FMRP Enhances the Translation of 4EBP2 mRNA during Neuronal Differentiation
Abstract
:1. Introduction
2. Results
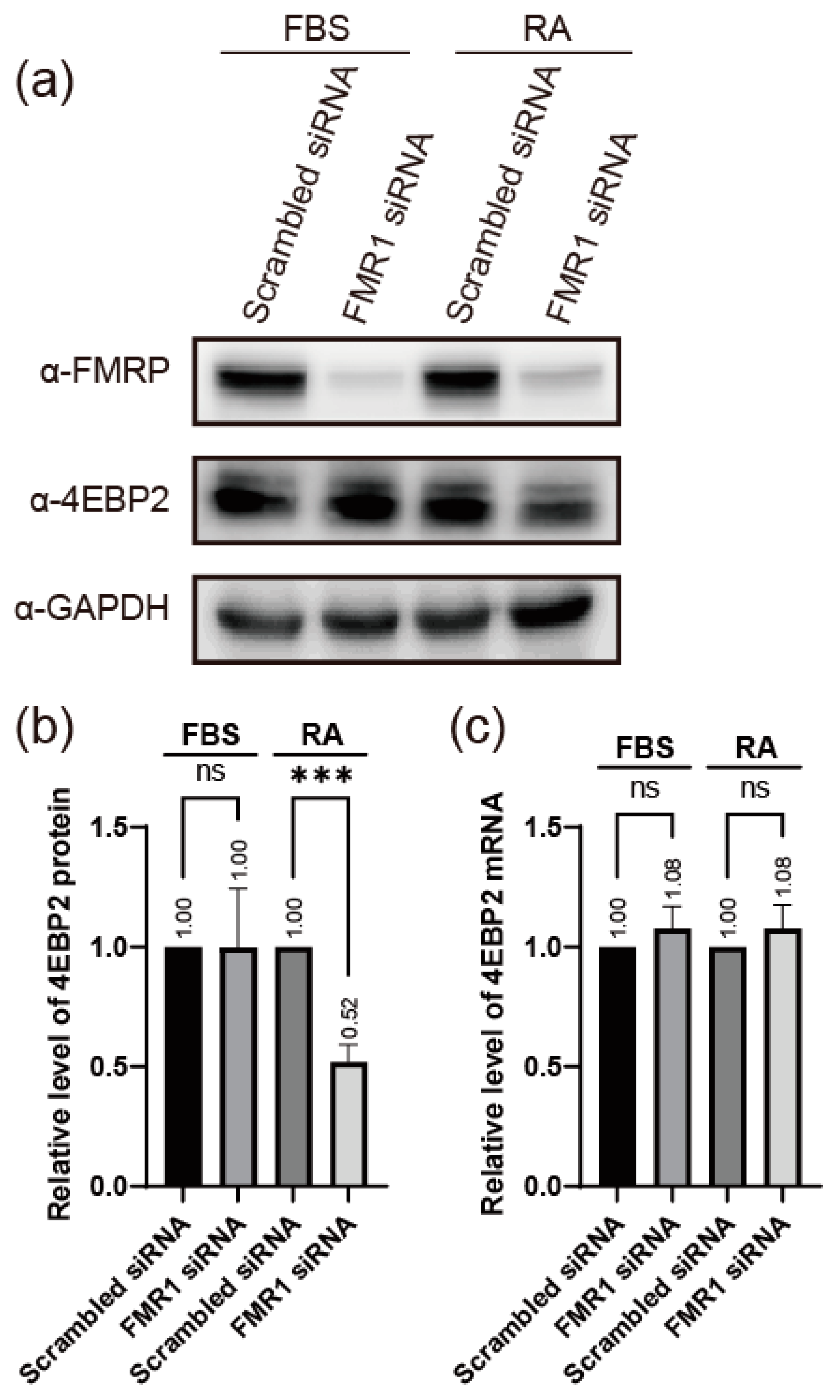
2.1. FMRP Enhances the Expression of 4EBP2 in Neuronal Cells
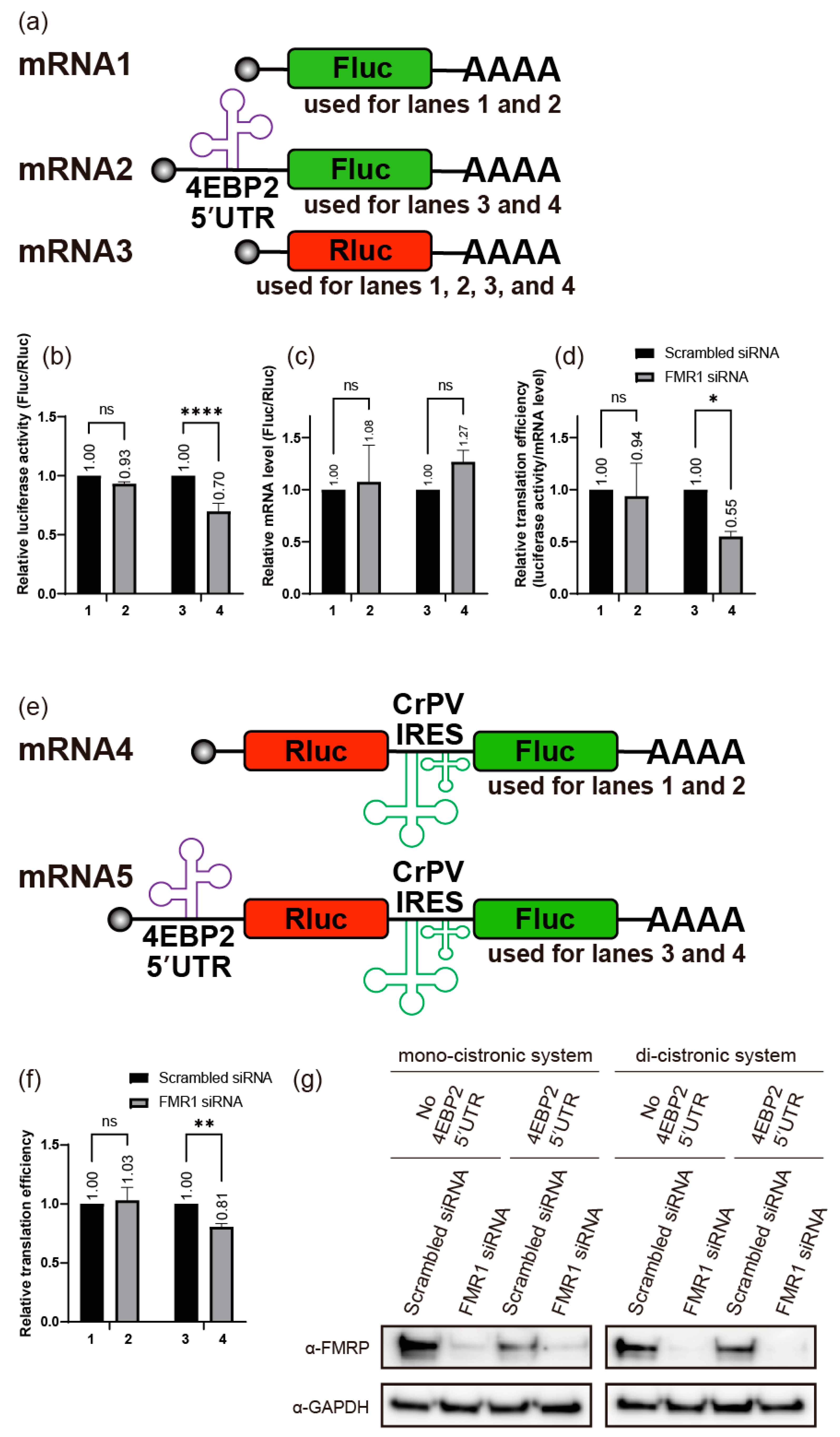
2.2. FMRP Enhances Translation of 4EBP2
2.3. FMRP Binds to the 5′UTR of 4EBP2 mRNA
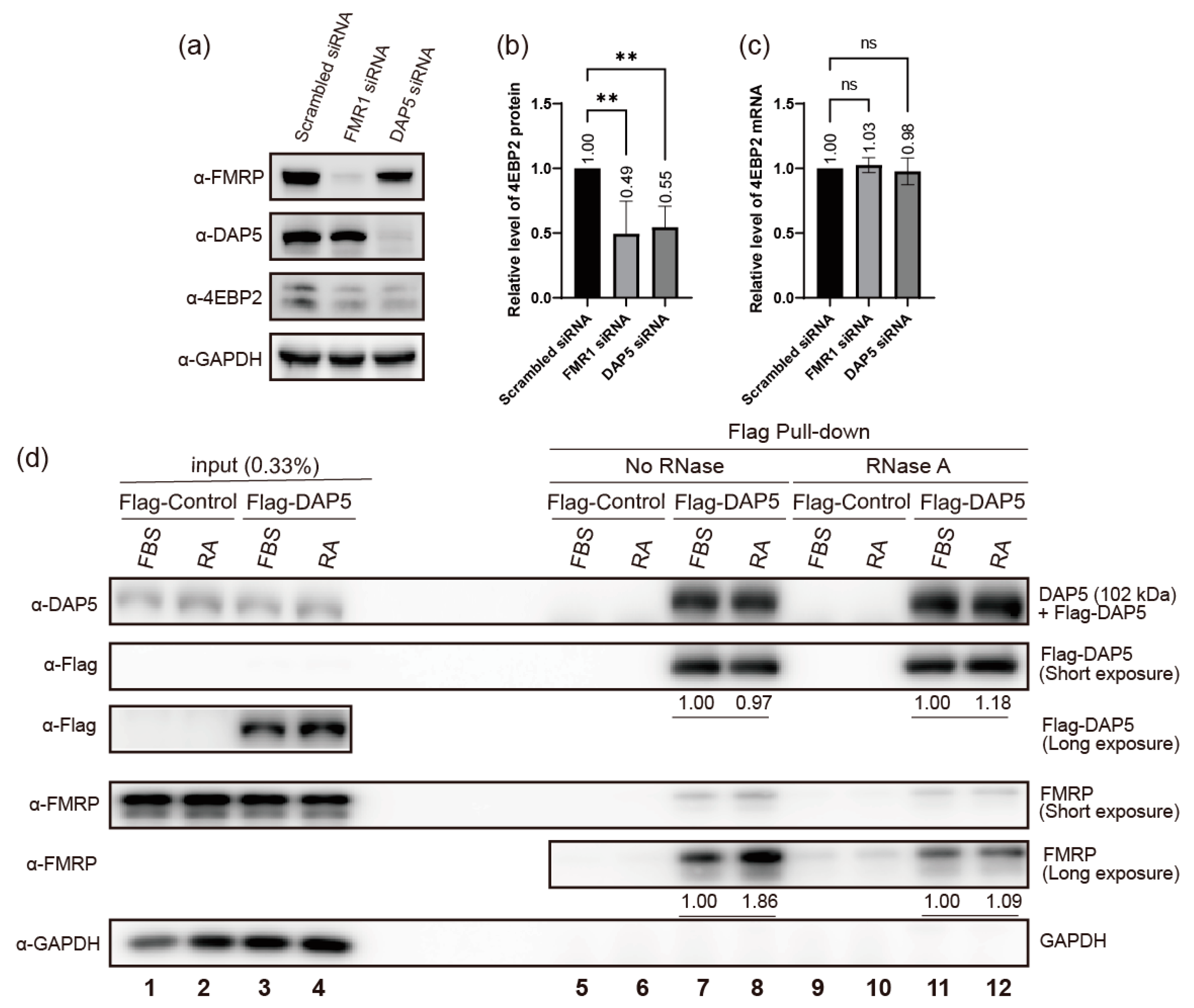
2.4. DAP5 Enhances Translation of 4EBP2
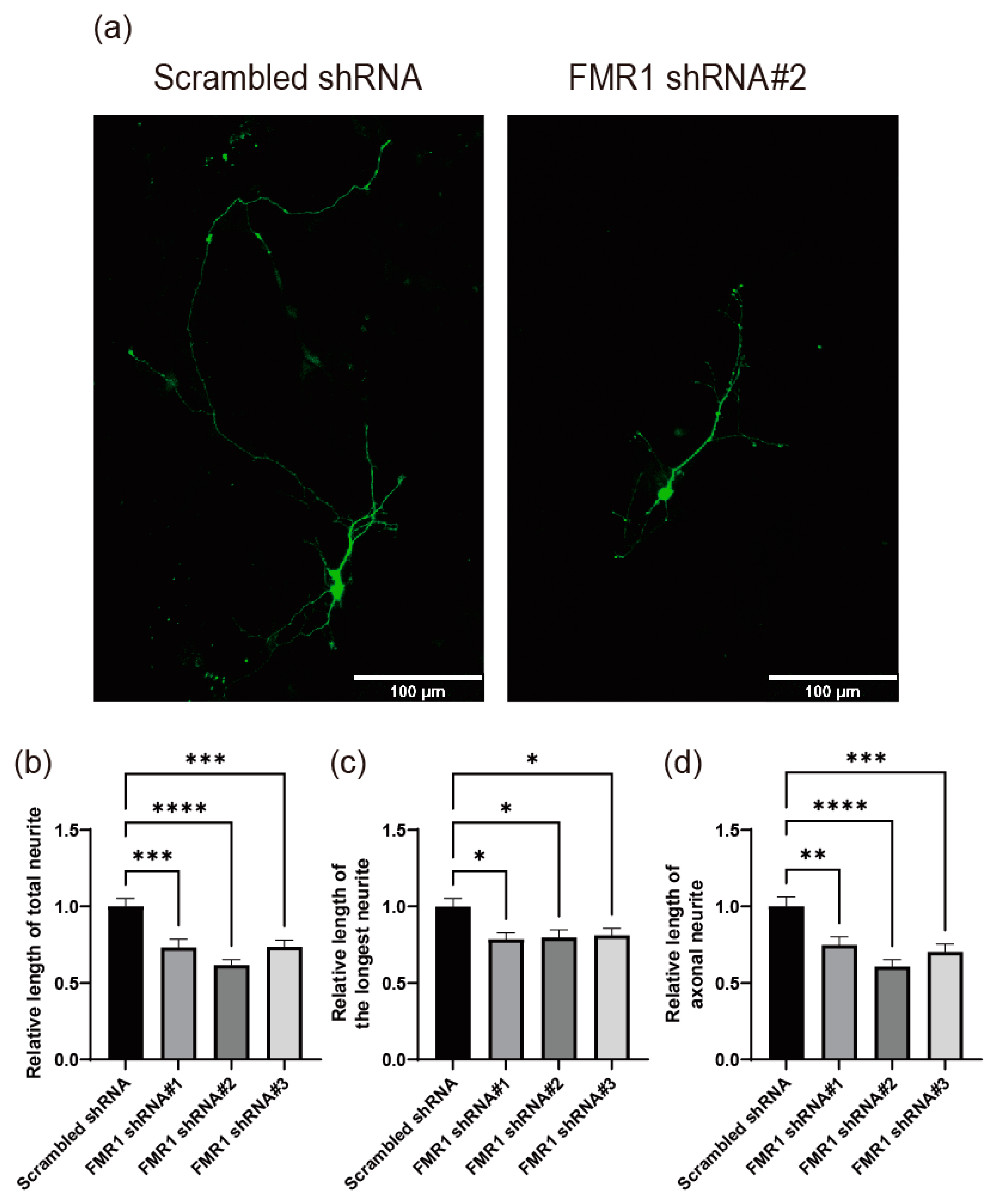
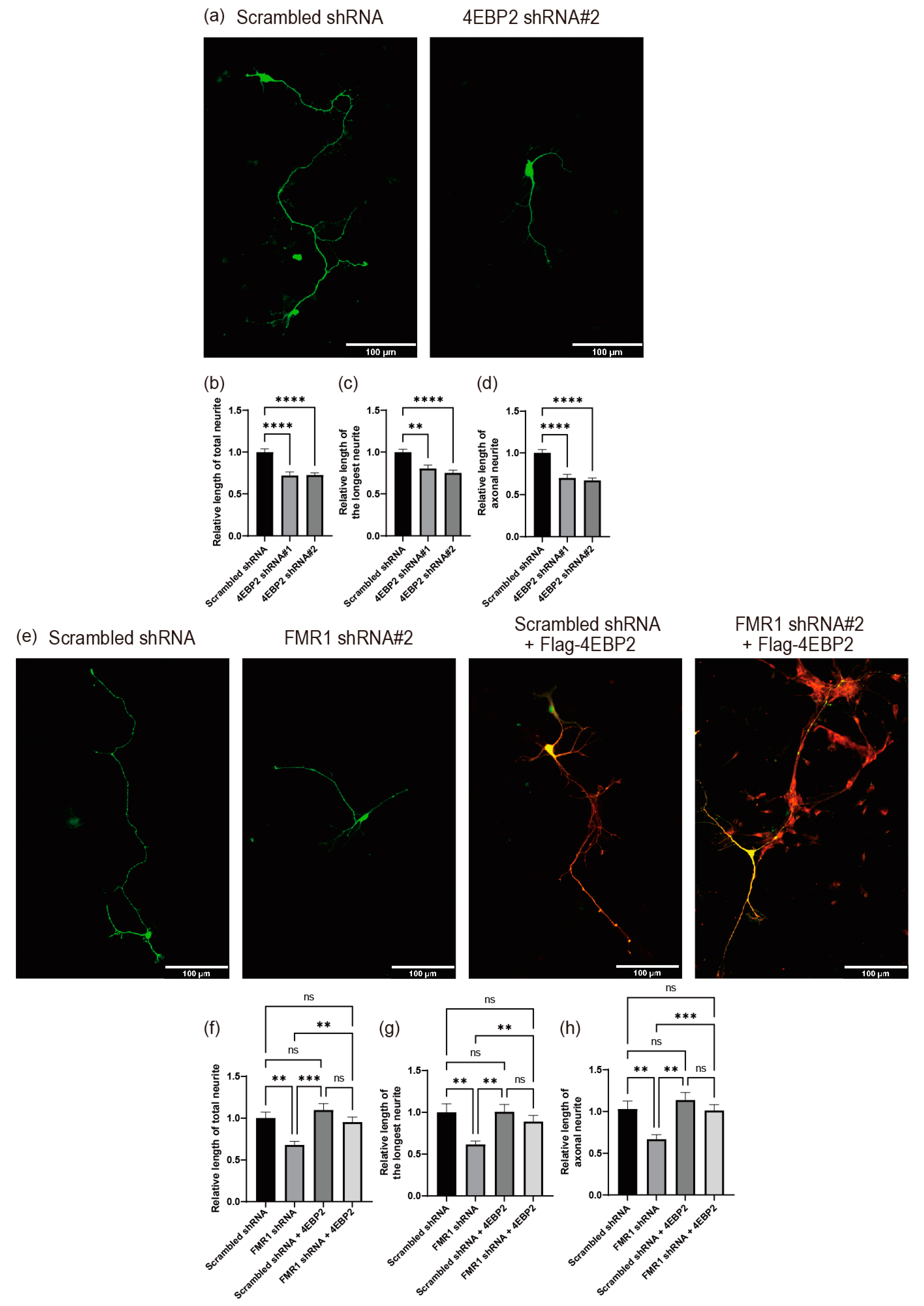
2.5. Depletion of FMRP Induces Defects in Neurite Outgrowth
2.6. 4EBP2 Is Epistatic to FMRP in the Modulation of Neurite Outgrowth
2.7. Inactivation of eIF4E Function Neutralizes the Defect in Neurite Outgrowth Caused by FMRP and/or 4EBP2 Deficiency
3. Discussion
4. Materials and Methods
4.1. Construction of Plasmids
4.2. Cell Culture and Differentiation
4.3. Animals
4.4. Immunocytochemistry
4.5. In Vitro Neurite Outgrowth Assay
4.6. Western Blotting and Antibodies
4.7. Luciferase Assay
4.8. Quantitative RT-PCR Analysis
4.9. In Vitro Transcription and RNA Pull-Down Experiments with Biotinylated RNAs
4.10. Flag-Immunoprecipitation
4.11. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Conflicts of Interest
References
- Richter, J.D.; Zhao, X. The molecular biology of FMRP: New insights into fragile X syndrome. Nat. Rev. Neurosci. 2021, 22, 209–222. [Google Scholar] [CrossRef] [PubMed]
- Hagerman, R.; Au, J.; Hagerman, P. FMR1 premutation and full mutation molecular mechanisms related to autism. J. Neurodev. Disord. 2011, 3, 211–224. [Google Scholar] [CrossRef] [PubMed]
- Verkerk, A.J.; Pieretti, M.; Sutcliffe, J.S.; Fu, Y.H.; Kuhl, D.P.; Pizzuti, A.; Reiner, O.; Richards, S.; Victoria, M.F.; Zhang, F.P.; et al. Identification of a gene (FMR-1) containing a CGG repeat coincident with a breakpoint cluster region exhibiting length variation in fragile X syndrome. Cell 1991, 65, 905–914. [Google Scholar] [CrossRef] [PubMed]
- Sztainberg, Y.; Zoghbi, H.Y. Lessons learned from studying syndromic autism spectrum disorders. Nat. Neurosci. 2016, 19, 1408–1417. [Google Scholar] [CrossRef]
- Hill, M.K.; Archibald, A.D.; Cohen, J.; Metcalfe, S.A. A systematic review of population screening for fragile X syndrome. Genet. Med. 2010, 12, 396–410. [Google Scholar] [CrossRef]
- Hersh, J.H.; Saul, R.A. Health supervision for children with fragile X syndrome. Pediatrics 2011, 127, 994–1006. [Google Scholar] [CrossRef]
- Bagni, C.; Tassone, F.; Neri, G.; Hagerman, R. Fragile X syndrome: Causes, diagnosis, mechanisms, and therapeutics. J. Clin. Investig. 2012, 122, 4314–4322. [Google Scholar] [CrossRef]
