Lyons, N.J.; Burns, R.; et al. Genetic and Transcriptional Evolution Alters Cancer Cell Line Response. Nature 2018, 560, 325–330. [Google Scholar] [CrossRef]
- Patrizii, M.; Bartucci, M.; Pine, S.R.; Sabaawy, H.E. Utility of Glioblastoma Patient-Derived Orthotopic Xenografts in drug Discovery and Personalized Therapy. Front. Oncol. 2018, 8, 23. [Google Scholar] [CrossRef]
- Friedrich, J.; Seidel, C.; Ebner, R.; Kunz-Schughart, L.A. Spheroid-Based Drug Screen: Considerations and Practical. Nat. Protoc. 2009, 4, 309–324. [Google Scholar] [CrossRef]
- Weiswald, L.-B.; Bellet, D.; Dangles-Marie, V. Spherical Cancer Models in Tumor Biology. Neoplasia 2015, 17, 1–15. [Google Scholar] [CrossRef]
- Svendsen, C.N.; ter Borg, M.G.; Armstrong, R.J.; Rosser, A.E.; Chandran, S.; Ostenfeld, T.; Caldwell, M.A. A New Method for the Rapid and Long Term Growth of Human Neural Cells. J. Neurosci. Methods 1998, 85, 141–152. [Google Scholar] [CrossRef]
- Reynolds, B.; Weiss, S. Generation of Neurons and Astrocytes from Isolated of the Adult Mammalian Central Nervous System. Science (1979) 1992, 255, 1707–1710. [Google Scholar] [CrossRef] [PubMed]
- Galli, R.; Binda, E.; Orfanelli, U.; Cipelletti, B.; Gritti, A.; De Vitis, S.; Fiocco, R.; Foroni, C.; Dimeco, F.; Vescovi, A. Isolation and Characterization of Tumorigenic, Stem-like Neural from Human Glioblastoma. Cancer Res. 2004, 64, 7011–7021. [Google Scholar] [CrossRef] [PubMed]
- Tunici, P.; Bissola, L.; Lualdi, E.; Pollo, B.; Cajola, L.; Broggi, G.; Sozzi, G.; Finocchiaro, G. Genetic Alterations and In Vivo Tumorigenicity of Neurospheres from an Adult Glioblastoma. Mol. Cancer 2004, 3, 25. [Google Scholar] [CrossRef] [PubMed]
- Vukicevic, V.; Jauch, A.; Dinger, T.C.; Gebauer, L.; Hornich, V.; Bornstein, S.R.; Ehrhart-Bornstein, M.; Müller, A.M. Genetic Instability and Diminished Differentiation Capacity In-Term Cultured Mouse Neurosphere Cells. Mech. Ageing Dev. 2010, 131, 124–132. [Google Scholar] [CrossRef]
- Pollard, S.M.; Yoshikawa, K.; Clarke, I.D.; Danovi, D.; Stricker, S.; Russell, R.; Bayani, J.; Head, R.; Lee, M.; Bernstein Mark and Squire, J.A.; et al. Glioma Stem Cell Lines Expanded in Adherent Culture Have-Specific Phenotypes and Are Suitable for Chemical and genetic Screens. Cell Stem Cell 2009, 4, 568–580. [Google Scholar] [CrossRef]
- Singh, S.K.; Clarke, I.D.; Terasaki, M.; Bonn, V.E.; Hawkins, C.; Squire, J.; Dirks, P.B. Identification of a Cancer Stem Cell in Human Brain Tumors. Cancer Res. 2003, 63, 5821–5828. [Google Scholar]
- Ruiz-Garcia, H.; Alvarado-Estrada, K.; Schiapparelli, P.; Quinones-Hinojosa, A.; Trifiletti, D.M. Engineering Three-Dimensional Tumor Models to Study Glioma Cancer Cells and Tumor Microenvironment. Front. Cell. Neurosci. 2020, 14, 558381. [Google Scholar] [CrossRef]
- Mirab, F.; Kang, Y.J.; Majd, S. Preparation and Characterization of Size-Controlled Glioma Using Agarose Hydrogel Microwells. PLoS ONE 2019, 14, e0211078. [Google Scholar] [CrossRef]
- Lee, J.; Kotliarova, S.; Kotliarov, Y.; Li, A.; Su, Q.; Donin, N.M.; Pastorino, S.; Purow, B.W.; Christopher, N.; Zhang, W.; et al. Tumor Stem Cells Derived from Glioblastomas Cultured in bFGF and EGF More Closely Mirror the Phenotype and Genotype of primary Tumors than Do Serum-Cultured Cell Lines. Cancer Cell 2006, 9, 391–403. [Google Scholar] [CrossRef] [PubMed]
- Claes, A.; Schuuring, J.; Boots-Sprenger, S.; Hendriks-Cornelissen, S.; Dekkers, M.; van der Kogel, A.J.; Leenders, W.P.; Wesseling Pieter and Jeuken, J.W. Phenotypic and Genotypic Characterization of Orthotopic Human Models and Its Relevance for the Study of Anti-Glioma. Brain Pathol. 2008, 18, 423–433. [Google Scholar] [CrossRef]
- Sutherland, R.M.; McCredie, J.A.; Inch, W.R. Growth of Multicell Spheroids in Tissue Culture as a Model of nodular Carcinomas. J. Natl. Cancer Inst. 1971, 46, 113–120. [Google Scholar] [PubMed]
- Mashiyama, S.; Katakura, R.; Takahashi, K.; Kitahara, M.; Suzuki, J.; Sasaki, T. Enhancement of the Effect of X-Irradiation against Cultured Human Glioblastoma Cells by Pretreatment with ACNU. Neurol. Med. Chir. 1989, 29, 1070–1077. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Bell, H.S.; Wharton, S.B.; Leaver, H.A.; Whittle, I.R. Effects of N-6 Essential Fatty Acids on Glioma Invasion and growth: Experimental Studies with Glioma Spheroids in Collagen. J. Neurosurg. 1999, 91, 989–996. [Google Scholar] [CrossRef]
- Oraiopoulou, M.-E.; Tampakaki, M.; Tzamali, E.; Tamiolakis, T.; Makatounakis, V.; Vakis, A.F.; Zacharakis, G.; Sakkalis, V.; Papamatheakis, J. A 3D Tumor Spheroid Model for the T98G Glioblastoma Cell Phenotypic Characterization. Tissue Cell 2019, 59, 39–43. [Google Scholar] [CrossRef]
- Yahyanejad, S.; van Hoof, S.J.; Theys, J.; Barbeau, L.M.O.; Granton, P.V.; Paesmans, K.; Verhaegen, F.; Vooijs, M. An Image Guided Small Animal Radiation Therapy Platform(SmART) to Monitor Glioblastoma Progression and Therapy. Radiother. Oncol. 2015, 116, 467–472. [Google Scholar] [CrossRef]
- Bell, H.S.; Whittle, I.R.; Walker, M.; Leaver, H.A.; Wharton, S.B. The Development of Necrosis and Apoptosis in Glioma: Experimental Findings Using Spheroid Culture Systems. Neuropathol. Appl. Neurobiol. 2001, 27, 291–304. [Google Scholar] [CrossRef]
- Watanabe, H.; Miura, M.; Sasaki, T. Differential Effects of the Insulin-like Growth Factor I on Radiosensitivity and Spontaneous Necrosis formation of Human Glioblastoma Cells Grown in Multicellular Spheroids. Exp. Cell Res. 1999, 250, 99–111. [Google Scholar] [CrossRef]
- Neufeld, L.; Yeini, E.; Reisman, N.; Shtilerman, Y.; Ben-Shushan, D.; Pozzi, S.; Madi, A.; Tiram, G.; Eldar-Boock, A.; Ferber, S.; et al. Microengineered Perfusable 3D-Bioprinted Glioblastoma Model for In Vivo Mimicry of Tumor Microenvironment. Sci. Adv. 2021, 7, eabi9119. [Google Scholar] [CrossRef] [PubMed]
- Preynat-Seauve, O.; Suter, D.M.; Tirefort, D.; Turchi, L.; Virolle, T.; Chneiweiss, H.; Foti, M.; Lobrinus, J.-A.; Stoppini, L.; Feki, A.; et al. Development of Human Nervous Tissue upon Differentiation of embryonic Stem Cells in Three-Dimensional Culture. Stem Cells 2009, 27, 509–520. [Google Scholar] [CrossRef]
- Azzarelli, R. Organoid Models of Glioblastoma to Study Brain Tumor Stem Cells. Front. Cell Dev. Biol. 2020, 8, 220. [Google Scholar] [CrossRef]
- Lancaster, M.A.; Knoblich, J.A. Generation of Cerebral Organoids from Human Pluripotent Stem. Nat. Protoc. 2014, 9, 2329–2340. [Google Scholar] [CrossRef]
- Bian, S.; Repic, M.; Guo, Z.; Kavirayani, A.; Burkard, T.; Bagley, J.A.; Krauditsch, C.; Knoblich, J.A. Genetically Engineered Cerebral Organoids Model Brain Tumor. Nat. Methods 2018, 15, 631–639. [Google Scholar] [CrossRef] [PubMed]
- Ogawa, J.; Pao, G.M.; Shokhirev, M.N.; Verma, I.M. Glioblastoma Model Using Human Cerebral Organoids. Cell Rep. 2018, 23, 1220–1229. [Google Scholar] [CrossRef]
- Nayernia, Z.; Turchi, L.; Cosset, E.; Peterson, H.; Dutoit, V.; Dietrich Pierre-Yves and Tirefort, D.; Chneiweiss, H.; Lobrinus, J.-A.; Krause, K.-H.; Virolle Thierry and Preynat-Seauve, O. The Relationship between Brain Tumor Cell Invasion of Engineered Tissues and In Vivo Features of Glioblastoma. Biomaterials 2013, 34, 8279–8290. [Google Scholar] [CrossRef] [PubMed]
- Linkous, A.; Balamatsias, D.; Snuderl Matija and Edwards, L.; Miyaguchi, K.; Milner, T.; Reich, B.; Cohen-Gould, L.; Storaska, A.; Nakayama, Y.; Schenkein, E.; et al. Modeling Patient-Derived Glioblastoma with Cerebral Organoids. Cell Rep. 2019, 26, 3203–3211.e5. [Google Scholar] [CrossRef]
- Fedorova, V.; Pospisilova, V.; Vanova Tereza and Amruz Cerna, K.; Abaffy, P.; Sedmik, J.; Raska, J.; Vochyanova, S.; Matusova, Z.; Houserova, J.; Valihrach, L.; et al. Glioblastoma and Cerebral Organoids: Development and Analysis of an In Vitro Model for Glioblastoma Migration. Mol. Oncol. 2023, 17, 647–663. [Google Scholar] [CrossRef]
- Plummer, S.; Wallace, S.; Ball, G.; Lloyd, R.; Schiapparelli, P.; Quiñones-Hinojosa, A.; Hartung, T.; Pamies, D. A Human IPSC-Derived 3D Platform Using Primary Brain Cancer to Study Drug Development and Personalized Medicine. Sci. Rep. 2019, 9, 1407. [Google Scholar] [CrossRef]
- Hubert, C.G.; Rivera, M.; Spangler Lisa, C.; Wu, Q.; Mack, S.C.; Prager, B.C.; Couce, M.; McLendon, R.E.; Sloan, A.E.; Rich, J.N. A Three-Dimensional Organoid Culture System Derived from Human Recapitulates the Hypoxic Gradients and Cancer Cell Heterogeneity of Tumors Found In Vivo. Cancer Res. 2016, 76, 2465–2477. [Google Scholar] [CrossRef]
- Jacob, F.; Salinas, R.D.; Zhang, D.Y.; Nguyen, P.T.T.; Schnoll, J.G.; Wong, S.Z.H.; Thokala, R.; Sheikh, S.; Saxena, D.; Prokop, S.; et al. A Patient-Derived Glioblastoma Organoid Model and Biobank Inter- and Intra-Tumoral Heterogeneity. Cell 2020, 180, 188–204.e22. [Google Scholar] [CrossRef] [PubMed]
- Tang, M.; Xie, Q.; Gimple, R.C.; Zhong, Z.; Tam, T.; Tian, J.; Kidwell, R.L.; Wu Qiulian and Prager, B.C.; Qiu, Z.; Yu, A.; et al. Three-Dimensional Bioprinted Glioblastoma Microenvironments Cellular Dependencies and Immune Interactions. Cell Res. 2020, 30, 833–853. [Google Scholar] [CrossRef] [PubMed]
- Jacob, F.; Ming, G.-L.; Song, H. Generation and Biobanking of Patient-Derived Glioblastoma and Their Application in CAR T Cell Testing. Nat. Protoc. 2020, 15, 4000–4033. [Google Scholar] [CrossRef]
- LeBlanc, V.G.; Trinh, D.L.; Aslanpour, S.; Hughes, M.; Livingstone, D.; Jin, D.; Ahn, B.Y.; Blough, M.D.; Cairncross, J.G.; Chan, J.A.; et al. Single-Cell Landscapes of Primary Glioblastomas and Matched and Cell Lines Show Variable Retention of Inter- and intratumor Heterogeneity. Cancer Cell 2022, 40, 379–392.e9. [Google Scholar] [CrossRef] [PubMed]
- Pedrosa, L.; Bedia, C.; Diao, D.; Mosteiro, A.; Ferres, A.; Stanzani, E.; Martinez-Soler, F.; Tortosa, A.; Pineda, E.; Aldecoa, I.; et al. Preclinical Studies with Glioblastoma Brain Organoid Co-Cultures Show Efficient 5-ALA Photodynamic Therapy. Cells 2023, 12, 1125. [Google Scholar] [CrossRef]
- Morelli, M.; Lessi, F.; Barachini, S.; Liotti, R.; Montemurro, N.; Perrini, P.; Santonocito, O.S.; Gambacciani, C.; Snuderl, M.; Pieri, F.; et al. Metabolic-Imaging of Human Glioblastoma Live Tumors: A New Precision-Medicine Approach to Predict Tumor Treatment Response Early. Front. Oncol. 2022, 12, 969812. [Google Scholar] [CrossRef] [PubMed]
- Mitchell, K.; Sprowls, S.A.; Arora, S.; Shakya, S.; Silver, D.J.; Goins, C.M.; Wallace, L.; Roversi, G.; Schafer, R.E.; Kay, K.; et al. WDR5 Represents a Therapeutically Exploitable Target for Cancer Stem Cells in Glioblastoma. Genes. Dev. 2023, 37, 86–102. [Google Scholar] [CrossRef] [PubMed]
- Yang, X.; Lu, D.; Sun, Y.; Wei, T.; Man, D.; Chen, A.; Luo, T.; Zhao, F.; Liu, X.; Cheng, B.; et al. Network Pharmacology and Experimental Verification Reveal the Mechanism of Safranal against Glioblastoma (GBM). Front. Oncol. 2023, 13, 1255164. [Google Scholar] [CrossRef] [PubMed]
- Kong, W.; Biddy, B.A.; Kamimoto, K.; Amrute, J.M.; Butka, E.G.; Morris, S.A. CellTagging: Combinatorial Indexing to Simultaneously Map and Identity at Single-Cell Resolution. Nat. Protoc. 2020, 15, 750–772. [Google Scholar] [CrossRef]
- Klein, E.; Hau, A.-C.; Oudin, A.; Golebiewska, A.; Niclou, S.P. Glioblastoma Organoids: Pre-Clinical Applications and challenges in the Context of Immunotherapy. Front. Oncol. 2020, 10, 604121. [Google Scholar] [CrossRef] [PubMed]
- Chongsathidkiet, P.; Jackson, C.; Koyama, S.; Loebel, F.; Cui, X.; Farber, S.H.; Woroniecka, K.; Elsamadicy, A.A.; Dechant, C.A.; Kemeny, H.R.; et al. Sequestration of T Cells in Bone Marrow in the Setting of glioblastoma and Other Intracranial Tumors. Nat. Med. 2018, 24, 1459–1468. [Google Scholar] [CrossRef] [PubMed]
- Woroniecka, K.I.; Rhodin, K.E.; Chongsathidkiet, P.; Keith, K.A.; Fecci, P.E. T-Cell Dysfunction in Glioblastoma: Applying a New Framework. Clin. Cancer Res. 2018, 24, 3792–3802. [Google Scholar] [CrossRef] [PubMed]
- Dijkstra, K.K.; Cattaneo, C.M.; Weeber, F.; Chalabi, M.; van de Haar, J.; Fanchi, L.F.; Slagter, M.; van der Velden, D.L.; Kaing Sovann and Kelderman, S.; van Rooij, N.; et al. Generation of Tumor-Reactive T Cells by Co-Culture of peripheral Blood Lymphocytes and Tumor Organoids. Cell 2018, 174, 1586–1598.e12. [Google Scholar] [CrossRef]
- Venkatesh, H.S.; Morishita, W.; Geraghty, A.C.; Silverbush, D.; Gillespie, S.M.; Arzt, M.; Tam, L.T.; Espenel, C.; Ponnuswami, A.; Ni, L.; et al. Electrical and Synaptic Integration of Glioma into Neural. Nature 2019, 573, 539–545. [Google Scholar] [CrossRef]
- Hausmann, D.; Hoffmann, D.C.; Venkataramani Varun and Jung, E.; Horschitz, S.; Tetzlaff, S.K.; Jabali, A.; Hai, L.; Kessler, T.; Azoŕin, D.D.; Weil, S.; et al. Autonomous Rhythmic Activity in Glioma Networks Drives Brain Growth. Nature 2023, 613, 179–186. [Google Scholar] [CrossRef]
- Wang, C.; Yu, M.; Zhang, W. Neoantigen Discovery and Applications in Glioblastoma: An Immunotherapy Perspective. Cancer Lett. 2022, 550, 215945. [Google Scholar] [CrossRef]
- Watson, D.C.; Bayik, D.; Storevik, S.; Moreino, S.S.; Sprowls, S.A.; Augustsson, M.T.H.J.; Lauko, A.; Røsland, G.V.S.P.; Troike, K.; Tronstad, K.J.; et al. GAP43-Dependent Mitochondria Transfer from Astrocytes Enhances Tumorigenicity. Nat. Cancer 2023, 4, 648–664. [Google Scholar] [CrossRef]
- Pinto, G.; Saenz-de-Santa-Maria, I.; Chastagner, P.; Perthame, E.; Delmas, C.; Toulas, C.; Moyal-Jonathan-Cohen, E.; Brou Christel and Zurzolo, C. Patient-Derived Glioblastoma Stem Cells Transfer mitochondria through Tunneling Nanotubes in Tumor Organoids. Biochem. J. 2021, 478, 21–39. [Google Scholar] [CrossRef] [PubMed]
- Mallick, K.K.; Cox, S.C. Biomaterial Scaffolds for Tissue Engineering. Front. Biosci. 2013, 5, 341–360. [Google Scholar] [CrossRef]
- Erickson, A.E.; Lan Levengood, S.K.; Sun, J.; Chang, F.-C.; Zhang, M. Fabrication and Characterization of Chitosan-Hyaluronic Acid with Varying Stiffness for Glioblastoma Cell Culture. Adv. Healthc. Mater. 2018, 7, e1800295. [Google Scholar] [CrossRef] [PubMed]