- Darnell, J.C.; Van Driesche, S.J.; Zhang, C.; Hung, K.Y.; Mele, A.; Fraser, C.E.; Stone, E.F.; Chen, C.; Fak, J.J.; Chi, S.W.; et al. FMRP stalls ribosomal translocation on mRNAs linked to synaptic function and autism. Cell 2011, 146, 247–261. [Google Scholar] [CrossRef]
- Darnell, J.C.; Mostovetsky, O.; Darnell, R.B. FMRP RNA targets: Identification and validation. Genes Brain Behav. 2005, 4, 341–349. [Google Scholar] [CrossRef]
- Napoli, I.; Mercaldo, V.; Boyl, P.P.; Eleuteri, B.; Zalfa, F.; De Rubeis, S.; Di Marino, D.; Mohr, E.; Massimi, M.; Falconi, M.; et al. The fragile X syndrome protein represses activity-dependent translation through CYFIP1, a new 4E-BP. Cell 2008, 134, 1042–1054. [Google Scholar] [CrossRef]
- Chen, E.; Sharma, M.R.; Shi, X.; Agrawal, R.K.; Joseph, S. Fragile X mental retardation protein regulates translation by binding directly to the ribosome. Mol. Cell 2014, 54, 407–417. [Google Scholar] [CrossRef] [PubMed]
- Maurin, T.; Bardoni, B. Fragile X Mental Retardation Protein: To Be or Not to Be a Translational Enhancer. Front. Mol. Biosci. 2018, 5, 113. [Google Scholar] [CrossRef]
- Greenblatt, E.J.; Spradling, A.C. Fragile X mental retardation 1 gene enhances the translation of large autism-related proteins. Science 2018, 361, 709–712. [Google Scholar] [CrossRef] [PubMed]
- Bechara, E.G.; Didiot, M.C.; Melko, M.; Davidovic, L.; Bensaid, M.; Martin, P.; Castets, M.; Pognonec, P.; Khandjian, E.W.; Moine, H.; et al. A novel function for fragile X mental retardation protein in translational activation. PLoS Biol. 2009, 7, e16. [Google Scholar] [CrossRef]
- Gross, C.; Yao, X.; Pong, D.L.; Jeromin, A.; Bassell, G.J. Fragile X mental retardation protein regulates protein expression and mRNA translation of the potassium channel Kv4.2. J. Neurosci. 2011, 31, 5693–5698. [Google Scholar] [CrossRef] [PubMed]
- Tabet, R.; Moutin, E.; Becker, J.A.; Heintz, D.; Fouillen, L.; Flatter, E.; Krężel, W.; Alunni, V.; Koebel, P.; Dembélé, D.; et al. Fragile X Mental Retardation Protein (FMRP) controls diacylglycerol kinase activity in neurons. Proc. Natl. Acad. Sci. USA 2016, 113, E3619–E3628. [Google Scholar] [CrossRef]
- Fähling, M.; Mrowka, R.; Steege, A.; Kirschner, K.M.; Benko, E.; Förstera, B.; Persson, P.B.; Thiele, B.J.; Meier, J.C.; Scholz, H. Translational regulation of the human achaete-scute homologue-1 by fragile X mental retardation protein. J. Biol. Chem. 2009, 284, 4255–4266. [Google Scholar] [CrossRef]