- Chaicharoenaudomrung, N.; Kunhorm, P.; Promjantuek, W.; Heebkaew, N.; Rujanapun, N.; Noisa, P. Fabrication of 3D Calcium-Alginate Scaffolds for Human Modeling and Anticancer Drug Response Evaluation. J. Cell. Physiol. 2019, 234, 20085–20097. [Google Scholar] [CrossRef] [PubMed]
- Xiao, Y.; Kim, D.; Dura, B.; Zhang, K.; Yan, R.; Li, H.; Han, E.; Ip, J.; Zou, P.; Liu, J.; et al. Ex Vivo Dynamics of Human Glioblastoma Cells in A-on-a-Chip System Correlates with Tumor and Subtypes. Adv. Sci 2019, 6, 1801531. [Google Scholar] [CrossRef] [PubMed]
- Lv, D.; Yu, S.-C.; Ping, Y.-F.; Wu, H.; Zhao, X.; Zhang, H.; Cui, Y.; Chen Bing and Zhang, X.; Dai, J.; Bian, X.-W.; et al. A Three-Dimensional Collagen Scaffold Cell Culture System for screening Anti-Glioma Therapeutics. Oncotarget 2016, 7, 56904–56914. [Google Scholar] [CrossRef] [PubMed]
- Hospodiuk, M.; Dey, M.; Sosnoski, D.; Ozbolat, I.T. The Bioink: A Comprehensive Review on Bioprintable Materials. Biotechnol. Adv. 2017, 35, 217–239. [Google Scholar] [CrossRef]
- Matai, I.; Kaur, G.; Seyedsalehi, A.; McClinton, A.; Laurencin, C.T. Progress in 3D Bioprinting Technology for Tissue/Organ Engineering. Biomaterials 2020, 226, 119536. [Google Scholar] [CrossRef]
- Dai, X.; Ma, C.; Lan, Q.; Xu, T. 3D Bioprinted Glioma Stem Cells for Brain Tumor Model and applications of Drug Susceptibility. Biofabrication 2016, 8, 45005. [Google Scholar] [CrossRef] [PubMed]
- Tasoglu, S.; Demirci, U. Bioprinting for Stem Cell Research. Trends Biotechnol. 2013, 31, 10–19. [Google Scholar] [CrossRef] [PubMed]
- Hermida, M.A.; Kumar, J.D.; Schwarz, D.; Laverty, K.G.; Di Bartolo, A.; Ardron, M.; Bogomolnijs, M.; Clavreul, A.; Brennan Paul, M.; Wiegand, U.K.; et al. Three Dimensional In Vitro Models of Cancer: Bioprinting Glioblastoma Models. Adv. Biol. Regul. 2020, 75, 100658. [Google Scholar] [CrossRef]
- Wang, X.; Li, X.; Dai, X.; Zhang Xinzhi and Zhang, J.; Xu, T.; Lan, Q. Bioprinting of Glioma Stem Cells Improves Their Endotheliogenic. Colloids Surf. B Biointerfaces 2018, 171, 629–637. [Google Scholar] [CrossRef]
- Yeini, E.; Ofek, P.; Pozzi, S.; Albeck, N.; Ben-Shushan, D.; Tiram, G.; Golan Sapir and Kleiner, R.; Sheinin, R.; Israeli Dangoor, S.; Reich-Zeliger, S.; et al. P-Selectin Axis Plays a Key Role in Microglia immunophenotype and Glioblastoma Progression. Nat. Commun. 2021, 12, 1912. [Google Scholar] [CrossRef] [PubMed]
- Heinrich, M.A.; Bansal, R.; Lammers Twan and Zhang, Y.S.; Michel Schiffelers, R.; Prakash, J. 3D-Bioprinted Mini-Brain: A Glioblastoma Model to Study Cellular and Therapeutics. Adv. Mater. 2019, 31, e1806590. [Google Scholar] [CrossRef]
- Ashammakhi, N.; Ahadian, S.; Zengjie, F.; Suthiwanich, K.; Lorestani, F.; Orive, G.; Ostrovidov, S.; Khademhosseini, A. Advances and Future Perspectives in 4D Bioprinting. Biotechnol. J. 2018, 13, e1800148. [Google Scholar] [CrossRef] [PubMed]
- Truong, D.; Fiorelli, R.; Barrientos, E.S.; Melendez, E.L.; Sanai, N.; Mehta, S.; Nikkhah, M. A Three-Dimensional (3D) Organotypic Microfluidic Model for glioma Stem Cells—Vascular Interactions. Biomaterials 2019, 198, 63–77. [Google Scholar] [CrossRef]
- Yang, G.H.; Yeo, M.; Koo, Y.W.; Kim, G.H. 4D Bioprinting: Technological Advances in Biofabrication. Macromol. Biosci. 2019, 19, e1800441. [Google Scholar] [CrossRef]
- Ruskowitz, E.; DeForest, C. Photoresponsive Biomaterials for Targeted Drug Delivery and 4D Cell Culture. Nat. Rev. Mater. 2018, 3, 17087. [Google Scholar] [CrossRef]
- Gao, B.; Yang, Q.; Zhao, X.; Jin, G.; Ma, Y.; Xu, F. 4D Bioprinting for Biomedical Applications. Trends Biotechnol. 2016, 34, 746–756. [Google Scholar] [CrossRef]
- Mehling, M.; Tay, S. Microfluidic Cell Culture. Curr. Opin. Biotechnol. 2014, 25, 95–102. [Google Scholar] [CrossRef]
- Novo, P.; Volpetti, F.; Chu, V.; Conde, J.P. Control of Sequential Fluid Delivery in a Fully Autonomous Microfluidic Device. Lab. Chip 2013, 13, 641–645. [Google Scholar] [CrossRef]
- Bennett, M.R.; Pang, W.L.; Ostroff Natalie, A.; Baumgartner, B.L.; Nayak, S.; Tsimring Lev, S.; Hasty, J. Metabolic Gene Regulation in a Dynamically Changing Environment. Nature 2008, 454, 1119–1122. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.-A.; King, A.D.; Shih, H.-C.; Peng, C.-C.; Wu, C.-Y.; Liao, W.-H.; Tung, Y.-C. Generation of Oxygen Gradients in Microfluidic Devices for Cell Using Spatially Confined Chemical Reactions. Lab. Chip 2011, 11, 3626–3633. [Google Scholar] [CrossRef]
- Chiu, D.T.; Jeon, N.L.; Huang, S.; Kane, R.S.; Wargo, C.J.; Choi, I.S.; Ingber, D.E.; Whitesides, G.M. Patterned Deposition of Cells and Proteins onto Surfaces by using Three-Dimensional Microfluidic Systems. Proc. Natl. Acad. Sci. USA 2000, 97, 2408–2413. [Google Scholar] [CrossRef] [PubMed]
- Gottwald, E.; Giselbrecht, S.; Augspurger Caroline and Lahni, B.; Dambrowsky, N.; Truckenmüller, R.; Piotter, V.; Gietzelt, T.; Wendt Oliver and Pfleging, W.; Welle, A.; Rolletschek Alexandra and Wobus, A.M.; et al. A Chip-Based Platform for the In Vitro Generation of Tissues in three-Dimensional Organization. Lab. Chip 2007, 7, 777–785. [Google Scholar] [CrossRef]
- Woodruff, K.; Fidalgo, L.M.; Gobaa, S.; Lutolf, M.P.; Maerkl, S.J. Live Mammalian Cell Arrays. Nat. Methods 2013, 10, 550–552. [Google Scholar] [CrossRef]
- Vyawahare, S.; Griffiths, A.D.; Merten, C.A. Miniaturization and Parallelization of Biological and Chemical in Microfluidic Devices. Chem. Biol. 2010, 17, 1052–1065. [Google Scholar] [CrossRef]
- Gómez-Sjöberg, R.; Leyrat, A.A.; Pirone, D.M.; Chen, C.S.; Quake, S.R. Versatile, Fully Automated, Microfluidic Cell Culture System. Anal. Chem. 2007, 79, 8557–8563. [Google Scholar] [CrossRef]
- Vedel, S.; Tay, S.; Johnston, D.M.; Bruus, H.; Quake, S.R. Migration of Cells in a Social Context. Proc. Natl. Acad. Sci. USA 2013, 110, 129–134. [Google Scholar] [CrossRef]
- Toepke, M.W.; Beebe, D.J. PDMS Absorption of Small Molecules and Consequences in microfluidic Applications. Lab. Chip 2006, 6, 1484–1486. [Google Scholar] [CrossRef] [PubMed]
- Lecault, V.; Vaninsberghe, M.; Sekulovic, S.; Knapp, D.J.H.F.; Wohrer, S.; Bowden, W.; Viel, F.; McLaughlin, T.; Jarandehei, A.; Miller, M.; et al. High-Throughput Analysis of Single Hematopoietic Stem Cell in Microfluidic Cell Culture Arrays. Nat. Methods 2011, 8, 581–586. [Google Scholar] [CrossRef] [PubMed]
- Melin, J.; Quake, S.R. Microfluidic Large-Scale Integration: The Evolution of Design for Biological Automation. Annu. Rev. Biophys. Biomol. Struct. 2007, 36, 213–231. [Google Scholar] [CrossRef] [PubMed]
- Paguirigan, A.L.; Beebe, D.J. From the Cellular Perspective: Exploring Differences in the cellular Baseline in Macroscale and Microfluidic Cultures. Integr. Biol. 2009, 1, 182–195. [Google Scholar] [CrossRef]
- Regehr, K.J.; Domenech, M.; Koepsel, J.T.; Carver, K.C.; Ellison-Zelski, S.J.; Murphy, W.L.; Schuler, L.A.; Alarid, E.T.; Beebe, D.J. Biological Implications of Polydimethylsiloxane-Based Cell Culture. Lab. Chip 2009, 9, 2132–2139. [Google Scholar] [CrossRef]