- Kwan, K.Y.; Lam, M.M.; Johnson, M.B.; Dube, U.; Shim, S.; Rašin, M.R.; Sousa, A.M.; Fertuzinhos, S.; Chen, J.G.; Arellano, J.I.; et al. Species-dependent posttranscriptional regulation of NOS1 by FMRP in the developing cerebral cortex. Cell 2012, 149, 899–911. [Google Scholar] [CrossRef]
- Sonenberg, N.; Rupprecht, K.M.; Hecht, S.M.; Shatkin, A.J. Eukaryotic mRNA cap binding protein: Purification by affinity chromatography on sepharose-coupled m7GDP. Proc. Natl. Acad. Sci. USA 1979, 76, 4345–4349. [Google Scholar] [CrossRef]
- Montanaro, L.; Pandolfi, P.P. Initiation of mRNA translation in oncogenesis: The role of eIF4E. Cell Cycle 2004, 3, 1387–1389. [Google Scholar] [CrossRef]
- Feoktistova, K.; Tuvshintogs, E.; Do, A.; Fraser, C.S. Human eIF4E promotes mRNA restructuring by stimulating eIF4A helicase activity. Proc. Natl. Acad. Sci. USA 2013, 110, 13339–13344. [Google Scholar] [CrossRef]
- Truitt, M.L.; Conn, C.S.; Shi, Z.; Pang, X.; Tokuyasu, T.; Coady, A.M.; Seo, Y.; Barna, M.; Ruggero, D. Differential Requirements for eIF4E Dose in Normal Development and Cancer. Cell 2015, 162, 59–71. [Google Scholar] [CrossRef] [PubMed]
- Amorim, I.S.; Kedia, S.; Kouloulia, S.; Simbriger, K.; Gantois, I.; Jafarnejad, S.M.; Li, Y.; Kampaite, A.; Pooters, T.; Romanò, N.; et al. Loss of eIF4E Phosphorylation Engenders Depression-like Behaviors via Selective mRNA Translation. J. Neurosci. 2018, 38, 2118–2133. [Google Scholar] [CrossRef] [PubMed]
- Kelleher, R.J., 3rd; Bear, M.F. The autistic neuron: Troubled translation? Cell 2008, 135, 401–406. [Google Scholar] [CrossRef] [PubMed]
- Gkogkas, C.G.; Khoutorsky, A.; Cao, R.; Jafarnejad, S.M.; Prager-Khoutorsky, M.; Giannakas, N.; Kaminari, A.; Fragkouli, A.; Nader, K.; Price, T.J.; et al. Pharmacogenetic inhibition of eIF4E-dependent Mmp9 mRNA translation reverses fragile X syndrome-like phenotypes. Cell Rep. 2014, 9, 1742–1755. [Google Scholar] [CrossRef]
- Moerke, N.J.; Aktas, H.; Chen, H.; Cantel, S.; Reibarkh, M.Y.; Fahmy, A.; Gross, J.D.; Degterev, A.; Yuan, J.; Chorev, M.; et al. Small-molecule inhibition of the interaction between the translation initiation factors eIF4E and eIF4G. Cell 2007, 128, 257–267. [Google Scholar] [CrossRef]
- Santini, E.; Huynh, T.N.; Longo, F.; Koo, S.Y.; Mojica, E.; D’Andrea, L.; Bagni, C.; Klann, E. Reducing eIF4E-eIF4G interactions restores the balance between protein synthesis and actin dynamics in fragile X syndrome model mice. Sci. Signal. 2017, 10, eaan0665. [Google Scholar] [CrossRef]
- Melancia, F.; Trezza, V. Modelling fragile X syndrome in the laboratory setting: A behavioral perspective. Behav. Brain Res. 2018, 350, 149–163. [Google Scholar] [CrossRef]
- Santini, E.; Huynh, T.N.; MacAskill, A.F.; Carter, A.G.; Pierre, P.; Ruggero, D.; Kaphzan, H.; Klann, E. Exaggerated translation causes synaptic and behavioural aberrations associated with autism. Nature 2013, 493, 411–415. [Google Scholar] [CrossRef]
- Gkogkas, C.G.; Khoutorsky, A.; Ran, I.; Rampakakis, E.; Nevarko, T.; Weatherill, D.B.; Vasuta, C.; Yee, S.; Truitt, M.; Dallaire, P.; et al. Autism-related deficits via dysregulated eIF4E-dependent translational control. Nature 2013, 493, 371–377. [Google Scholar] [CrossRef]
- Banko, J.L.; Poulin, F.; Hou, L.; DeMaria, C.T.; Sonenberg, N.; Klann, E. The translation repressor 4E-BP2 is critical for eIF4F complex formation, synaptic plasticity, and memory in the hippocampus. J. Neurosci. 2005, 25, 9581–9590. [Google Scholar] [CrossRef] [PubMed]
- Pause, A.; Belsham, G.J.; Gingras, A.C.; Donzé, O.; Lin, T.A.; Lawrence, J.C., Jr.; Sonenberg, N. Insulin-dependent stimulation of protein synthesis by phosphorylation of a regulator of 5’-cap function. Nature 1994, 371, 762–767. [Google Scholar] [CrossRef] [PubMed]
- Gingras, A.C.; Raught, B.; Sonenberg, N. eIF4 initiation factors: Effectors of mRNA recruitment to ribosomes and regulators of translation. Annu. Rev. Biochem. 1999, 68, 913–963. [Google Scholar] [CrossRef] [PubMed]
- Meldolesi, J. Neurite outgrowth: This process, first discovered by Santiago Ramon y Cajal, is sustained by the exocytosis of two distinct types of vesicles. Brain Res. Rev. 2011, 66, 246–255. [Google Scholar] [CrossRef] [PubMed]
- Banker, G. The Development of Neuronal Polarity: A Retrospective View. J. Neurosci. 2018, 38, 1867–1873. [Google Scholar] [CrossRef]
- Lin, Y.C.; Frei, J.A.; Kilander, M.B.; Shen, W.; Blatt, G.J. A Subset of Autism-Associated Genes Regulate the Structural Stability of Neurons. Front. Cell. Neurosci. 2016, 10, 263. [Google Scholar] [CrossRef]
- Utami, K.H.; Skotte, N.H.; Colaço, A.R.; Yusof, N.; Sim, B.; Yeo, X.Y.; Bae, H.G.; Garcia-Miralles, M.; Radulescu, C.I.; Chen, Q.; et al. Integrative Analysis Identifies Key Molecular Signatures Underlying Neurodevelopmental Deficits in Fragile X Syndrome. Biol. Psychiatry 2020, 88, 500–511. [Google Scholar] [CrossRef]
- Gross, C.; Berry-Kravis, E.M.; Bassell, G.J. Therapeutic strategies in fragile X syndrome: Dysregulated mGluR signaling and beyond. Neuropsychopharmacology 2012, 37, 178–195. [Google Scholar] [CrossRef]