- McDonald, J.C.; Duffy, D.C.; Anderson, J.R.; Chiu, D.T.; Wu, H.; Schueller, O.J.; Whitesides, G.M. Fabrication of Microfluidic Systems in Poly(Dimethylsiloxane). Electrophoresis 2000, 21, 27–40. [Google Scholar] [CrossRef]
- Sia, S.K.; Whitesides, G.M. Microfluidic Devices Fabricated in Poly(Dimethylsiloxane) for biological Studies. Electrophoresis 2003, 24, 3563–3576. [Google Scholar] [CrossRef] [PubMed]
- Zhao, W.; Cheng, R.; Miller, J.R.; Mao, L. Label-Free Microfluidic Manipulation of Particles and Cells in magnetic Liquids. Adv. Funct. Mater. 2016, 26, 3916–3932. [Google Scholar] [CrossRef] [PubMed]
- Gossett, D.R.; Weaver, W.M.; Mach, A.J.; Hur, S.C.; Tse, H.T.K.; Lee, W.; Amini, H.; Di Carlo, D. Label-Free Cell Separation and Sorting in Microfluidic Systems. Anal. Bioanal. Chem. 2010, 397, 3249–3267. [Google Scholar] [CrossRef] [PubMed]
- Didar, T.F.; Tabrizian, M. Adhesion Based Detection, Sorting and Enrichment of Cells in microfluidic Lab-on-Chip Devices. Lab. Chip 2010, 10, 3043–3053. [Google Scholar] [CrossRef]
- Sullivan, J.P.; Nahed, B.V.; Madden, M.W.; Oliveira, S.M.; Springer, S.; Bhere, D.; Chi, A.S.; Wakimoto, H.; Rothenberg, S.M.; Sequist, L.V.; et al. Brain Tumor Cells in Circulation Are Enriched for Mesenchymal Expression. Cancer Discov. 2014, 4, 1299–1309. [Google Scholar] [CrossRef]
- Wan, Y.; Tan, J.; Asghar, W.; Kim Young-Tae and Liu, Y.; Iqbal, S.M. Velocity Effect on Aptamer-Based Circulating Tumor Cell in Microfluidic Devices. J. Phys. Chem. B 2011, 115, 13891–13896. [Google Scholar] [CrossRef]
- Zhang, Y.; Wong, C.Y.; Lim, C.Z.J.; Chen, Q.; Yu, Z.; Natalia, A.; Wang, Z.; Pang, Q.Y.; Lim, S.W.; Loh, T.P.; et al. Multiplexed RNA Profiling by Regenerative Catalysis Enables Blood-Based Subtyping of Brain Tumors. Nat. Commun. 2023, 14, 4278. [Google Scholar] [CrossRef]
- Logun, M.; Zhao, W.; Mao, L.; Karumbaiah, L. Microfluidics in Malignant Glioma Research and Precision Medicine. Adv. Biosyst. 2018, 2, 1700221. [Google Scholar] [CrossRef] [PubMed]
- Chang, T.C.; Mikheev, A.M.; Huynh, W.; Monnat, R.J.; Rostomily, R.C.; Folch, A. Parallel Microfluidic Chemosensitivity Testing on Individual Cultures. Lab. Chip 2014, 14, 4540–4551. [Google Scholar] [CrossRef] [PubMed]
- Jonas, O.; Landry, H.M.; Fuller, J.E.; Santini Jr, J.T.; Baselga, J.; Tepper, R.I.; Cima, M.J.; Langer, R. An Implantable Microdevice to Perform High-Throughput In Vivo Sensitivity Testing in Tumors. Sci. Transl. Med. 2015, 7, 284ra57. [Google Scholar] [CrossRef]
- Sun, Z.; Fu, X.; Zhang, L.; Yang, X.; Liu, F.; Hu, G. A Protein Chip System for Parallel Analysis of Multi-Tumor and Its Application in Cancer Detection. Anticancer. Res. 2004, 24, 1159–1165. [Google Scholar]
- Shao, H.; Chung, J.; Lee, K.; Balaj, L.; Min, C.; Carter, B.S.; Hochberg, F.H.; Breakefield, X.O.; Lee, H.; Weissleder, R. Chip-Based Analysis of Exosomal MRNA Mediating Drug resistance in Glioblastoma. Nat. Commun. 2015, 6, 6999. [Google Scholar] [CrossRef] [PubMed]
- Sun, J.; Masterman-Smith, M.D.; Graham Nicholas, A.; Jiao, J.; Mottahedeh, J.; Laks, D.R.; Ohashi, M.; DeJesus, J.; Kamei, K.-I.; Lee, K.-B.; et al. A Microfluidic Platform for Systems Pathology: Multiparameter-Cell Signaling Measurements of Clinical Brain Tumor. Cancer Res. 2010, 70, 6128–6138. [Google Scholar] [CrossRef]
- Liga, A.; Vliegenthart, A.D.B.; Oosthuyzen, W.; Dear J W and Kersaudy-Kerhoas, M. Exosome Isolation: A Microfluidic Road-Map. Lab. Chip 2015, 15, 2388–2394. [Google Scholar] [CrossRef]
- Shao, H.; Chung, J.; Balaj, L.; Charest, A.; Bigner, D.D.; Carter, B.S.; Hochberg, F.H.; Breakefield, X.O.; Weissleder, R.; Lee, H. Protein Typing of Circulating Microvesicles Allows Real-Time of Glioblastoma Therapy. Nat. Med. 2012, 18, 1835–1840. [Google Scholar] [CrossRef] [PubMed]
- Bauer, M.; Su, G.; Beebe, D.J.; Friedl, A. 3D Microchannel Co-Culture: Method and Biological Validation. Integr. Biol. 2010, 2, 371–378. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Altiok, S.; Mezzadra, H.; Jagannath, S.; Tsottles, N.; Rudek, M.A.; Abdallah, N.; Berman, D.; Forastiere, A.; Gibson, M.K. A Novel Pharmacodynamic Approach to Assess and Predict Tumor to the Epidermal Growth Factor Receptor Inhibitor in Patients with Esophageal Cancer. Int. J. Oncol. 2010, 36, 19–27. [Google Scholar] [CrossRef]
- Olubajo, F.; Achawal, S.; Greenman, J. Development of a Microfluidic Culture Paradigm for Ex Vivo Maintenance of Human Glioblastoma Tissue: A New Glioblastoma Model? Transl. Oncol. 2020, 13, 1–10. [Google Scholar] [CrossRef]
- Huang, Y.; Agrawal, B.; Clark, P.A.; Williams, J.C.; Kuo, J.S. Evaluation of Cancer Stem Cell Migration Using Microfluidic Devices and Live Cell Imaging. J. Vis. Exp. 2011, 58, e3297. [Google Scholar]
- Walsh, C.L.; Babin, B.M.; Kasinskas, R.W.; Foster, J.A.; McGarry, M.J.; Forbes, N.S. A Multipurpose Microfluidic Device Designed to Mimic Gradients and Develop Targeted Cancer. Lab. Chip 2009, 9, 545–554. [Google Scholar] [CrossRef] [PubMed]
- Ozturk, M.S.; Lee, V.K.; Zou, H.; Friedel, R.H.; Intes, X.; Dai, G. High-Resolution Tomographic Analysis of In Vitro 3D Glioblastoma Tumor Model under Long-Term Drug Treatment. Sci. Adv. 2020, 6, eaay7513. [Google Scholar] [CrossRef]
- Akay, M.; Hite, J.; Avci, N.G.; Fan, Y.; Akay, Y.; Lu, G.; Zhu, J.J. Drug Screening of Human GBM Spheroids in Brain Cancer Chip. Sci. Rep. 2018, 8, 15423. [Google Scholar] [CrossRef] [PubMed]
- Nizar, R.; Cazacu, S.; Xiang, C.; Krasner, M.; Barbiro-Michaely, E.; Gerber, D.; Schwartz, J.; Fried, I.; Yuval, S.; Brodie, A.; et al. Propofol Inhibits Glioma Stem Cell Growth and Migration and Their Interaction with Microglia via BDNF-AS and Extracellular Vesicles. Cells 2023, 12, 1921. [Google Scholar] [CrossRef]
- Han, J.; Jun, Y.; Kim, S.H.; Hoang, H.-H.; Jung, Y.; Kim, S.; Kim, J.; Austin, R.H.; Lee, S.; Park, S. Rapid Emergence and Mechanisms of Resistance by U87 Cells to Doxorubicin in an In Vitro Tumor Ecology. Proc. Natl. Acad. Sci. USA 2016, 113, 14283–14288. [Google Scholar] [CrossRef]
- Zhang, Q.; Lambert, G.; Liao, D.; Kim, H.; Robin, K.; Tung, C.-K.; Pourmand, N.; Austin, R.H. Acceleration of Emergence of Bacterial Antibiotic Resistance in connected Microenvironments. Science (1979) 2011, 333, 1764–1767. [Google Scholar] [CrossRef]
- Aref, A.R.; Huang, R.Y.-J.; Yu, W.; Chua, K.-N.; Sun, W.; Tu, T.-Y.; Bai, J.; Sim, W.-J.; Zervantonakis, I.K.; Thiery, J.P.; et al. Screening Therapeutic EMT Blocking Agents in A-Dimensional Microenvironment. Integr. Biol. 2013, 5, 381–389. [Google Scholar] [CrossRef]
- Booth, R.; Kim, H. Characterization of a Microfluidic In Vitro Model of The-Brain Barrier (ΜBBB). Lab. Chip 2012, 12, 1784–1792. [Google Scholar] [CrossRef]
- Cucullo, L.; Marchi, N.; Hossain, M.; Janigro, D. A Dynamic In Vitro BBB Model for the Study of Immune Cell into the Central Nervous System. J. Cereb. Blood Flow. Metab. 2011, 31, 767–777. [Google Scholar] [CrossRef]
- Prabhakarpandian, B.; Shen, M.-C.; Nichols, J.B.; Mills, I.R.; Sidoryk-Wegrzynowicz, M.; Aschner, M.; Pant, K. SyM-BBB: A Microfluidic Blood Brain Barrier Model. Lab. Chip 2013, 13, 1093–1101. [Google Scholar] [CrossRef]