- Doers, M.E.; Musser, M.T.; Nichol, R.; Berndt, E.R.; Baker, M.; Gomez, T.M.; Zhang, S.C.; Abbeduto, L.; Bhattacharyya, A. iPSC-derived forebrain neurons from FXS individuals show defects in initial neurite outgrowth. Stem. Cells Dev. 2014, 23, 1777–1787. [Google Scholar] [CrossRef]
- Telias, M.; Kuznitsov-Yanovsky, L.; Segal, M.; Ben-Yosef, D. Functional Deficiencies in Fragile X Neurons Derived from Human Embryonic Stem Cells. J. Neurosci. 2015, 35, 15295–15306. [Google Scholar] [CrossRef]
- Kuznitsov-Yanovsky, L.; Shapira, G.; Gildin, L.; Shomron, N.; Ben-Yosef, D. Transcriptomic Analysis of Human Fragile X Syndrome Neurons Reveals Neurite Outgrowth Modulation by the TGFβ/BMP Pathway. Int. J. Mol. Sci. 2022, 23, 9278. [Google Scholar] [CrossRef] [PubMed]
- Castrén, M.; Tervonen, T.; Kärkkäinen, V.; Heinonen, S.; Castrén, E.; Larsson, K.; Bakker, C.E.; Oostra, B.A.; Akerman, K. Altered differentiation of neural stem cells in fragile X syndrome. Proc. Natl. Acad. Sci. USA 2005, 102, 17834–17839. [Google Scholar] [CrossRef] [PubMed]
- Constantinescu, R.; Constantinescu, A.T.; Reichmann, H.; Janetzky, B. Neuronal differentiation and long-term culture of the human neuroblastoma line SH-SY5Y. J. Neural. Transm. Suppl. 2007, 72, 17–28. [Google Scholar] [CrossRef]
- Shipley, M.M.; Mangold, C.A.; Szpara, M.L. Differentiation of the SH-SY5Y Human Neuroblastoma Cell Line. J. Vis. Exp. 2016, 108, 53193. [Google Scholar] [CrossRef]
- Wilson, J.E.; Powell, M.J.; Hoover, S.E.; Sarnow, P. Naturally occurring dicistronic cricket paralysis virus RNA is regulated by two internal ribosome entry sites. Mol. Cell. Biol. 2000, 20, 4990–4999. [Google Scholar] [CrossRef]
- Deniz, N.; Lenarcic, E.M.; Landry, D.M.; Thompson, S.R. Translation initiation factors are not required for Dicistroviridae IRES function in vivo. Rna 2009, 15, 932–946. [Google Scholar] [CrossRef]
- Neelagandan, N.; Gonnella, G.; Dang, S.; Janiesch, P.C.; Miller, K.K.; Küchler, K.; Marques, R.F.; Indenbirken, D.; Alawi, M.; Grundhoff, A.; et al. TDP-43 enhances translation of specific mRNAs linked to neurodegenerative disease. Nucleic Acids Res. 2019, 47, 341–361. [Google Scholar] [CrossRef]
- Wang, G.; Guo, X.; Floros, J. Differences in the translation efficiency and mRNA stability mediated by 5′-UTR splice variants of human SP-A1 and SP-A2 genes. Am. J. Physiol. Lung Cell. Mol. Physiol. 2005, 289, L497–L508. [Google Scholar] [CrossRef]
- Silva, J.; Nina, P.; Romão, L. Translation of ABCE1 Is Tightly Regulated by Upstream Open Reading Frames in Human Colorectal Cells. Biomedicines 2021, 9, 911. [Google Scholar] [CrossRef]