- Marino, A.; Battaglini, M.; Carmignani, A.; Pignatelli, F.; De Pasquale, D.; Tricinci, O.; Ciofani, G. Magnetic Self-Assembly of 3D Multicellular Microscaffolds: A Biomimetic Brain Tumor-on-a-Chip for Drug Delivery and Selectivity Testing. APL Bioeng. 2023, 7, 36103. [Google Scholar] [CrossRef]
- Chen, D.; Love, K.T.; Chen, Y.; Eltoukhy, A.A.; Kastrup, C.; Sahay, G.; Jeon, A.; Dong, Y.; Whitehead, K.A.; Anderson, D.G. Rapid Discovery of Potent SiRNA-Containing Lipid Nanoparticles by Controlled Microfluidic Formulation. J. Am. Chem. Soc. 2012, 134, 6948–6951. [Google Scholar] [CrossRef]
- Mendanha, D.; Gimondi, S.; Costa, B.M.; Ferreira, H.; Neves, N.M. Microfluidic-Derived Docosahexaenoic Acid Liposomes for Glioblastoma Therapy. Nanomedicine 2023, 53, 102704. [Google Scholar] [CrossRef]
- Brown, D.V.; Filiz, G.; Daniel, P.M.; Hollande, F.; Dworkin, S.; Amiridis, S.; Kountouri, N.; Ng, W.; Morokoff, A.P.; Mantamadiotis, T. Expression of CD133 and CD44 in Glioblastoma Stem Cells with Cell Proliferation, Phenotype Stability and intra-Tumor Heterogeneity. PLoS ONE 2017, 12, e0172791. [Google Scholar] [CrossRef]
- Yi, H.-G.; Jeong, Y.H.; Kim, Y.; Choi, Y.-J.; Moon, H.E.; Park, S.H.; Kang, K.S.; Bae, M.; Jang, J.; Youn, H.; et al. A Bioprinted Human-Glioblastoma-on-a-Chip for the identification of Patient-Specific Responses to Chemoradiotherapy. Nat. Biomed. Eng. 2019, 3, 509–519. [Google Scholar] [CrossRef] [PubMed]
- Aijian, A.P.; Garrell, R.L. Digital Microfluidics for Automated Hanging Drop Cell Spheroid. J. Lab. Autom. 2015, 20, 283–295. [Google Scholar] [CrossRef] [PubMed]
- Nguyen, D.-H.T.; Stapleton, S.C.; Yang, M.T.; Cha, S.S.; Choi, C.K.; Galie, P.A.; Chen, C.S. Biomimetic Model to Reconstitute Angiogenic Sprouting In Vitro. Proc. Natl. Acad. Sci. USA 2013, 110, 6712–6717. [Google Scholar] [CrossRef]
- Chaw, K.C.; Manimaran, M.; Tay, E.H.; Swaminathan, S. Multi-Step Microfluidic Device for Studying Cancer Metastasis. Lab. Chip 2007, 7, 1041–1047. [Google Scholar] [CrossRef] [PubMed]
- Friedl, P.; Alexander, S. Cancer Invasion and the Microenvironment: Plasticity and reciprocity. Cell 2011, 147, 992–1009. [Google Scholar] [CrossRef]
- Zhong, J.; Paul, A.; Kellie, S.J.; O’Neill, G.M. Mesenchymal Migration as a Therapeutic Target in Glioblastoma. J. Oncol. 2010, 2010, 430142. [Google Scholar] [CrossRef]
- Rao, J.S. Molecular Mechanisms of Glioma Invasiveness: The Role of proteases. Nat. Rev. Cancer 2003, 3, 489–501. [Google Scholar] [CrossRef] [PubMed]
- Irimia, D.; Toner, M. Spontaneous Migration of Cancer Cells under Conditions of mechanical Confinement. Integr. Biol. 2009, 1, 506–512. [Google Scholar] [CrossRef]
- Mak, M.; Reinhart-King, C.A.; Erickson, D. Elucidating Mechanical Transition Effects of Invading Cancer with a Subnucleus-Scaled Microfluidic Serial Dimensional Device. Lab. Chip 2013, 13, 340–348. [Google Scholar] [CrossRef]
- Kievit, F.M.; Florczyk, S.J.; Leung, M.C.; Wang, K.; Wu, J.D.; Silber, J.R.; Ellenbogen, R.G.; Lee, J.S.H.; Zhang, M. Proliferation and Enrichment of CD133(+) Glioblastoma Cancer Cells on 3D Chitosan-Alginate Scaffolds. Biomaterials 2014, 35, 9137–9143. [Google Scholar] [CrossRef]
- Kravchenko-Balasha, N.; Wang, J.; Remacle Francoise and Levine, R.D.; Heath, J.R. Glioblastoma Cellular Architectures Are Predicted through the characterization of Two-Cell Interactions. Proc. Natl. Acad. Sci. USA 2014, 111, 6521–6526. [Google Scholar] [CrossRef]
- Nguyen, D.X.; Bos, P.D.; Massagué, J. Metastasis: From Dissemination to Organ-Specific Colonization. Nat. Rev. Cancer 2009, 9, 274–284. [Google Scholar] [CrossRef] [PubMed]
- Hong, X.; Sin, W.C.; Harris, A.L.; Naus, C.C. Gap Junctions Modulate Glioma Invasion by Direct Transfer of microRNA. Oncotarget 2015, 6, 15566–15577. [Google Scholar] [CrossRef]
- Geribaldi-Doldan, N.; Fernandez-Ponce, C.; Quiroz, R.N.; Sanchez-Gomar, I.; Escorcia, L.G.; Velasquez, E.P.; Quiroz, E.N. The Role of Microglia in Glioblastoma. Front. Oncol. 2020, 10, 603495. [Google Scholar] [CrossRef] [PubMed]
- Gu, R.; Zhang, X.; Zhang, G.; Tao, T.; Yu, H.; Liu, L.; Dou, Y.; Li, A.; Qin, J. Probing the Bi-Directional Interaction Between Microglia and Gliomas in a Tumor Microenvironment on a Microdevice. Neurochem. Res. 2017, 42, 1478–1487. [Google Scholar] [CrossRef] [PubMed]
- Hong, S.; You, J.Y.; Paek, K.; Park, J.; Kang, S.J.; Han, E.H.; Choi, N.; Chung, S.; Rhee, W.J.; Kim, J.A. Inhibition of Tumor Progression and M2 Microglial Polarization by Extracellular Vesicle-Mediated MicroRNA-124 in a 3D Microfluidic Glioblastoma Microenvironment. Theranostics 2021, 11, 9687–9704. [Google Scholar] [CrossRef] [PubMed]
- Adjei-Sowah, E.A.; O’Connor, S.A.; Veldhuizen, J.; Lo Cascio, C.; Plaisier, C.; Mehta, S.; Nikkhah, M. Investigating the Interactions of Glioma Stem Cells in the Perivascular Niche at Single-Cell Resolution Using a Microfluidic Tumor Microenvironment Model. Adv. Sci. 2022, 9, e2201436. [Google Scholar] [CrossRef]
- Heldin, C.-H.; Rubin, K.; Pietras Kristian and Ostman, A. High Interstitial Fluid Pressure—An Obstacle in Cancer Therapy. Nat. Rev. Cancer 2004, 4, 806–813. [Google Scholar] [CrossRef]
- Huynh, G.H.; Deen, D.F.; Szoka Jr, F.C. Barriers to Carrier Mediated Drug and Gene Delivery to Brain. J. Control. Release 2006, 110, 236–259. [Google Scholar] [CrossRef] [PubMed]
- Hsu, Y.-H.; Moya, M.L.; Hughes Christopher, C.W.; George, S.C.; Lee, A.P. A Microfluidic Platform for Generating Large-Scale Nearly Human Microphysiological Vascularized Tissue Arrays. Lab. Chip 2013, 13, 2990–2998. [Google Scholar] [CrossRef]
- van Duinen, V.; Trietsch, S.J.; Joore Jos and Vulto, P.; Hankemeier, T. Microfluidic 3D Cell Culture: From Tools to Tissue Models. Curr. Opin. Biotechnol. 2015, 35, 118–126. [Google Scholar] [CrossRef]
- Rong, Y.; Durden, D.L.; Van Meir, E.G.; Brat, D.J. “Pseudopalisading” Necrosis in Glioblastoma: A Familiar Feature That Links Vascular Pathology, Hypoxia, and angiogenesis. J. Neuropathol. Exp. Neurol. 2006, 65, 529–539. [Google Scholar] [CrossRef]
- Polacheck, W.J.; Charest, J.L.; Kamm, R.D. Interstitial Flow Influences Direction of Tumor Cell migration through Competing Mechanisms. Proc. Natl. Acad. Sci. USA 2011, 108, 11115–11120. [Google Scholar] [CrossRef]
- Chen, M.B.; Whisler, J.A.; Fröse, J.; Yu, C.; Shin, Y.; Kamm, R.D. On-Chip Human Microvasculature Assay for Visualization and quantification of Tumor Cell Extravasation Dynamics. Nat. Protoc. 2017, 12, 865–880. [Google Scholar] [CrossRef]
- Stroock, A.D.; Fischbach, C. Microfluidic Culture Models of Tumor Angiogenesis. Tissue Eng. Part. A 2010, 16, 2143–2146. [Google Scholar] [CrossRef]
- Oppegard, S.C.; Eddington, D.T. A Microfabricated Platform for Establishing Oxygen Gradients In-D Constructs. Biomed. Microdevices 2013, 15, 407–414. [Google Scholar] [CrossRef] [PubMed]
- Funamoto, K.; Zervantonakis, I.K.; Liu Yuchun and Ochs, C.J.; Kim, C.; Kamm, R.D. A Novel Microfluidic Platform for High-Resolution Imaging of a three-Dimensional Cell Culture under a Controlled Hypoxic. Lab. Chip 2012, 12, 4855–4863. [Google Scholar] [CrossRef]
- Cui, X.; Ma, C.; Vasudevaraja, V.; Serrano, J.; Tong, J.; Peng, Y.; Delorenzo, M.; Shen, G.; Frenster, J.; Morales, R.T.; et al. Dissecting the Immunosuppressive Tumor Microenvironments in Glioblastoma-on-a-Chip for Optimized PD-1 Immunotherapy. eLife 2020, 9, e52253. [Google Scholar] [CrossRef] [PubMed]
- Feder-Mengus, C.; Ghosh, S.; Reschner, A.; Martin, I.; Spagnoli, G.C. New Dimensions in Tumor Immunology: What Does 3D Culture? Trends Mol. Med. 2008, 14, 333–340. [Google Scholar] [CrossRef]
- Labelle, M.; Begum, S.; Hynes, R.O. Direct Signaling between Platelets and Cancer Cells Induces An-Mesenchymal-like Transition and Promotes Metastasis. Cancer Cell 2011, 20, 576–590. [Google Scholar] [CrossRef]
- Kim, H.J.; Huh, D.; Hamilton, G.; Ingber, D.E. Human Gut-on-a-Chip Inhabited by Microbial Flora That Intestinal Peristalsis-like Motions and Flow. Lab. Chip 2012, 12, 2165–2174. [Google Scholar] [CrossRef] [PubMed]
- Nesmith, A.P.; Agarwal, A.; McCain, M.L.; Parker, K.K. Human Airway Musculature on a Chip: An In Vitro Model of allergic Asthmatic Bronchoconstriction and Bronchodilation. Lab. Chip 2014, 14, 3925–3936. [Google Scholar] [CrossRef]
- Huang, J.; Li, Y.B.; Charlebois, C.; Nguyen, T.; Liu, Z.; Bloemberg, D.; Zafer, A.; Baumann, E.; Sodja, C.; Leclerc, S.; et al. Application of Blood Brain Barrier Models in Pre-Clinical Assessment of Glioblastoma-Targeting CAR-T Based Immunotherapies. Fluids Barriers CNS 2022, 19, 38. [Google Scholar] [CrossRef]
- Xie, Z.; Chen, M.; Lian, J.; Wang, H.; Ma, J. Glioblastoma-on-a-Chip Construction and Therapeutic Applications. Front. Oncol. 2023, 13, 1183059. [Google Scholar] [CrossRef]
- Greene, H.S.N.; Arnold, H. The Homologous and Heterologous Transplantation of Brain and Brain Tumors. J. Neurosurg. 1945, 2, 315–331. [Google Scholar] [CrossRef]
- Huszthy, P.C.; Daphu, I.; Niclou, S.P.; Stieber, D.; Nigro, J.M.; Sakariassen, P.Ø.; Miletic, H.; Thorsen, F.; Bjerkvig, R. In Vivo Models of Primary Brain Tumors: Pitfalls and Perspectives. Neuro Oncol. 2012, 14, 979–993. [Google Scholar] [CrossRef]
- Simeonova, I.; Huillard, E. In Vivo Models of Brain Tumors: Roles of Genetically Engineered Mouse Models in Understanding Tumor Biology and Use in Preclinical Studies. Cell Mol. Life Sci. 2014, 71, 4007–4026. [Google Scholar] [CrossRef] [PubMed]
- Hicks, W.H.; Bird, C.E.; Traylor, J.I.; Shi, D.D.; El Ahmadieh, T.Y.; Richardson, T.E.; McBrayer, S.K.; Abdullah, K.G. Contemporary Mouse Models in Glioma Research. Cells 2021, 10, 712. [Google Scholar] [CrossRef]
- Dagle, G.E.; Zwicker, G.M.; Renne, R.A. Morphology of Spontaneous Brain Tumors in the Rat. Vet. Pathol. 1979, 16, 318–324. [Google Scholar] [CrossRef] [PubMed]
- Lantos, P.L.; Pilkington, G.J. Neuroblasts in Cerebral Tumors Induced by Ethylnitrosourea in Rats. A Fine Structrual Study. Virchows Arch. B Cell Pathol. 1977, 25, 243–259. [Google Scholar] [CrossRef]
- Bulnes, S.; Murueta-Goyena, A.; Lafuente, J.V. Differential Exposure to N-Ethyl N-Nitrosourea during Pregnancy Is Relevant to the Induction of Glioma and PNSTs in the Brain. Neurotoxicol. Teratol. 2021, 86, 106998. [Google Scholar] [CrossRef]
- Laerum, O.D.; Rajewsky, M.F. Neoplastic Transformation of Fetal Rat Brain Cells in Culture after Exposure to Ethylnitrosourea In Vivo. J. Natl. Cancer Inst. 1975, 55, 1177–1187. [Google Scholar] [CrossRef]
- Louis, D.N.; Perry, A.; Wesseling, P.; Brat, D.J.; Cree, I.A.; Figarella-Branger, D.; Hawkins, C.; Ng, H.K.; Pfister, S.M.; Reifenberger, G.; et al. The 2021 WHO Classification of Tumors of the Central Nervous System: A Summary. Neuro Oncol. 2021, 23, 1231–1251. [Google Scholar] [CrossRef] [PubMed]
- Gómez-Oliva, R.; Domínguez-García, S.; Carrascal, L.; Abalos-Martínez, J.; Pardillo-Díaz, R.; Verástegui, C.; Castro, C.; Nunez-Abades, P.; Geribaldi-Doldán, N. Evolution of Experimental Models in the Study of Glioblastoma: Toward Finding Efficient Treatments. Front. Oncol. 2020, 10, 614295. [Google Scholar] [CrossRef]
- Kersten, K.; de Visser, K.E.; van Miltenburg, M.H.; Jonkers, J. Genetically Engineered Mouse Models in Oncology Research and Cancer Medicine. EMBO Mol. Med. 2017, 9, 137–153. [Google Scholar] [CrossRef]
- Shih, D.J.; Northcott, P.A.; Remke, M.; Korshunov, A.; Ramaswamy, V.; Kool, M.; Luu, B.; Yao, Y.; Wang, X.; Dubuc, A.M.; et al. Cytogenetic Prognostication within Medulloblastoma Subgroups. J. Clin. Oncol. 2014, 32, 886–896. [Google Scholar] [CrossRef]
- Al Dahhan, N.Z.; De Felice, F.G.; Munoz, D.P. Potentials and Pitfalls of Cross-Translational Models of Cognitive Impairment. Front. Behav. Neurosci. 2019, 13, 48. [Google Scholar] [CrossRef]
- Wechsler, W.; Ramadan, M.A.; Pfeiffer, S.E. Morphologic and Biochemical Characteristics of Transplantable Neurogenic Tumors Induced by N-Ethyl-N-Nitrosourea in Inbred BD IX Rats2. JNCI J. Natl. Cancer Inst. 1979, 62, 811–817. [Google Scholar] [CrossRef]
- LeBlanc, A.K.; Mazcko, C.N. Improving Human Cancer Therapy through the Evaluation of Pet Dogs. Nat. Rev. Cancer 2020, 20, 727–742. [Google Scholar] [CrossRef]
- Fernández, F.; Deviers, A.; Dally, C.; Mogicato, G.; Delverdier, M.; Cauzinille, L.; Gnirs, K.; Añor, S.; de la Fuente, C.; Fondevila, D.; et al. Presence of Neural Progenitors in Spontaneous Canine Gliomas: A Histopathological and Immunohistochemical Study of 20 Cases. Vet. J. 2016, 209, 125–132. [Google Scholar] [CrossRef]
- LeBlanc, A. A Report from the NCI Comparative Brain Tumor Consortium (CBTC) Glioma Pathology Board: A Revised Diagnostic Classification in Support of Validation of the Canine Glioma Patient as a Model for Humans. Vet. Pathol. 2019, 56, 642–643. [Google Scholar] [CrossRef] [PubMed]
- Lonser, R.R.; Walbridge, S.; Vortmeyer, A.O.; Pack, S.D.; Nguyen, T.T.; Gogate, N.; Olson, J.J.; Akbasak, A.; Bobo, R.H.; Goffman, T.; et al. Induction of Glioblastoma Multiforme in Nonhuman Primates after Therapeutic Doses of Fractionated Whole-Brain Radiation Therapy. J. Neurosurg. 2002, 97, 1378–1389. [Google Scholar] [CrossRef] [PubMed]
- Lowenstine, L.J. Neoplasms and Proliferative Disorders in Nonhuman Primates. In Primates; Benirschke, K., Ed.; Springer: New York, NY, USA, 1986; pp. 781–814. [Google Scholar]
- Neff, E.P. Cancer Modeling Thinks Big with the Pig. Lab. Anim. 2019, 48, 75–78. [Google Scholar] [CrossRef]
- Schook, L.B.; Collares, T.V.; Darfour-Oduro, K.A.; De, A.K.; Rund, L.A.; Schachtschneider, K.M.; Seixas, F.K. Unraveling the Swine Genome: Implications for Human Health. Annu. Rev. Anim. Biosci. 2015, 3, 219–244. [Google Scholar] [CrossRef] [PubMed]
- Lind, N.M.; Moustgaard, A.; Jelsing, J.; Vajta, G.; Cumming, P.; Hansen, A.K. The Use of Pigs in Neuroscience: Modeling Brain Disorders. Neurosci. Biobehav. Rev. 2007, 31, 728–751. [Google Scholar] [CrossRef]