- Grentzmann, G.; Ingram, J.A.; Kelly, P.J.; Gesteland, R.F.; Atkins, J.F. A dual-luciferase reporter system for studying recoding signals. Rna 1998, 4, 479–486. [Google Scholar]
- Lee, K.H.; Woo, K.C.; Kim, D.Y.; Kim, T.D.; Shin, J.; Park, S.M.; Jang, S.K.; Kim, K.T. Rhythmic interaction between Period1 mRNA and hnRNP Q leads to circadian time-dependent translation. Mol. Cell. Biol. 2012, 32, 717–728. [Google Scholar] [CrossRef] [PubMed]
- Kikin, O.; D’Antonio, L.; Bagga, P.S. QGRS Mapper: A web-based server for predicting G-quadruplexes in nucleotide sequences. Nucleic Acids Res. 2006, 34, W676–W682. [Google Scholar] [CrossRef]
- Darnell, J.C.; Jensen, K.B.; Jin, P.; Brown, V.; Warren, S.T.; Darnell, R.B. Fragile X mental retardation protein targets G quartet mRNAs important for neuronal function. Cell 2001, 107, 489–499. [Google Scholar] [CrossRef] [PubMed]
- Alard, A.; Marboeuf, C.; Fabre, B.; Jean, C.; Martineau, Y.; Lopez, F.; Vende, P.; Poncet, D.; Schneider, R.J.; Bousquet, C.; et al. Differential Regulation of the Three Eukaryotic mRNA Translation Initiation Factor (eIF) 4Gs by the Proteasome. Front. Genet. 2019, 10, 254. [Google Scholar] [CrossRef] [PubMed]
- Imataka, H.; Olsen, H.S.; Sonenberg, N. A new translational regulator with homology to eukaryotic translation initiation factor 4G. EMBO J. 1997, 16, 817–825. [Google Scholar] [CrossRef]
- de la Parra, C.; Ernlund, A.; Alard, A.; Ruggles, K.; Ueberheide, B.; Schneider, R.J. A widespread alternate form of cap-dependent mRNA translation initiation. Nat. Commun. 2018, 9, 3068. [Google Scholar] [CrossRef]
- Hundsdoerfer, P.; Thoma, C.; Hentze, M.W. Eukaryotic translation initiation factor 4GI and p97 promote cellular internal ribosome entry sequence-driven translation. Proc. Natl. Acad. Sci. USA 2005, 102, 13421–13426. [Google Scholar] [CrossRef]
- Yamanaka, S.; Zhang, X.Y.; Maeda, M.; Miura, K.; Wang, S.; Farese, R.V., Jr.; Iwao, H.; Innerarity, T.L. Essential role of NAT1/p97/DAP5 in embryonic differentiation and the retinoic acid pathway. EMBO J. 2000, 19, 5533–5541. [Google Scholar] [CrossRef]
- Seo, J.Y.; Jung, Y.; Kim, D.Y.; Ryu, H.G.; Lee, J.; Kim, S.W.; Kim, K.T. DAP5 increases axonal outgrowth of hippocampal neurons by enhancing the cap-independent translation of DSCR1.4 mRNA. Cell Death Dis. 2019, 10, 49. [Google Scholar] [CrossRef]
- Sugiyama, H.; Takahashi, K.; Yamamoto, T.; Iwasaki, M.; Narita, M.; Nakamura, M.; Rand, T.A.; Nakagawa, M.; Watanabe, A.; Yamanaka, S. Nat1 promotes translation of specific proteins that induce differentiation of mouse embryonic stem cells. Proc. Natl. Acad. Sci. USA 2017, 114, 340–345. [Google Scholar] [CrossRef]
- Jang, S.K.; Kräusslich, H.G.; Nicklin, M.J.; Duke, G.M.; Palmenberg, A.C.; Wimmer, E. A segment of the 5′ nontranslated region of encephalomyocarditis virus RNA directs internal entry of ribosomes during in vitro translation. J. Virol. 1988, 62, 2636–2643. [Google Scholar] [CrossRef] [PubMed]
- Godet, A.C.; David, F.; Hantelys, F.; Tatin, F.; Lacazette, E.; Garmy-Susini, B.; Prats, A.C. IRES Trans-Acting Factors, Key Actors of the Stress Response. Int. J. Mol. Sci. 2019, 20, 924. [Google Scholar] [CrossRef] [PubMed]
- Lee, A.S.; Kranzusch, P.J.; Doudna, J.A.; Cate, J.H. eIF3d is an mRNA cap-binding protein that is required for specialized translation initiation. Nature 2016, 536, 96–99. [Google Scholar] [CrossRef]
- Weber, R.; Kleemann, L.; Hirschberg, I.; Chung, M.Y.; Valkov, E.; Igreja, C. DAP5 enables main ORF translation on mRNAs with structured and uORF-containing 5’ leaders. Nat. Commun. 2022, 13, 7510. [Google Scholar] [CrossRef] [PubMed]
- Chen, J.; Brunner, A.D.; Cogan, J.Z.; Nuñez, J.K.; Fields, A.P.; Adamson, B.; Itzhak, D.N.; Li, J.Y.; Mann, M.; Leonetti, M.D.; et al. Pervasive functional translation of noncanonical human open reading frames. Science 2020, 367, 1140–1146. [Google Scholar] [CrossRef] [PubMed]
- Ingolia, N.T.; Brar, G.A.; Rouskin, S.; McGeachy, A.M.; Weissman, J.S. The ribosome profiling strategy for monitoring translation in vivo by deep sequencing of ribosome-protected mRNA fragments. Nat. Protoc. 2012, 7, 1534–1550. [Google Scholar] [CrossRef]
- Paek, K.Y.; Hong, K.Y.; Ryu, I.; Park, S.M.; Keum, S.J.; Kwon, O.S.; Jang, S.K. Translation initiation mediated by RNA looping. Proc. Natl. Acad. Sci. USA 2015, 112, 1041–1046. [Google Scholar] [CrossRef]
- Vasudevan, S.; Steitz, J.A. AU-rich-element-mediated upregulation of translation by FXR1 and Argonaute 2. Cell 2007, 128, 1105–1118. [Google Scholar] [CrossRef]
- Ingolia, N.T.; Brar, G.A.; Stern-Ginossar, N.; Harris, M.S.; Talhouarne, G.J.; Jackson, S.E.; Wills, M.R.; Weissman, J.S. Ribosome profiling reveals pervasive translation outside of annotated protein-coding genes. Cell Rep. 2014, 8, 1365–1379. [Google Scholar] [CrossRef]
- Brummelkamp, T.R.; Bernards, R.; Agami, R. A system for stable expression of short interfering RNAs in mammalian cells. Science 2002, 296, 550–553. [Google Scholar] [CrossRef]
- Hong, J.H.; Kwak, Y.; Woo, Y.; Park, C.; Lee, S.A.; Lee, H.; Park, S.J.; Suh, Y.; Suh, B.K.; Goo, B.S.; et al. Regulation of the actin cytoskeleton by the Ndel1-Tara complex is critical for cell migration. Sci. Rep. 2016, 6, 31827. [Google Scholar] [CrossRef] [PubMed]