- Sauleau, P.; Lapouble, E.; Val-Laillet, D.; Malbert, C.H. The Pig Model in Brain Imaging and Neurosurgery. Animal 2009, 3, 1138–1151. [Google Scholar] [CrossRef]
- Shahzad, U.; Taccone, M.S.; Kumar, S.A.; Okura, H.; Krumholtz, S.; Ishida, J.; Mine, C.; Gouveia, K.; Edgar, J.; Smith, C.; et al. Modeling Human Brain Tumors in Flies, Worms, and Zebrafish: From Proof of Principle to Novel Therapeutic Targets. Neuro Oncol. 2021, 23, 718–731. [Google Scholar] [CrossRef] [PubMed]
- Zon, L.I. Zebrafish: A New Model for Human Disease. Genome Res. 1999, 9, 99–100. [Google Scholar] [CrossRef]
- Reimunde, P.; Pensado-Lopez, A.; Carreira Crende, M.; Lombao Iglesias, V.; Sanchez, L.; Torrecilla-Parra, M.; Ramirez, C.M.; Anfray, C.; Torres Andon, F. Cellular and Molecular Mechanisms Underlying Glioblastoma and Zebrafish Models for the Discovery of New Treatments. Cancers 2021, 13, 1087. [Google Scholar] [CrossRef]
- Liu, P.; Griffiths, S.; Veljanoski, D.; Vaughn-Beaucaire, P.; Speirs, V.; Bruning-Richardson, A. Preclinical Models of Glioblastoma: Limitations of Current Models and the Promise of New Developments. Expert. Rev. Mol. Med. 2021, 23, e20. [Google Scholar] [CrossRef] [PubMed]
- Wainwright, D.A.; Horbinski, C.M.; Hashizume, R.; James, C.D. Therapeutic Hypothesis Testing with Rodent Brain Tumor Models. Neurotherapeutics 2017, 14, 385–392. [Google Scholar] [CrossRef] [PubMed]
- Reardon, D.A.; Gokhale, P.C.; Klein, S.R.; Ligon, K.L.; Rodig, S.J.; Ramkissoon, S.H.; Jones, K.L.; Conway, A.S.; Liao, X.; Zhou, J.; et al. Glioblastoma Eradication Following Immune Checkpoint Blockade in an Orthotopic, Immunocompetent Model. Cancer Immunol. Res. 2016, 4, 124–135. [Google Scholar] [CrossRef]
- Akbasak, A.; Oldfield, E.H.; Saris, S.C. Expression and Modulation of Major Histocompatibility Antigens on Murine Primary Brain Tumor In Vitro. J. Neurosurg. 1991, 75, 922–929. [Google Scholar] [CrossRef]
- Kong, Y.; Feng, Z.C.; Zhang, Y.L.; Liu, X.F.; Ma, Y.; Zhao, Z.M.; Huang, B.; Chen, A.J.; Zhang, D.; Thorsen, F.; et al. Identification of Immune-Related Genes Contributing to the Development of Glioblastoma Using Weighted Gene Co-Expression Network Analysis. Front. Immunol. 2020, 11, 1281. [Google Scholar] [CrossRef]
- Ito, M.; Hiramatsu, H.; Kobayashi, K.; Suzue, K.; Kawahata, M.; Hioki, K.; Ueyama, Y.; Koyanagi, Y.; Sugamura, K.; Tsuji, K.; et al. NOD/SCID/Gamma(c)(Null) Mouse: An Excellent Recipient Mouse Model for Engraftment of Human Cells. Blood 2002, 100, 3175–3182. [Google Scholar] [CrossRef]
- Jin, F.; Jin-Lee, H.J.; Johnson, A.J. Mouse Models of Experimental Glioblastoma. In Gliomas; Debinski, W., Ed.; Exon: Brisbane, Australia, 2021; ISBN 978-0-6450017-4-7. [Google Scholar]
- Bao, S.; Wu, Q.; McLendon, R.E.; Hao, Y.; Shi, Q.; Hjelmeland, A.B.; Dewhirst, M.W.; Bigner, D.D.; Rich, J.N. Glioma Stem Cells Promote Radioresistance by Preferential Activation of the DNA Damage Response. Nature 2006, 444, 756–760. [Google Scholar] [CrossRef]
- Robertson, F.L.; Marques-Torrejon, M.A.; Morrison, G.M.; Pollard, S.M. Experimental Models and Tools to Tackle Glioblastoma. Dis. Model. Mech. 2019, 12, dmm040386. [Google Scholar] [CrossRef]
- Joo, K.M.; Kim, J.; Jin, J.; Kim, M.; Seol, H.J.; Muradov, J.; Yang, H.; Choi, Y.L.; Park, W.Y.; Kong, D.S.; et al. Patient-Specific Orthotopic Glioblastoma Xenograft Models Recapitulate the Histopathology and Biology of Human Glioblastomas in Situ. Cell Rep. 2013, 3, 260–273. [Google Scholar] [CrossRef] [PubMed]
- Ben-David, U.; Ha, G.; Tseng, Y.Y.; Greenwald, N.F.; Oh, C.; Shih, J.; McFarland, J.M.; Wong, B.; Boehm, J.S.; Beroukhim, R.; et al. Patient-Derived Xenografts Undergo Mouse-Specific Tumor Evolution. Nat. Genet. 2017, 49, 1567–1575. [Google Scholar] [CrossRef] [PubMed]
- deCarvalho, A.C.; Kim, H.; Poisson, L.M.; Winn, M.E.; Mueller, C.; Cherba, D.; Koeman, J.; Seth, S.; Protopopov, A.; Felicella, M.; et al. Discordant Inheritance of Chromosomal and Extrachromosomal DNA Elements Contributes to Dynamic Disease Evolution in Glioblastoma. Nat. Genet. 2018, 50, 708–717. [Google Scholar] [CrossRef] [PubMed]
- Garralda, E.; Paz, K.; Lopez-Casas, P.P.; Jones, S.; Katz, A.; Kann, L.M.; Lopez-Rios, F.; Sarno, F.; Al-Shahrour, F.; Vasquez, D.; et al. Integrated Next-Generation Sequencing and Avatar Mouse Models for Personalized Cancer Treatment. Clin. Cancer Res. 2014, 20, 2476–2484. [Google Scholar] [CrossRef]
- Chen, J.; Liao, S.; Xiao, Z.; Pan, Q.; Wang, X.; Shen, K.; Wang, S.; Yang, L.; Guo, F.; Liu, H.F.; et al. The Development and Improvement of Immunodeficient Mice and Humanized Immune System Mouse Models. Front. Immunol. 2022, 13, 1007579. [Google Scholar] [CrossRef]
- Ashizawa, T.; Iizuka, A.; Nonomura, C.; Kondou, R.; Maeda, C.; Miyata, H.; Sugino, T.; Mitsuya, K.; Hayashi, N.; Nakasu, Y.; et al. Antitumor Effect of Programmed Death-1 (PD-1) Blockade in Humanized the NOG-MHC Double Knockout Mouse. Clin. Cancer Res. 2017, 23, 149–158. [Google Scholar] [CrossRef] [PubMed]
- Carlson, B.L.; Pokorny, J.L.; Schroeder, M.A.; Sarkaria, J.N. Establishment, Maintenance and In Vitro and In Vivo Applications of Primary Human Glioblastoma Multiforme (GBM) Xenograft Models for Translational Biology Studies and Drug Discovery. Curr. Protoc. Pharmacol. 2011, 52, 14–16. [Google Scholar] [CrossRef]
- Parsons, D.W.; Jones, S.; Zhang, X.; Lin, J.C.; Leary, R.J.; Angenendt, P.; Mankoo, P.; Carter, H.; Siu, I.M.; Gallia, G.L.; et al. An Integrated Genomic Analysis of Human Glioblastoma Multiforme. Science (1979) 2008, 321, 1807–1812. [Google Scholar] [CrossRef]
- Akter, F.; Simon, B.; de Boer, N.L.; Redjal, N.; Wakimoto, H.; Shah, K. Pre-Clinical Tumor Models of Primary Brain Tumors: Challenges and Opportunities. Biochim. Biophys. Acta Rev. Cancer 2021, 1875, 188458. [Google Scholar] [CrossRef] [PubMed]
- Kim, H.; Kim, M.; Im, S.K.; Fang, S. Mouse Cre-LoxP System: General Principles to Determine Tissue-Specific Roles of Target Genes. Lab. Anim. Res. 2018, 34, 147–159. [Google Scholar] [CrossRef] [PubMed]
- Hara, T.; Verma, I.M. Modeling Gliomas Using Two Recombinases. Cancer Res. 2019, 79, 3983–3991. [Google Scholar] [CrossRef] [PubMed]
- Ivics, Z.; Izsvak, Z. The Expanding Universe of Transposon Technologies for Gene and Cell Engineering. Mob. DNA 2010, 1, 25. [Google Scholar] [CrossRef] [PubMed]
- Kosaka, A.; Ohkuri, T.; Okada, H. Combination of an Agonistic Anti-CD40 Monoclonal Antibody and the COX-2 Inhibitor Celecoxib Induces Anti-Glioma Effects by Promotion of Type-1 Immunity in Myeloid Cells and T-Cells. Cancer Immunol. Immunother. 2014, 63, 847–857. [Google Scholar] [CrossRef]
- Wiesner, S.M.; Decker, S.A.; Larson, J.D.; Ericson, K.; Forster, C.; Gallardo, J.L.; Long, C.; Demorest, Z.L.; Zamora, E.A.; Low, W.C.; et al. De Novo Induction of Genetically Engineered Brain Tumors in Mice Using Plasmid DNA. Cancer Res. 2009, 69, 431–439. [Google Scholar] [CrossRef]
- Miyai, M.; Tomita, H.; Soeda, A.; Yano, H.; Iwama, T.; Hara, A. Current Trends in Mouse Models of Glioblastoma. J. Neurooncol. 2017, 135, 423–432. [Google Scholar] [CrossRef]