- Kwon, O.S.; An, S.; Kim, E.; Yu, J.; Hong, K.Y.; Lee, J.S.; Jang, S.K. An mRNA-specific tRNAi carrier eIF2A plays a pivotal role in cell proliferation under stress conditions: Stress-resistant translation of c-Src mRNA is mediated by eIF2A. Nucleic Acids Res. 2017, 45, 296–310. [Google Scholar] [CrossRef] [PubMed]
- Luo, M.; Huang, Z.; Yang, X.; Chen, Y.; Jiang, J.; Zhang, L.; Zhou, L.; Qin, S.; Jin, P.; Fu, S.; et al. PHLDB2 Mediates Cetuximab Resistance via Interacting With EGFR in Latent Metastasis of Colorectal Cancer. Cell. Mol. Gastroenterol. Hepatol. 2022, 13, 1223–1242. [Google Scholar] [CrossRef]
- Zhou, Z.; Cao, M.; Guo, Y.; Zhao, L.; Wang, J.; Jia, X.; Li, J.; Wang, C.; Gabriel, G.; Xue, Q.; et al. Fragile X mental retardation protein stimulates ribonucleoprotein assembly of influenza A virus. Nat. Commun. 2014, 5, 3259. [Google Scholar] [CrossRef]
- Schindelin, J.; Arganda-Carreras, I.; Frise, E.; Kaynig, V.; Longair, M.; Pietzsch, T.; Preibisch, S.; Rueden, C.; Saalfeld, S.; Schmid, B.; et al. Fiji: An open-source platform for biological-image analysis. Nat. Methods 2012, 9, 676–682. [Google Scholar] [CrossRef] [PubMed]
- Gu, S.; Jeon, H.M.; Nam, S.W.; Hong, K.Y.; Rahman, M.S.; Lee, J.B.; Kim, Y.; Jang, S.K. The flip-flop configuration of the PABP-dimer leads to switching of the translation function. Nucleic Acids Res. 2022, 50, 306–321. [Google Scholar] [CrossRef] [PubMed]


Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yu, J.; Woo, Y.; Kim, H.; An, S.; Park, S.K.; Jang, S.K. FMRP Enhances the Translation of 4EBP2 mRNA during Neuronal Differentiation. Int. J. Mol. Sci. 2023, 24, 16319. https://doi.org/10.3390/ijms242216319
Yu J, Woo Y, Kim H, An S, Park SK, Jang SK. FMRP Enhances the Translation of 4EBP2 mRNA during Neuronal Differentiation. International Journal of Molecular Sciences. 2023; 24(22):16319. https://doi.org/10.3390/ijms242216319
Chicago/Turabian StyleYu, Jinbae, Youngsik Woo, Heesun Kim, Sihyeon An, Sang Ki Park, and Sung Key Jang. 2023. "FMRP Enhances the Translation of 4EBP2 mRNA during Neuronal Differentiation" International Journal of Molecular Sciences 24, no. 22: 16319. https://doi.org/10.3390/ijms242216319
APA StyleYu, J., Woo, Y., Kim, H., An, S., Park, S. K., & Jang, S. K. (2023). FMRP Enhances the Translation of 4EBP2 mRNA during Neuronal Differentiation. International Journal of Molecular Sciences, 24(22), 16319. https://doi.org/10.3390/ijms242216319