- Hambardzumyan, D.; Amankulor, N.M.; Helmy, K.Y.; Becher, O.J.; Holland, E.C. Modeling Adult Gliomas Using RCAS/t-va Technology. Transl. Oncol. 2009, 2, 89–95. [Google Scholar] [CrossRef]
- Rankin, S.L.; Zhu, G.; Baker, S.J. Review: Insights Gained from Modelling High-Grade Glioma in the Mouse. Neuropathol. Appl. Neurobiol. 2012, 38, 254–270. [Google Scholar] [CrossRef]
- Marumoto, T.; Tashiro, A.; Friedmann-Morvinski, D.; Scadeng, M.; Soda, Y.; Gage, F.H.; Verma, I.M. Development of a Novel Mouse Glioma Model Using Lentiviral Vectors. Nat. Med. 2009, 15, 110–116. [Google Scholar] [CrossRef] [PubMed]
- Wei, Q.; Clarke, L.; Scheidenhelm, D.K.; Qian, B.; Tong, A.; Sabha, N.; Karim, Z.; Bock, N.A.; Reti, R.; Swoboda, R.; et al. High-Grade Glioma Formation Results from Postnatal Pten Loss or Mutant Epidermal Growth Factor Receptor Expression in a Transgenic Mouse Glioma Model. Cancer Res. 2006, 66, 7429–7437. [Google Scholar] [CrossRef] [PubMed]
- Majc, B.; Novak, M.; Kopitar-Jerala, N.; Jewett, A.; Breznik, B. Immunotherapy of Glioblastoma: Current Strategies and Challenges in Tumor Model Development. Cells 2021, 10, 265. [Google Scholar] [CrossRef]
- Xu, C.; Yuan, X.; Hou, P.; Li, Z.; Wang, C.; Fang, C.; Tan, Y. Development of Glioblastoma Organoids and Their Applications in Personalized Therapy. Cancer Biol. Med. 2023, 20, 353–368. [Google Scholar] [CrossRef] [PubMed]
- Cai, X.; Briggs, R.G.; Homburg, H.B.; Young, I.M.; Davis, E.J.; Lin, Y.H.; Battiste, J.D.; Sughrue, M.E. Application of Microfluidic Devices for Glioblastoma Study: Current Status and Future Directions. Biomed. Microdevices 2020, 22, 60. [Google Scholar] [CrossRef]
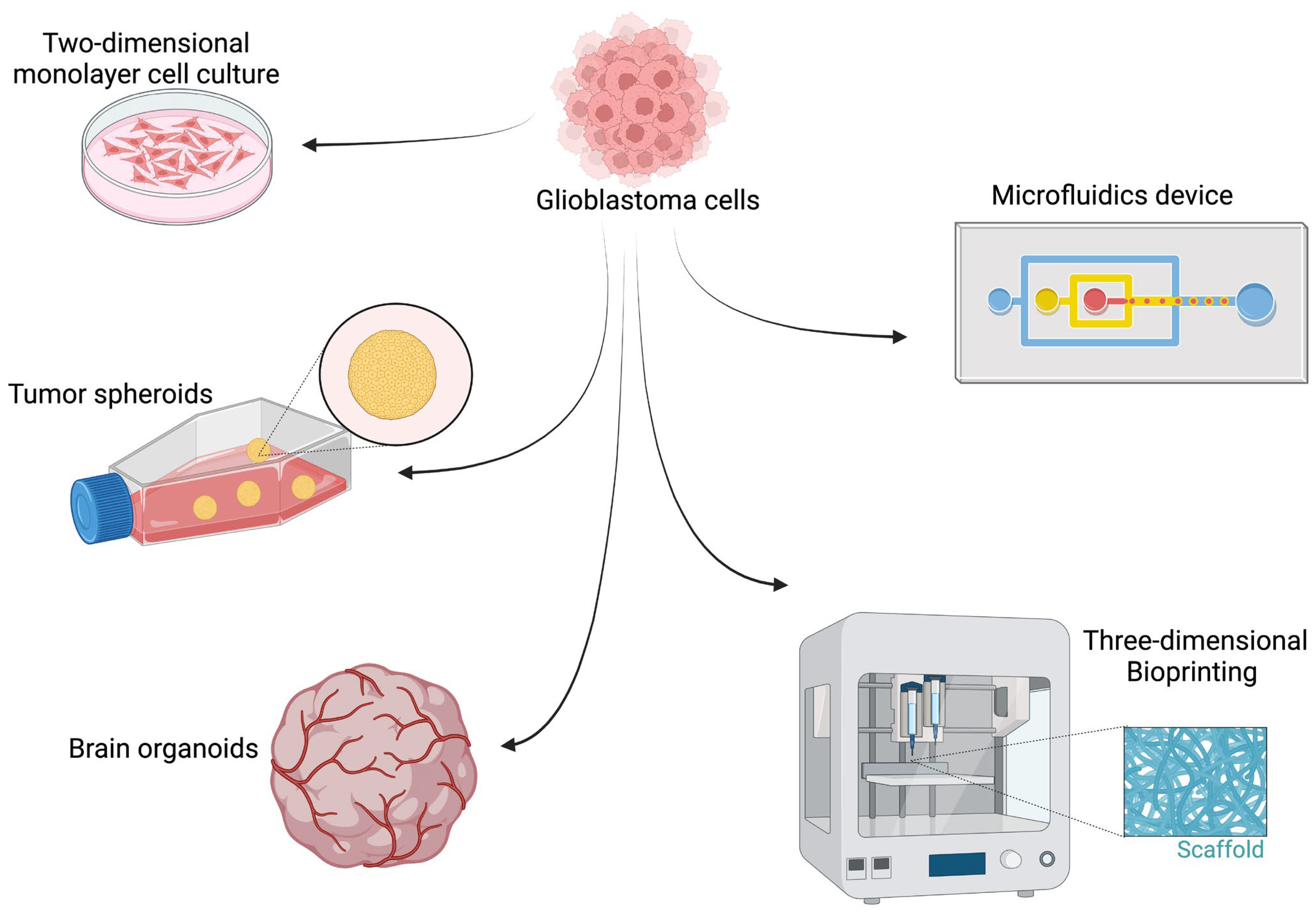

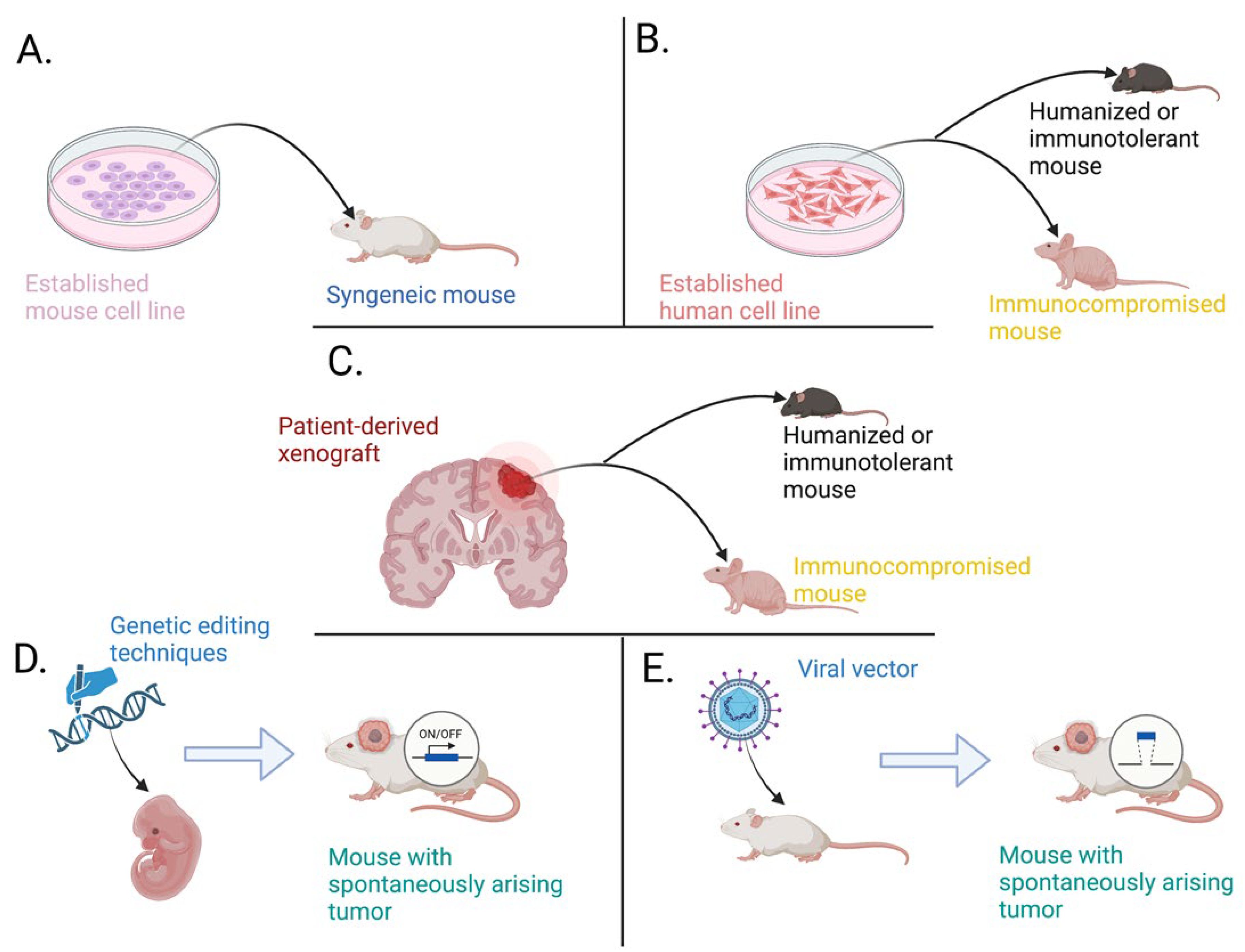
| C6 | 9L | F98 | RG2 | GL261 | CT2A | U87 | U251 | |
|---|---|---|---|---|---|---|---|---|
| Origin | Wistar Rat | Fischer Rat | Fischer Rat | Fischer Rat | C57BL/6 mice | C57BL/6 mice | Patient | Patient |
| P14ARF mut. | − | − | NA | + | + | NA | + | + |
| P16 mut. | + | − | + | + | + | NA | + | + |
| P53 mut. | − | + | − | − | + | − | − | + |
| PTEN mut. | − | − | NA | NA | + | + | + | + |
| RAS mut./overexpression | + | NA | + | + | + | NA | + | + |
| IDH1 mut. | − | NA | NA | NA | − | − | − | − |
| IDH2 mut. | − | NA | NA | NA | NA | − | NA | NA |
| EGFR overexpression | + | + | + | − | + | NA | − | + |
| PDGF-β amplification | + | − | + | + | NA | NA | NA | NA |
| GFAP | + | + | + | NA | − | + | − | + |
| S100 | + | + | − | NA | NA | NA | − | + |
| Vimentin | − | NA | + | NA | + | NA | + | + |
| Nuclear pleomorphism | + | + | + | + | + | + | + | + |
| Mitotic index | high | high | high | high | high | high | high | high |
| Tumor necrosis | moderate | low | low | high | moderate | high | low | high |
| Invasiveness | moderate | moderate | high | high | moderate | low | low | high |
| Angiogenesis | high | high | high | moderate | moderate | high | low | high |
| Immunogenicity | high | high | low | low | low | high | high | high |
| References | [13,14,15,16,17,18,19,20,21,22,23,24,25] | [13,14,15,16,17,18,19,20,21,22,23,24,25] | [13,14,15,16,17,18,19,20,21,22,23,24,25] | [13,14,15,16,17,18,19,20,21,22,23,24,25] | [15,26,27,28,29,30,31,32,33,34,35,36] | [15,26,27,28,29,30,31,32,33,34,35,36] | [37,38,39,40,41,42,43,44] | [37,38,39,40,41,42,43,44] |
| Model | Brief Description | Advantages | Limitations | References |
|---|---|---|---|---|
| In vitro | ||||
| Two-dimensional cell culture | The simplest form of growing cell lines in an appropriate medium and passaging them over several generations |
|
| [300,345,346] |
| Spherical cancer models | Three-dimensional spheroids that originate from glioblastoma cancer stem cells |
|
| [150,334,345,346] |
| Brain organotypic models | This model aims to act as a miniature of the brain and is derived using pluripotent stem cells. Glioblastoma is introduced via genetic editing or coculturing with patient-derived cancer stem cells. |
|
| [150,334,345,346] |
| Scaffolds (3D bioprinting) | This technology refers to the construction of three-dimensional networks of various cells and biomaterials that resemble the constituents of glioblastoma and its microenvironment. |
|
| [150,334,345,346] |
| Microfluidics | This technology utilizes microchips and micromechanical valves that enable researchers to accurately control the medium constituents and the flow of fluid at the nanoliter level. |
|
| [150,231,290,347] |
| In vivo | ||||
| Allografts in syngeneic models | This model involves the implantation of established cancer cell lines that were generated in a certain species into strain-matched hosts. |
|
| [15,300,334,345] |
| Patient-derived xenografts in immunocompromised models | This model is created by harvesting glioblastoma cells from cancer patients and implanting them into animal hosts that were engineered to develop deficient immune systems |
|
| [15,300,334,345] |
| Patient-derived xenografts in humanized/immunotolerant models | This model is also generated using glioblastoma cells originating from human patients; however, these cells are implanted into mice that were engrafted with hematopoietic stem cells that can generate a human-like immune system (humanized mice). Alternatively, immunocompetent mice are implanted with tumor cells and treated with immunosuppressive drugs until the tumors are well established (immunotolerant mice). |
|
| [15,300,334,345] |
| Genetically engineered and viral vector-induced models | These models rely on the genetic modification of animals in order to turn off/on specific tumor suppressor/oncogenes. The result is an animal model that develops glioblastoma in a “spontaneous” manner. |
|
| [15,300,334,345] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Slika, H.; Karimov, Z.; Alimonti, P.; Abou-Mrad, T.; De Fazio, E.; Alomari, S.; Tyler, B. Preclinical Models and Technologies in Glioblastoma Research: Evolution, Current State, and Future Avenues. Int. J. Mol. Sci. 2023, 24, 16316. https://doi.org/10.3390/ijms242216316
Slika H, Karimov Z, Alimonti P, Abou-Mrad T, De Fazio E, Alomari S, Tyler B. Preclinical Models and Technologies in Glioblastoma Research: Evolution, Current State, and Future Avenues. International Journal of Molecular Sciences. 2023; 24(22):16316. https://doi.org/10.3390/ijms242216316
Chicago/Turabian StyleSlika, Hasan, Ziya Karimov, Paolo Alimonti, Tatiana Abou-Mrad, Emerson De Fazio, Safwan Alomari, and Betty Tyler. 2023. "Preclinical Models and Technologies in Glioblastoma Research: Evolution, Current State, and Future Avenues" International Journal of Molecular Sciences 24, no. 22: 16316. https://doi.org/10.3390/ijms242216316
APA StyleSlika, H., Karimov, Z., Alimonti, P., Abou-Mrad, T., De Fazio, E., Alomari, S., & Tyler, B. (2023). Preclinical Models and Technologies in Glioblastoma Research: Evolution, Current State, and Future Avenues. International Journal of Molecular Sciences, 24(22), 16316. https://doi.org/10.3390/ijms242216316






